Abstract
The spi1 (pu.1) gene has recently been identified as a useful marker of early myeloid cells in zebrafish. To enhance the versatility of this organism as a model for studying myeloid development, the promoter of this gene has been isolated and characterized. Transient transgenesis revealed that a 5.3 kilobase promoter fragment immediately upstream of the spi1 coding sequence was sufficient to drive expression of enhanced green fluorescent protein (EGFP) in injected embryos in a manner that largely recapitulated the native spi1 gene expression pattern. This fragment was successfully used to produce a germ line transgenic line of zebrafish with EGFP-expressing myeloid cells. These TG(spi1:EGFP)pA301 transgenic zebrafish represent a valuable tool for further studies of myeloid development and its perturbation.
Introduction
The zebrafish (Danio rerio) is an efficient genetic system for the study of vertebrate development and its perturbation,1 including hematopoiesis.2,3 Zebrafish erythropoiesis has been particularly well studied, largely augmented by hemoglobin pigment that readily flags erythroid cells in this transparent organism. Numerous mutants with defects in erythroid development have been identified,4,5 some of which present molecular and phenotypic copies of congenital human hematopoietic diseases.6,7 Both macrophage and 2 distinct granulocytic lineages have been identified.8-10 However, the lack of an obvious visual marker has limited studies of zebrafish myelopoiesis. Instead, investigators have largely resorted to using in situ histochemistry and hybridization with myeloid-specific markers, including l-plastin,8 mpx,9,10 c/ebp1,11 and spi1.12
The SPI1 (PU.1) gene encodes a member of the E26-transformation–specific (Ets) family of transcription factors that possesses a glutamine-rich transactivating domain at its N-terminus, a central proline–glutamic acid–serine–threonine (PEST) domain involved in protein-protein interactions, and a carboxyl-terminal domain that binds to canonical GAGGAA sequences found in the promoters of many hematopoietic genes.13-16 Targeted disruption of murine Spi1 has revealed that it plays a key role in the development of monocytes, B lymphocytes, and granulocytes.17,18 While the gene is expressed in a range of hematopoietic cells, myeloid expression is pronounced.13,19
Expression of spi1 marks the early compartment of zebrafish myelopoiesis.9,12 It is first expressed at 12 hours after fertilization (hpf) in cells of the lateral plate mesoderm (LPM) that subsequently radiate out across the yolk sac in the period 14 to 20 hpf, while a second site of expression appears in the caudal LPM, which disappears by 20 to 22 hpf. After 22 hpf, spi1 expression is observed in the posterior intermediate cell mass (pICM), and ultimately in the adult kidney, the site of zebrafish hematopoiesis.9,12 Given this expression pattern, we have isolated a functional spi1 promoter as a useful reagent for studies in zebrafish myeloid development. Transient expression studies showed that a 5.3 kilobase (kb) promoter fragment produced a pattern of enhanced green fluorescent protein (EGFP) expression that largely recapitulated native spi1 expression. This sequence was used to produce a viable transgenic line of zebrafish with EGFP-marked myeloid cells.
Study design
Molecular analysis
Techniques for library screening, sequence analysis, reverse transcription–polymerase chain reaction (RT-PCR), and whole-mount in situ hybridization were as described.10,12 Standard cloning techniques were used to subclone a 5.3-kb spi1 promoter fragment into pBluescript (Stratagene, La Jolla, CA). The start codon of spi1 was mutagenized to an NcoI site and an NcoI-AflIII fragment from pEGFP1 (ClonTech, Palo Alto, CA), containing EGFP and a downstream simian virus 40 (SV40) polyA site, cloned downstream to generate an spi1:EGFP fusion (pA301). Simple internal restriction-mediated deletions were used to create pA302-pA306.
Zebrafish and their manipulation
Zebrafish embryos were microinjected at the one-cell stage by means of a finely drawn capillary with approximately 1 nL linearized DNA (100 ng/μL in 0.25 M KCl) and allowed to develop as described.10 EGFP expression was visualized on anesthetized embryos by means of a Leica FL111 Zoom stereo fluorescence microscope (Heidelberg, Germany) or a Bio-Rad MRC1024 confocal microscope (Hercules, CA).
Results and discussion
Characterization of full-length zebrafish spi1 cDNA
We previously reported the sequence of a 1034-bp fragment of spi1 cDNA.12 To facilitate analysis of the spi1 gene locus, additional spi1 cDNA clones were isolated and analyzed as described,12 which enabled an extended sequence of 2064 base pair (bp) to be assembled (GenBank accession no. AY293624). The first 7 bases of our original sequence were found to be artifactual, to be replaced by 21 bp of additional 5′ sequence encoding 6 extra amino acids at the N-terminus (MLHPYR), identical to those present in the spi1 protein from the related cichlid fish (GenBank accession no. AF247366). There was also an additional 1016 bp of 3′ untranslated region (3′ UTR) sequence.
Characterization of the zebrafish spi1 genomic locus
Genomic bacterial artificial chromosome (BAC) clones embracing the zebrafish spi1 locus were identified by hybridization with the 1034-bp spi1 cDNