Abstract
The passage of leukocytes through basement membranes involves proteolytic degradation of extracellular matrix (ECM) components executed by focalized proteolysis. We have investigated whether the migration of leukocytes through 3-dimensional collagenous tissue scaffolds requires similar ECM breakdown. Human T blasts and SupT1 lymphoma cells expressed mRNA of MMP-9, MT1-MMP, MT4-MMP, cathepsin L, uPA, and uPAR as well as ADAM-9, -10, -11, -15, and -17. Upon long-term migration within 3-dimensional collagen matrices, however, no in situ collagenolysis was obtained by sensitive fluorescein isothiocyanate–collagen fragmentation analysis and confocal fluorescence/backscatter microscopy. Consistent with nonproteolytic migration, T-cell crawling and path generation were not impaired by protease inhibitor cocktail targeting MMPs, serine proteases, cysteine proteases, and cathepsins. Dynamic imaging of cell-ECM interactions showed T-cell migration as an amoeba-like process driven by adaptive morphology, crawling along collagen fibrils (contact guidance) and squeezing through pre-existing matrix gaps by vigorous shape change. The concept of nonproteolytic amoeboid migration was confirmed for multicomponent collagen lattices containing hyaluronan and chondroitin sulfate and for other migrating leukocytes including CD8+ T blasts, monocyte-derived dendritic cells, and U937 monocytic cells. Together, amoeboid shape change and contact guidance provide constitutive protease-independent mechanisms for leukocyte trafficking through interstitial tissues that are insensitive toward pharmacologic protease inhibitors.
Introduction
The migration and recirculation of T lymphocytes through the tissues is a multistep process that requires cell adhesion coupled with cellular strategies to overcome physical tissue barriers.1 The principles of T-cell migration follow the paradigm of “amoeboid” movement established for the single cell state of the lower eucaryote Dictyostelium discoideum.2,3 Amoeboid movement is a fast low-affinity migration type driven by a roundish yet flexible cell morphology, dynamic and polarized pseudopod protrusions and retractions, which are independent of focal contact formation and stress fibers.2 Similar to Dictyostelium, migrating T lymphocytes show an elliptoid polarized shape with a smooth yet dynamically ruffling leading edge, and an additional trailing uropod.4 This shape generates fast (5-25 μm/min) low-affinity gliding across surfaces and through 3-dimensional (3D) collagenous scaffolds4 driven by a diffuse cortical actin cytoskeleton along the inner leaflet of the plasma membrane devoid of focal contacts at substrate interactions and stress fibers.5,6 Amoeboid T-cell migration within extracellular matrix (ECM) occurs independently of β1 integrin–mediated adhesion both in vitro6 and in vivo,7 suggesting differences of the molecular basis between the movement of lymphocytes and other mobile cells, such as fibroblasts and tumor cells.8,9
After transendothelial migration, T cells and other leukocytes enter the fibrillar and reticular networks of the extracellular matrix in peripheral and lymphatic tissues, predominantly composed of fibrillar type I and III collagens.10,11 In many cell types, adhesion and cytoskeletal dynamics in collagenous tissues are coordinated with proteolytic strategies to lower the physical matrix resistance. Fibroblasts and tumor cells, as well as neutrophils, migrate across ECM proteins such as gelatin and fibronectin while simultaneously generating localized tracks of proteolytic substrate degradation,12-15 prompting concepts on pericellular proteolysis as a cellular strategy to overcome matrix barriers.16,17 Pericellular matrix degradation is mediated by secreted as well as cell-bound matrix proteases, including matrix metalloproteinases (MMPs), serine proteases, and cathepsins.13,14,18 In tumor cells interacting with ECM, cell surface MMPs and serine proteases become enriched at the surface of leading pseudopods (“invadopodia”) to cooperate with integrins and other adhesion receptors in a focalized manner.12,17,18 Focalization of ECM cleavage can occur by 2 principal mechanisms: membrane-bound proteases such as MT-MMPs coclustering with β1 or β3 integrins at contacts to substrate,18,19 or heterophilic binding of soluble MMPs to cell surface receptors, such as MMP-2 binding to MT1-MMP or urokinase-type plasminogen activator (uPA) to its receptor uPAR.20,21 Whereas focal contacts recruit proteases to substrate interactions in fibroblasts and tumor cells,18,19,22 it remains unresolved how pericellular proteolysis is spatially and temporally achieved by short-lived and molecular diffuse interactions to ECM substrate, as developed by rapidly moving leukocytes that generate migration velocities that exceed those of fibroblasts and tumor cells by 20- to 40-fold.8
In monocytes, dendritic cells, and neutrophils, proteases from different classes are constitutively expressed, including MMPs, sheddases (eg, a disintegrin and a metalloproteinase [ADAMs]), cysteine, and aspartatic proteases (eg, cathepsins), as well as serine proteases (eg, cathepsin G, urokinase-type plasminogen activator, human leukocyte elastase [HLE]).17,23,24 Resting peripheral T lymphocytes express a more restricted spectrum of proteases and only upon activation do T cells up-regulate or de novo express MMP-2, MMP-9, cathepsins, members of the PA/plasmin-system, and HLE.25-27 The penetration of T cells and other leukocytes through basement membrane equivalents (matrigel) in vitro is facilitated by active MMP-2 and MMP-9,26,28 however it remains subject to debate in which tissue compartments and under which conditions matrix proteases contribute to transendothelial migration in vitro and in vivo.29-32
Although focalized proteolysis, as established for stromal cells, is an important mechanism supporting cell migration through tissue scaffolds, alternative pathways for bypassing ECM barriers might exist, depending on cell type and ECM environment. In the present study, we used rapidly moving T cells and other leukocytes to study the expression of MMPs and additional proteases and their function in migration through 3D fibrillar collagen matrices. Although we initially aimed at the mechanisms of focalized proteolysis, we followed up on the unexpected finding of undiminished leukocyte migration in the presence of protease inhibitors and show how amoeboid migration provides a nonproteolytic mechanism to overcome 3D collagenous tissue barriers.
Materials and methods
Antibodies, cells, and cell culture
For flow cytometry, rabbit IgG-polyclonal anti–MT1-MMP Ab (Chemicon, Hofheim, Germany) and isotypic rabbit IgG (Sigma, Taufenkirchen, Germany) were used.
Human peripheral blood mononuclear cells (PBMCs) were obtained from peripheral blood from different healthy donors (by density centrifugation on a 1.077 g/mL ficoll gradient (Lymphoprep; Axis-Shield, Oslo, Norway), washed, and stimulated for 3 days with 1.25 μg/mL Concavalin A (Oncogene, Schwalbach, Germany) and 100 U/mL IL-2 (Strathman Biotech, Hamburg, Germany) in RPMI medium. CD4+ and CD8+ T cells from stimulated PBMC cultures were obtained by positive immunomagnetic selection.33,34 Purity of isolated CD4+ and CD8+ populations was 95%-99%, as determined by flow cytometry.34 Human monocyte–derived dendritic cells (DCs) were generated from peripheral blood of different donors by cultivation in the presence of 5% autologous serum, granulocyte-macrophage colony-stimulating factor (GM-CSF), and interleukin-4 (IL-4) for 10 days and terminal maturation using tumor necrosis factor α (TNFα), IL-1β, IL-6, and prostaglandin E2 (PGE2) after 7 days.33
The human lymphoma CD4+ T-cell line SupT126 was obtained from the Deutsche Sammlung für Mikroorganismen und Zellkulturen (DSMZ, Braunschweig, Germany). SupT1 cells and U937 monocytic cells were cultured as suspension in RPMI medium (PAN, Heidenheim, Germany). HT-1080/MT1 fibrosarcoma cells overtransfected with MT1-MMP21 were used as collagenolytic positive controls (kindly provided by Dr A. J. Strongin, The Burnham Institute, La Jolla, CA). If not stated otherwise, all primary cells and cell lines were cultured in medium containing 50 U/mL penicillin and 50 μg/mL streptomycin (PAN, Verwiers, Belgium) and 10% heat-inactivated fetal calf serum (FCS) (Biowhittaker, Verwiers, Belgium) at 37°C in humidified 5% CO2 atmosphere. HT-1080/MT1 cells were cultured in DMEM (PAN) containing 0.2 mg/mL G418 (Oncogene).
Collagen matrix migration assay, cell tracking, and cell viability
3D collagen matrix cultures (collagen concentration: 1.67 mg/mL) of spontaneously migrating cells were monitored by time-lapse videomicroscopy.34 The collagen (Vitrogen, Cohesion, Palo Alto, CA) was resistant to trypsin and sensitive to degradation by MT1-MMP, confirming its native state as shown by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and silver staining (C. Overall, E. Tam, unpublished, May 2002). In some experiments, multicomponent lattices were generated by copolymerizing dermal collagen (1.67 mg/mL), human umbilical high-molecular-weight cord hyaluronan (1 mg/mL; Sigma) and chondroitin-4-sulfate (20 mg/mL; Sigma), as described.35 For all cell types spontaneous migration was investigated except U937 cells, which were stimulated by lysophosphatidic acid (0.5 μM; Sigma).
Locomotor parameters were obtained by computer-assisted cell tracking and reconstruction of the xy coordinates of cell paths for a step interval of 1 (lymphocytes; Sup T1 cells)34 or 3 minutes (U937 cells; DCs). The average velocity and percentage of locomoting cells in a population were calculated from each step interval of randomly selected cells divided by the number of cells.34
Cell viability was routinely assessed after migration experiments. Cells were released from the collagen lattice by collagenase digestion (Clostridium histolyticum collagenase type I; Sigma), stained by propidium iodide (Sigma), and analyzed by flow cytometry.
Reverse transcriptase–polymerase chain reaction
Total RNA from ConA-blasts and Sup-T1 cells grown in liquid culture was isolated using the RNeasy kit (Qiagen, Hilden, Germany). One microgram of total RNA was reversely transcribed (1st Strand cDNA Synthesis Kit; Roche Diagnostics, Mannheim, Germany) to cDNA using AMV Reverse Transcriptase (1000 U/mL), dNTPs (1 mM each), MgCl2 (5 mM), random primer p(dN)6 (80 μg/mL), gelatin (10 μg/mL), RNase inhibitor (2500 U/mL), and reaction buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]; 50 mM KCl; pH8,3). Each 0.25 μg cDNA was amplified by PCR in a reaction using bacterial recombinant Taq DNA polymerase (12.5 U/mL), dNTPs (0.2 mM), MgCl2 (1.5 mM), sense and antisense primer (0.2 μM each) (primer sequences and references can be viewed in Supplemental Table S1 on the Blood website; see the Supplemental Materials link at the top of the online article) and reaction buffer (10 mM Tris-HCl, pH 8.8; 50 mM KCl; 0.08% NP40; Life Technologies, Karlsruhe, Germany). Uniqueness of primer region was confirmed using the National Center for Biotechnology Information blastn program. PCR was performed for 30 cycles. Absence of DNA contamination was confirmed by subjecting total cell lysates (0.25 μg total RNA) to PCR in the absence of reverse transcriptase. The resulting PCR products were analyzed by electrophoresis in a 2% agarose gel and stained with ethidium bromide (1 μg/mL). Specificity was determined by the length of each PCR product.
Protease inhibitors
Broad-spectrum MMP inhibitor BB-2516 (marimastat) was kindly provided by British Biotech (British Biotech, Oxford, United Kingdom). BB-2516 blocks most MMPs and to some extent ADAMs, at nM or low μM ranges.36 Further protease inhibitors were aprotinin, trans-epoxysucciny-l-leucylamide-(4-guanidino)butane (E-64), pepstatin A (all from Sigma), and leupeptin (Molecular Probes, Leiden, The Netherlands). Inhibitor cocktail was used at the following concentrations: marimastat, E-64, and pepstatin A at 20 μM; aprotinin 0.7 μM; leupeptin 2 μM (for inhibitor specificity, established inhibitory concentration range, and references, see Supplemental Table S2).
Detection of collagenolysis
The efficiency of MMP inhibitor marimastat was controlled by zymography using recombinant MT1-MMP (Invitek, Berlin, Germany), MMP-2, and MMP-9 from supernatants conditioned by HT-1080/MT1 cells. Gelatin zymograms were obtained overnight in the absence or presence of marimastat (0.01-1 μM) in enzyme buffer and developed by Coomassie blue staining.
Collagenolysis generated by live cells while migrating through the native 3D collagen substrate was quantified by a novel fluorescein isothiocyanate (FITC)–release assay. Two percent FITC-collagen type I monomers from bovine skin (Molecular Probes) were copolymerized with rat-tail collagen (Becton Dickinson, Heidelberg, Germany; final concentration 1.65 mg/mL)18 and cells under phenol red–free conditions (3 × 105cells/l00 μL medium/ well). After 40 hours' culture in the absence or presence of protease inhibitor cocktail, solid-phase collagen including the cells was pelleted (15 000 × g, 10 minutes, 4°C), and the supernatant was analyzed for FITC contents by spectrofluorometry. 100% values represent total FITC recovery after collagenase digestion of cell-free collagen lattices. Background fluorescence was obtained for supernatants from pelleted nondigested cell-free lattices. In contrast to lymphocytes and monocytic cells, DCs generated high levels of unspecific FITC release that was not sensitive to protease inhibitors nor associated with the structural remodeling of collagen fibers. Compared to cells isolated directly after matrix polymerization, no increased cell-bound or endocytic FITC signal was observed after 24 hours culture in FITC-collagen lattices in untreated or protease inhibitor–treated cells using flow cytometry.
FITC-collagen monomers were degraded by recombinant MT1-MMP, MMP-2, as well as trypsin as shown by SDS-PAGE and silver staining (C. Overall and E. Tam, unpublished, May 2002). Whereas FITC release detected classical collagenase, gelatinase, and trypsinlike activity, it was most sensitive to MMP-inhibitor BB-2516 (reduction by 80% in HT-1080 cells18 ), indicating MMPs as primary yet not sole enzyme family in degrading the fibrillar collagen migration substrate. In contrast to detection of collagen fragments by SDS-PAGE and silver staining, FITC-collagen release was a more sensitive approach to detect collagen degradation in live cell samples (K.W., unpublished observations, April 2003).
Focalized cell-associated collagen degradation in situ was visualized by copolymerizing 5% quenched (q)FITC-collagen type I monomers from bovine skin (Molecular Probes) with nonlabeled collagen. As assessed by confocal microscopy, the fluorescence intensity of qFITC in polymerized collagen fibers strongly increased upon focalized proteolysis generated by proteolytic HT-1080 cells, which colocalized with the appearance of collagen cleavage-site specific neo-epitope mAb COL2 3/4 C (K.W. and P.F., unpublished, April 2003).
Confocal laser-scanning microscopy
3D confocal backscatter microscopy of fixed samples was carried out on a Leica TCS 4D system (Leica, Bensheim, Germany).37 Dynamic sequences from viable samples were obtained on the SP2 system (Leica) using a temperature-controlled stage (37°C). Cells within the collagen lattice were labeled with 1 μM calcein-AM (Molecular Probes) and scanned at 20-second time intervals as 3D stacks of 4 to 8 z-sections. For dynamic reconstruction, fluorescence, reflection and transmission signal were collected simultaneously. Movies were obtained from 3-dimensionally reconstructed image stacks over time. For digital image analysis, the National Institutes of Health image program (software version 1.62) was used.
Results
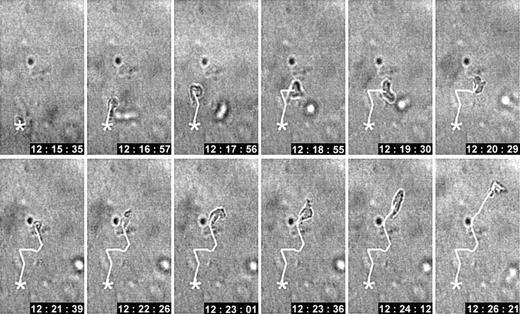
In 3D collagen matrices, ConA-activated T cells spontaneously developed an amoeboid migration type characterized by flexible elliptoid morphology. The rapid and frequent shape change and oscillatory stop-and-go patterns resulted in a nonlinear path structure (Figure 1). We have investigated which matrix-degrading enzymes are expressed by T cells and how these proteases contribute to migration and associated remodeling of the collagen fiber network.
Spontaneous locomotion of an activated CD4+T cell within 3D collagen lattice. Changes in morphology and oscillatory path development. CD4+ ConA blast migrating in a 3D collagen matrix in the absence of a chemotactic gradient. The average velocity was 6 μm/min. Time is indicated in hh:mm:ss (11 minutes total).
Spontaneous locomotion of an activated CD4+T cell within 3D collagen lattice. Changes in morphology and oscillatory path development. CD4+ ConA blast migrating in a 3D collagen matrix in the absence of a chemotactic gradient. The average velocity was 6 μm/min. Time is indicated in hh:mm:ss (11 minutes total).
Protease mRNA and surface expression
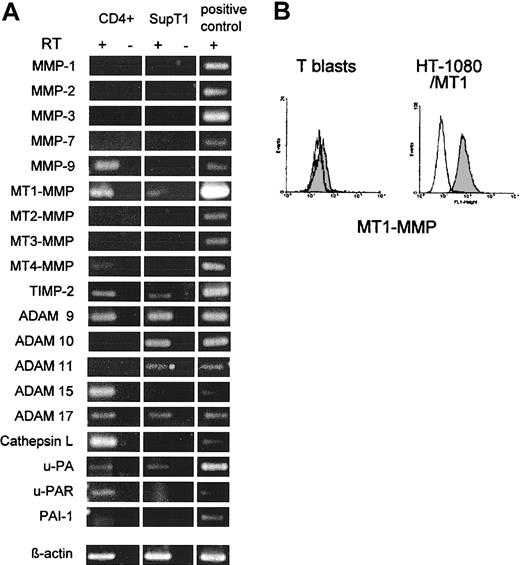
Primary human CD4+ ConA-T blasts and, for confirmation, SupT1 lymphoma cells26 were investigated for mRNA expression from proteases of different classes, including MMPs, serine proteases, and cathepsins (Figure 2 and Supplemental Table S1). Activated CD4+ T cells expressed MMP-9, MT1-MMP, MT4-MMP, ADAM-9, -15, and -17, cathepsin L, urokinase-type plasminogen activator (uPA) and its receptor uPAR as well as endogenous protease inhibitors TIMP-2 and PAI-1 (Figure 2A). SupT1 cells expressed MT1-MMP, ADAM-9, -10, -11, -17, uPA, and TIMP-2 (Figure 2A). MT1-MMP and cathepsin L were the only collagenases expressed by either cell type. ConA-stimulated CD4+ T blasts showed minor MT1-MMP surface expression (Figure 2B), while MT1-MMP surface levels were identical to the isotypic antibody in resting cells (not shown). As positive control, significant MT1-MMP surface staining was detected on HT-1080/MT1 cells (Figure 2B). However, even at low levels of surface protease expression, T cells might be able to focalize protease activity to substrate contacts and thereby generate local collagenolysis for tissue penetration.
mRNA and cell surface expression of proteases and endogenous protease inhibitors in CD4+T cells and SupT1 cells. (A) Total RNA from CD4+ T cells stimulated with ConA for 3 days and SupT1 lymphoma cells was subjected to RT-PCR (30 cycles). Fragment lengths conformed to the expected size. No mRNA was detected for MMP-8, -13, cathepsin B and K in either cell type (not shown). Positive controls were performed from HT-1080/MT1 or HT-1080/neo cells (MMP-7 and -9). RT indicates reverse transcriptase reaction. (B) Flow cytometry of MT1-MMP surface expression (gray histogram) in ConA-T blasts and positive control cells (HT-1080/MT1) as compared with isotype control (white histogram).
mRNA and cell surface expression of proteases and endogenous protease inhibitors in CD4+T cells and SupT1 cells. (A) Total RNA from CD4+ T cells stimulated with ConA for 3 days and SupT1 lymphoma cells was subjected to RT-PCR (30 cycles). Fragment lengths conformed to the expected size. No mRNA was detected for MMP-8, -13, cathepsin B and K in either cell type (not shown). Positive controls were performed from HT-1080/MT1 or HT-1080/neo cells (MMP-7 and -9). RT indicates reverse transcriptase reaction. (B) Flow cytometry of MT1-MMP surface expression (gray histogram) in ConA-T blasts and positive control cells (HT-1080/MT1) as compared with isotype control (white histogram).
Lack of in situ collagenolysis
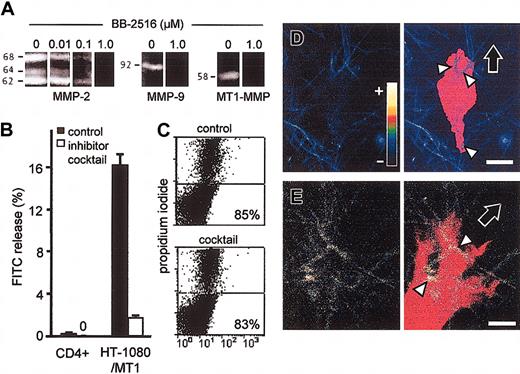
Initial dose-response studies on broad-spectrum MMP-inhibitor BB-2516 (marimastat) showed complete inhibition of gelatin degradation by MMP-2, MMP-9, and MT1-MMP at low μM concentration (Figure 3A). However, the topographic arrangement and focalization of proteolysis executed by live cells within the collagen substrate are not appropriately detected by zymography. Therefore, in situ collagen degradation caused by T cells in the process of migration was quantified as the FITC release from FITC-labeled fibrillar collagen lattices. After 40 hours of culture, ConA-stimulated CD4+ T blasts did not cause FITC release into the supernatant above background levels. In contrast, collagenolytic HT-1080/MT1 fibrosarcoma cells released high levels of soluble FITC into the supernatant (Figure 3B). For inhibition of proteases, a broad-spectrum inhibitor cocktail was used to simultaneously target MMPs, MT-MMPs, ADAMs, cathepsins, serine, and cysteine proteases.18 Near-total abrogation of collagenolysis by inhibitor cocktail was confirmed for HT-1080/MT1 cells as positive control, while no effect was obtained for CD4+ cells (Figure 3B). The lack of FITC release was not caused by a decrease in T-cell viability, as detected by flow cytometry after cell isolation by collagenase digestion after 40 hours (Figure 3C).
Lack of in situ collagen degradation by T blasts. (A) Dose-dependent inhibition of MMP-2, MMP-9, and recombinant MT1-MMP activity by BB-2615 (marimastat) in gelatin zymography. (B) Lack of FITC release from FITC-labeled collagen matrices by T cells, but not by HT-1080/MT1 cells (positive control). ConA-stimulated CD4+ T cells or HT-1080/MT1 cells were cultured in 3D collagen lattices containing 2% FITC-collagen in the absence or presence of protease inhibitor cocktail for 40 hours. Data show the means + SDs for 3 independent experiments. (C) The percentage of viable CD4+ cells remained unaffected after 40 hours of culture in collagen. (D) CD4+ T cell and (E) HT-1080/MT1 cell incorporated in a 3D collagen lattice containing 5% quenched FITC-collagen. The false-color encoded confocal FITC channel (color code inset: fluorescence channel for lowest (–) and highest (+) intensity, respectively) of the 3D reconstructed in focus sections (left) was superimposed on the shape of the cell body (right; red false color) obtained from the transmission image. In CD4+ T cell, physical contact with collagen fibers (arrowheads) did not result in increased cleavage-related fluorescence (blue false color; arrowheads), whereas HT-1080/MT1 cell (positive control) generated focal FITC-fluorescence at fiber binding sites (yellow false color; arrowheads). Fiber cleavage by HT-1080 cells in situ was confirmed by staining with cleavage-site specific antibody (not shown). Images are representative for 10 to 15 cells. Bars, 5 μm. Black arrows, direction of migration.
Lack of in situ collagen degradation by T blasts. (A) Dose-dependent inhibition of MMP-2, MMP-9, and recombinant MT1-MMP activity by BB-2615 (marimastat) in gelatin zymography. (B) Lack of FITC release from FITC-labeled collagen matrices by T cells, but not by HT-1080/MT1 cells (positive control). ConA-stimulated CD4+ T cells or HT-1080/MT1 cells were cultured in 3D collagen lattices containing 2% FITC-collagen in the absence or presence of protease inhibitor cocktail for 40 hours. Data show the means + SDs for 3 independent experiments. (C) The percentage of viable CD4+ cells remained unaffected after 40 hours of culture in collagen. (D) CD4+ T cell and (E) HT-1080/MT1 cell incorporated in a 3D collagen lattice containing 5% quenched FITC-collagen. The false-color encoded confocal FITC channel (color code inset: fluorescence channel for lowest (–) and highest (+) intensity, respectively) of the 3D reconstructed in focus sections (left) was superimposed on the shape of the cell body (right; red false color) obtained from the transmission image. In CD4+ T cell, physical contact with collagen fibers (arrowheads) did not result in increased cleavage-related fluorescence (blue false color; arrowheads), whereas HT-1080/MT1 cell (positive control) generated focal FITC-fluorescence at fiber binding sites (yellow false color; arrowheads). Fiber cleavage by HT-1080 cells in situ was confirmed by staining with cleavage-site specific antibody (not shown). Images are representative for 10 to 15 cells. Bars, 5 μm. Black arrows, direction of migration.
To exclude minor local collagen fiber cleavage exerted by T cells that might escape detection by FITC release, collagen lattices containing quenched FITC-labeled collagen monomers were used for confocal fluorescence and backscatter microscopy. At physical contacts of polarized CD4+ blasts to collagen fibers containing quenched FITC, no focal in situ fluorescence above background fluorescence was detected (Figure 3D, arrowheads), while significant focal fluorescence was developed by HT-1080/MT1 cells at interactions to these fibers (Figure 3E, arrowheads). Thus, MT1-MMP and other proteases, although expressed by CD4+, did not degrade the collagenous migration substrate.
Persisting migration in the presence of protease inhibitors
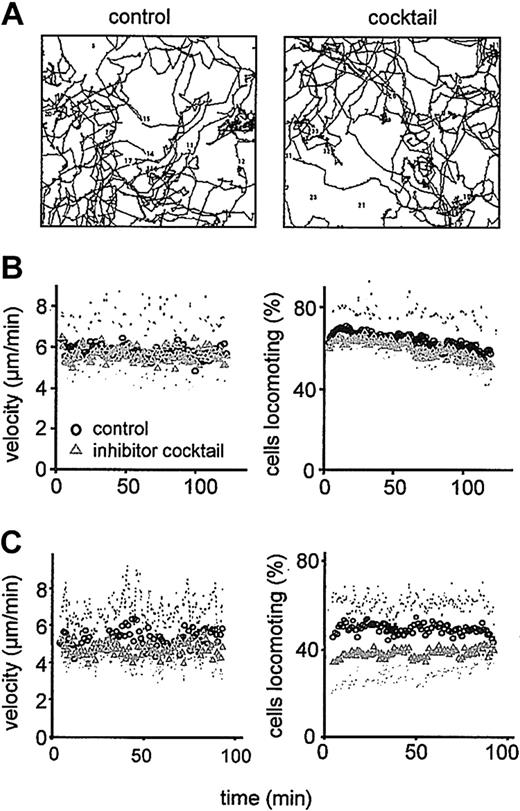
The contribution of enzymatic protease activity to T-cell migration in 3D collagen lattices was investigated in the absence or presence of protease inhibitor cocktail (Figure 4). Addition of inhibitor cocktail neither changed the amoeboid morphodynamics of CD4+ T cells (Supplemental Video 1) nor the oscillatory structure of the cell paths (Figure 4A). Also, the steady-state migration velocity was approximately 6 μm/min for both nontreated and inhibitor cocktail-exposed cells (Figure 4B, left). The number of migrating cells including their stop-and-go-pattern remained unchanged (Figure 4B, right). Likewise, T-cell migration within multicomponent matrices containing collagen, hyaluronan, and the cross-linker chondroitin sulfate did not depend on the function of matrix-degrading enzymes (Figure 4C). Thus, activated CD4+ T cells do not engage the enzymatic activity of expressed collagenases for their migration through 3D fibrillar collagen matrices.
Persisting migration of CD4+blasts in the presence of protease inhibitors. Migration in 3D collagen (A-B) and multicomponent matrices (C) in the absence or presence of protease inhibitor cocktail. (A) Digitized paths of 40 cells at orthotopic position (2-hour tracking period; 1 representative of 3 independent experiments). (B-C) Steady-state velocity and percentage of migrating CD4+ T cells in collagen (B) and collagen copolymerized with hyaluronan and chondroitin sulfate (C). Data show the means of time-dependent population parameters ± SDs for 3 independent experiments (120 cells).
Persisting migration of CD4+blasts in the presence of protease inhibitors. Migration in 3D collagen (A-B) and multicomponent matrices (C) in the absence or presence of protease inhibitor cocktail. (A) Digitized paths of 40 cells at orthotopic position (2-hour tracking period; 1 representative of 3 independent experiments). (B-C) Steady-state velocity and percentage of migrating CD4+ T cells in collagen (B) and collagen copolymerized with hyaluronan and chondroitin sulfate (C). Data show the means of time-dependent population parameters ± SDs for 3 independent experiments (120 cells).
Biophysics of nonproteolytic amoeboid T-cell migration
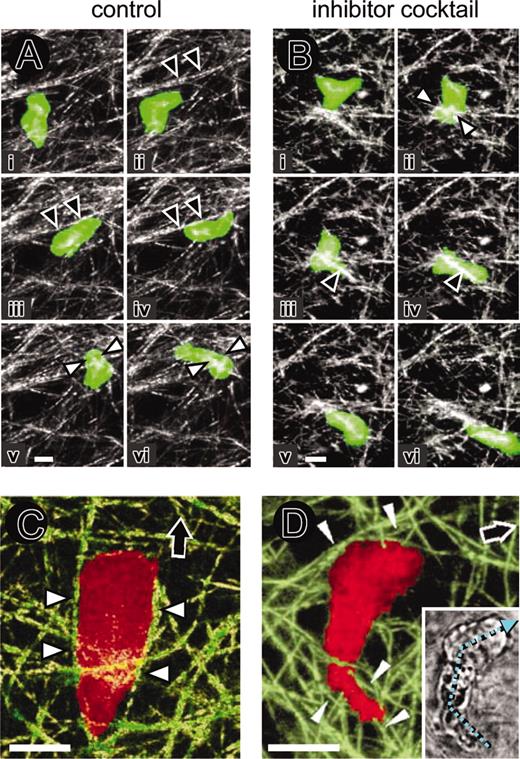
Because T-cell migration was neither accompanied by proteolytic collagen fiber degradation or sensitive to protease inhibitor cocktail, T-cell trafficking through fibrillar collagen should occur through a nonproteolytic mechanism. The biophysics of T-cell migration within the collagen network were reconstructed at high resolution by 4D confocal backscatter and fluorescence microscopy (Figure 5). In both, control and inhibitor cocktail–treated cultures, migrating T cells exhibited periods of alignment along matrix fibers (Figure 5A-B, black arrowheads; Supplemental Video 2). Propulsive squeezing through regions of narrow space prompted the formation of a narrow zone of the cell body, termed constriction ring38 (Figure 5Av-vi; Bii; white arrowheads). Whereas forward movement resulted from alignment of the cell body in parallel to more linear fiber strands (Figure 5C, black arrowheads), directional changes were frequent at regions of narrow space, forcing the cell to circummigrate rather than degrade the obstacle (Supplemental Video 3). Such migratory turns were caused by cell deflection along adjacent collagen fibers (Figure 5D, arrowheads) and resulted in angle changes along the length axis between leading edge and trailing uropod (Figure 5D, inset; cyan arrow). As was apparent from the movies that any change in cell shape, including cell extension and fiber pushing, cell contraction and fiber pulling, as well as outward dislocation of constraining fibers upon cell squeezing, resulted in temporary deformation of the collagen network structure (dislocation up to 2 μm; movies 2-3). These contact-dependent and temporary changes did not, however, generate long-lasting structural changes in matrix structure after the cell had detached.
Amoeboid T-cell migration, physical cell-fiber interaction, and contact guidance within 3D collagen matrix in the absence or presence of protease inhibitor cocktail. (A) Untreated and (B) protease inhibitor cocktail–teated calcein-stained T cells. Crawling, alignment along fiber strands (black arrowheads) and formation of constriction rings (white arrowheads). Image sequences (i-vi) represent as a time series 8 (A) and 3 (B) minutes of observation time. (C) T-cell alignment in parallel to matrix fibers (white arrowheads) upon forward migration (black arrow). (D) Change in migration direction. Angle turns along the cellular length axis (cyan arrow, small inset) and physical confinement of the cell body and uropod along guiding collagen fibers (white arrowheads), reflecting the direction change. Bars, 5 μm.
Amoeboid T-cell migration, physical cell-fiber interaction, and contact guidance within 3D collagen matrix in the absence or presence of protease inhibitor cocktail. (A) Untreated and (B) protease inhibitor cocktail–teated calcein-stained T cells. Crawling, alignment along fiber strands (black arrowheads) and formation of constriction rings (white arrowheads). Image sequences (i-vi) represent as a time series 8 (A) and 3 (B) minutes of observation time. (C) T-cell alignment in parallel to matrix fibers (white arrowheads) upon forward migration (black arrow). (D) Change in migration direction. Angle turns along the cellular length axis (cyan arrow, small inset) and physical confinement of the cell body and uropod along guiding collagen fibers (white arrowheads), reflecting the direction change. Bars, 5 μm.
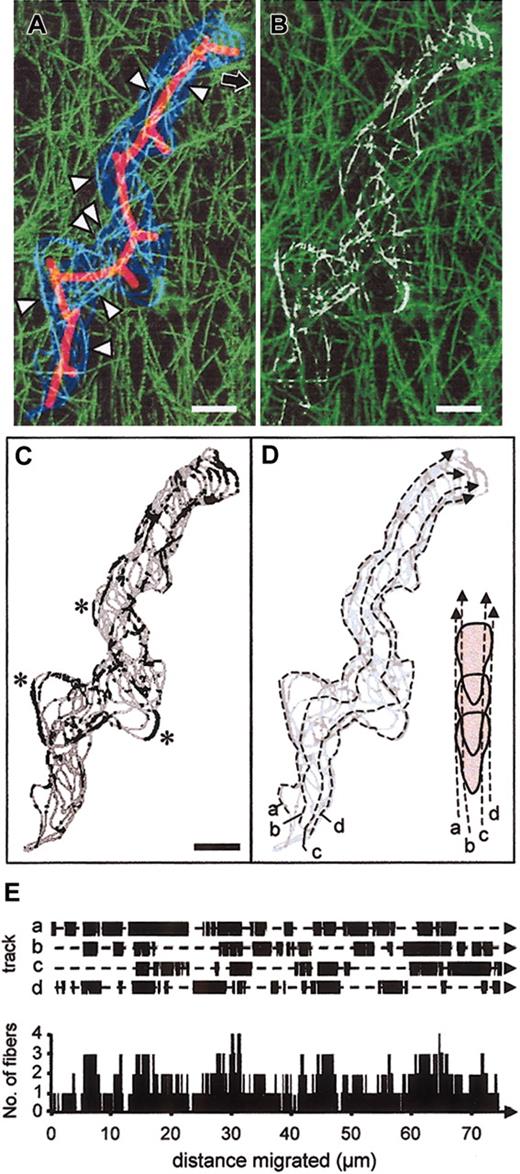
To analyze the dynamic nature of cellular interactions with collagen fibers quantitatively, the cell boundary of a migrating T cell (depicted in Figure 1) was obtained as 2D outline every 24 seconds (Figure 6A, dark blue) and superimposed onto the 3D reconstruction of the transmigrated collagen matrix (Figure 6A, green). Colocalization of outline and collagen fibers resulted in bright pixels (Figure 6A, cyan; arrowheads) at different position along the migration track (Figure 6A, red solid line). The position of collagen fibers that were touched upon migration shows that the volume of the cell follows precisely the predefined collagen scaffold (Figure 6B, white fiber zones). The location and frequency of cell-fiber guidance along the extracted cell outlines and their regions in parallel contact with fibers (Figure 6C, black segments) were quantified along 4 virtual tracks placed into the cell shape from step to step in order to represent the geometry of both maximum cell width (Figure 6D, a and d) and more narrow regions near the uropod (Figure 6D, b and c). Along the migration path, each of these tracks showed alternating contacts with collagen fibers ranging from 1 to 10 μm in length (Figure 6E, top). Although each interaction segment was of short-lived nature, together they generated a multidimensional physical scaffold for cell alignment to at least one, frequently 2 or more, fibers simultaneously (Figure 6E, bottom). By means of elongation and, presumably, by volume (which was not considered in this analysis yet apparent from the movies), polarized T cells simultaneously aligned at several positions to different fibers resulting in near-always guiding contact by outside cues (see also movies 1-3). Because such contact guidance determined both forward movement as well as migratory turns (Figure 6C, asterisks), no bundling, structural degradation, or remodeling of matrix fibers were required for fast T-cell crawling within collagenous scaffolds. These findings contrast with the migration-associated proteolytic degradation and remodeling of the same collagen substrate by fibroblasts and tumor cells (Friedl et al,8 Wolf et al,18 Maaser et al,35 and movies therein).
Reconstruction of contact guidance of migrating T cell by collagen fibers. (A) The outline of the cell depicted in Figure 1 was obtained every 24 seconds and superimposed on the 3D reconstructed backscatter signal of the collagen fibers. Cell boundary (blue), 3D collagen matrix (green; 10 μm in depth), and colocalization of cell boundary and individual collagen fibers (cyan, arrowheads). The mean path and pseudopodal extensions are shown by the solid red line. Migration occurred toward the top right corner (black arrow). (B) False color representation of the physics of the transmigrated path (white segments), calculated from pixel colocalization between T-cell boundary and collagen fibers. (C) Extracted cell boundary (gray line) and segments colocalized with collagen fibers (black sections). Asterisks indicate turns guided by outside fiber cues. (D) Definition of 4 virtual tracks along the migration path to approximate 4 major morphological regions of a polarized T cell; for instance, lateral portions of the anterior head (a, d) and posterior cell parts near the uropod (b, c). (E) Calculation of pixel series that represent black segments in (C) for each track along the migration path (E, top graph), resulting in the cumulative number of fibers that were simultaneously aligned along the forward moving cell edge (E, bottom graph). These reconstruction data are representative for more than 5 independent cells reconstructed by dynamic confocal backscatter imaging (compare movies 1-3). Bars, 5 μm.
Reconstruction of contact guidance of migrating T cell by collagen fibers. (A) The outline of the cell depicted in Figure 1 was obtained every 24 seconds and superimposed on the 3D reconstructed backscatter signal of the collagen fibers. Cell boundary (blue), 3D collagen matrix (green; 10 μm in depth), and colocalization of cell boundary and individual collagen fibers (cyan, arrowheads). The mean path and pseudopodal extensions are shown by the solid red line. Migration occurred toward the top right corner (black arrow). (B) False color representation of the physics of the transmigrated path (white segments), calculated from pixel colocalization between T-cell boundary and collagen fibers. (C) Extracted cell boundary (gray line) and segments colocalized with collagen fibers (black sections). Asterisks indicate turns guided by outside fiber cues. (D) Definition of 4 virtual tracks along the migration path to approximate 4 major morphological regions of a polarized T cell; for instance, lateral portions of the anterior head (a, d) and posterior cell parts near the uropod (b, c). (E) Calculation of pixel series that represent black segments in (C) for each track along the migration path (E, top graph), resulting in the cumulative number of fibers that were simultaneously aligned along the forward moving cell edge (E, bottom graph). These reconstruction data are representative for more than 5 independent cells reconstructed by dynamic confocal backscatter imaging (compare movies 1-3). Bars, 5 μm.
Nonproteolytic amoeboid migration in other leukocytes
Similar to T cells, a spectrum of matrix-degrading proteases is expressed in other leukocytes, such as SupT1 cells26 (Figure 2B), DCs,23 and monocytic cells.24 Similar to T cells, however, only background release of FITC was obtained from FITC-collagen for these cells upon migration over an extended time period of 40 hours (Figure 7A), and no reduction of FITC release was achieved by protease inhibitor cocktail (Figure 7A). The protease-independence of migration in these cells was shown as an undiminished velocity as well as number of moving cells in the presence of protease inhibitor cocktail (Figure 7B; Supplemental Video 4). Although the size and polarized morphology differ between lymphocytes, monocytes, and DCs, each cell type followed the principles of amoeboid movement, as indicated by rapid shape change (movie 4), the formation of constriction rings (Figure 7C and Supplemental Video 5), and concomitant cytoplasmic streaming through regions of narrow space (Supplemental Video 5).
Protease-independent migration of other leukocytes. (A) Release of FITC from FITC-containing 3D collagen matrix by migrating SupT1 lymphoma cells, CD8+ T blasts, U937 monocytic cells, and monocyte-derived human dendritic cells. Cells were cultured in FITC-collagen for 40 hours in the absence or presence of protease inhibitor cocktail. Data represent the means ± SD from 3 independent experiments. High nonspecific fluorescence of unclear origin was obtained for supernatants from dendritic cells only. This fluorescence was neither sensitive to protease inhibitors nor did it reflect fiber degradation, as previously shown by confocal analysis.46 (B) Mean velocity and percentage of locomoting cells in the absence or presence of inhibitor cocktail. Data show the mean activities over time (>1 hour) and standard deviations from 3 independent experiments (120 cells) obtained from cell tracking. (C) Amoeboid movement of U937 cell resulting in the formation of a constriction ring (arrow) that remains unchanged in position. Bar, 10 μm.
Protease-independent migration of other leukocytes. (A) Release of FITC from FITC-containing 3D collagen matrix by migrating SupT1 lymphoma cells, CD8+ T blasts, U937 monocytic cells, and monocyte-derived human dendritic cells. Cells were cultured in FITC-collagen for 40 hours in the absence or presence of protease inhibitor cocktail. Data represent the means ± SD from 3 independent experiments. High nonspecific fluorescence of unclear origin was obtained for supernatants from dendritic cells only. This fluorescence was neither sensitive to protease inhibitors nor did it reflect fiber degradation, as previously shown by confocal analysis.46 (B) Mean velocity and percentage of locomoting cells in the absence or presence of inhibitor cocktail. Data show the mean activities over time (>1 hour) and standard deviations from 3 independent experiments (120 cells) obtained from cell tracking. (C) Amoeboid movement of U937 cell resulting in the formation of a constriction ring (arrow) that remains unchanged in position. Bar, 10 μm.
Discussion
In contrast to fibroblasts39 and tumor cells,9,18 the migration of T cells and other leukocytes within 3D collagen matrices is independent of proteolytic ECM remodeling and therefore insensitive to protease inhibitor treatment. Our data provide a mechanism on how amoeboid crawling supports nondestructive leukocyte trafficking through interstitial tissues.8
Lack of migration-associated collagenolysis in migrating T cells
In this study, T lymphocytes were used as prototype cells for amoeboid movement in collagen. Although several matrix-degrading proteases are expressed by activated CD4+ T cells and SupT1 lymphoma cells, including MT1-MMP, MT4-MMP, cathepsin L, ADAM-9, and -1526,40 and although cathepsin L and MT1-MMP are potent collagenases toward native type I collagen,41,42 no degradation of the fibrillar collagen substrate was caused by them upon migration. We used a combination of physical and biochemical approaches to detect collagenolysis in situ and inhibitor efficiency: (1) gelatin zymography, (2) FITC-collagen release from FITC-labeled collagen matrices, (3) focal in-situ fluorescence at T-cell interactions to collagen fibers containing quenched FITC molecules, and (4) structural fiber remodeling using high-resolution confocal backscatter microscopy from live cell samples. Specific FITC release from FITC-collagen fibrils was at low levels for CD4+ and CD8+ blasts, SupT1 lymphoma cells, and U937 monocytic cells and largely insensitive to the multicomponent protease inhibitor cocktail. These values were 20- to 100-fold lower than seen in HT-1080/MT1 fibrosarcoma cells used as positive controls.18,21 As exception, DCs generated significant FITC release by mechanisms that were independent of the protease classes targeted by our inhibitor cocktail, thereby currently precluding a meaningful use of the FITC release assay for DCs. Besides collagenases, other enzymes generated by DCs, such as amino hydrolases,43 could dissolve the amid bond between FITC and protein carrier without cleaving the collagenous backbone.
Migrating T cells neither bundled, extensively pulled, nor reorganized collagen fibers nor generated focal cleavage of quenched FITC-collagen or proteolytic matrix defects, in contrast to collagenolytic HT-1080 cells.18 The protease inhibitor cocktail reduced collagenolysis by 95%-98% and abrogated fiber remodeling and proteolytic path generation in tumor cells.18 In leukocytes, however, the protease inhibitor cocktail did not alter any of the migration parameters assessed by sensitive cell tracking, including percentage of migrating cells, their velocity or stop-and-go-pattern, nor their path profiles and the underlying interaction dynamics to collagen fibers. These findings suggest that leukocytes do not need to structurally modify the fibrillar collagen network for their migration, thereby extending previous indirect evidence on T cells penetrating radioactive-labeled collagen matrices4 and neutrophils migrating within amnionic membrane.44 We cannot completely exclude minor focal cleavage of collagen by biochemical means. However, collagen backscatter imaging shows that the physical fiber structure and position remain intact during and after T-cell migration, whereas migrating tumor cells generate 3D matrix defects using the same collagen substrate.18,37 Together, these data argue against a biophysically significant change in matrix architecture by leukocytes along their migration tracks and further strengthen concepts on differences of molecular migration strategies between leukocytes and stromal cells.8,9
Amoeboid shape change: a nonproteolytic, biophysical migration mechanism bypassing matrix barriers
Previous dynamic reconstruction on nonlabeled neutrophils, T cells, and dendritic cells show leukocyte movement in collagen or amnionic membrane as a contact guidance-dependent process that appears to follow pre-existing matrix scaffolds.4,6,44-46 We have used improved dynamic real-time reconstruction of fluorescently labeled cells together with high-resolution imaging of the 3D matrix architecture. The typical random paths and turns developed by T cells in 3D collagen lattices result from stringent alignment of the cell body in parallel to fibrils and fiber bundles that border random gaps and trails pre-existing in the lattice. While following fibrillar guidance tracks, the volume of the cell body reflects the preformed space of trails of least resistance within the matrix. These reconstruction data support previous work that suggests physical contact guidance mechanisms to direct leukocyte migration in scaffolds independently of proteolytic ECM breakdown.6,44,45 In regions of narrow space, depending on the pore size, 2 different mechanisms sustain nonproteolytic movement. First, squeezing through narrow gaps by adapting the cell shape and the formation of a constriction ring,38 followed by cytoplasmic streaming through this immobile external confinement zone. Alternatively, if matrix barriers are too dense relative to the deformation ability of the cell (below 1-2 μm in diameter; compare movie 3), a change in migration direction provide movement around the obstacle. Hence, nonpenetrable regions are circummigrated at near-undiminished velocity.
These hallmarks of amoeboid biomechanics are clearly retained in migrating T cells, lymphoma cells, and monocytes. In DCs, the ubiquitous dendrites and an even more striking morphologic flexibility allow for sometimes grotesque deformation of the cell body (movie 4). Although the principal elliptoid amoeboid shape appears concealed in DCs, other amoeboid characteristics, such as shape change in response to outside cues, cell constriction and movement independent of proteases and β1 integrins remain retained throughout migration.46 Together, the data suggest that, similar to neutrophils,2 amoeboid features are retained in migrating T cells, monocytes, and DCs, likewise.
Nonproteolytic leukocyte migration in vivo
Whereas direct in vivo measurements on protease function in leukocyte migration are currently lacking, circumstantial evidence suggests the contribution of protease-independent processes to T-cell trafficking through the body. First, no obvious changes in peripheral T-cell counts and redistribution are apparent in vivo during MMP and other protease inhibitor–based therapy.31,32,47 Second, resting T cells are capable of entering lymphatic tissues through basement membranes underlying blood vessels and recirculating, although they produce little or no proteases. Third, resting as well as activated T cells generate high migration velocities up to 30 μm/min in vivo.48 As in 3D collagen, the resulting contact times toward individual collagen fibers in vivo are in the range of only 5 to 60 seconds (similar to in vitro kinetics; compare Figure 6E), thereby greatly exceeding interaction dynamics achieved by stromal cells that execute pericellular proteolysis (contact time in the range of minutes to hours).8 Lastly, a lack of matrix degradation by migrating T cells is obvious from in vivo imaging in the intact lymph node.48
Besides leukocyte shape change and contact guidance, additional (patho)physiologic mechanisms are likely to support nonproteolytic lymphocyte trafficking through 3D tissues. In vivo, regions of loose fibrillar collagen networks support oedematous swelling and reversible widening of matrix gaps in response to vasodilatation and inflammation. Local edema is likely to generate biomechanically suitable pathways for rapid leukocyte trafficking independent of proteolytic matrix degradation. In the dermis, such loose connective tissue zones, which are closely mimicked by collagen matrices, include perivascular spaces located along blood vessels, adjacent to basement membranes, and in the dermal papillae.10,11,18 These “trafficking highways” are preferentially used by passenger leukocytes upon acute and chronic inflammation, such as in eczema.10 Although the basic migration program in T cells and other leukocytes does not require proteases for removing matrix barriers, the next steps leading toward inflammatory connective tissue turnover and destruction remain to be investigated. These likely require additional microenvironmental changes, including the up-regulation and function of proteases in bystanding fibroblasts, endothelial cells, and/or other leukocytes.
Implications
Amoeboid shape change and guidance along pre-existing matrix structures provide supramolecular strategies for protease-independent movement through different fibrillar tissue matrices. Because rapidly moving leukocytes and, in particular, T cells establish only short-lived substrate contacts coupled to diffusely distributed integrins and actin cytoskeleton, it is reasonable to assume that their particular membrane architecture may not support sufficient receptor focalization required for contact-mediated proteolysis seen in slower moving cell types, such as fibroblasts and tumor cells.18 Amoeboid leukocyte movement, hence, represents a specialized migration type that is resistant to pharmacotherapeutic targeting of MMPs and other proteases, thereby explaining why clinical trials have not been complicated by leukocyte recirculation and immune defects. These characteristics may provide a mechanism for persistent tissue integrity despite abundant constitutive lymphocyte influx and recirculation.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2002-12-3791.
Supported by the Deutsche Forschungsgemeinschaft (FR 5511/2-2 and 2-3). K.W. was additionally supported by the foundation Evangelisches Studienwerk e.V., Haus Villigst.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We acknowledge Margit Ott for invaluable technical assistance, Bong-Seok Song for assistance with confocal microscopy, and Jörg Geiger for expert help in fluorometric analysis. Eric Tam and Chris Overall are acknowledged for collagen cleavage analysis.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal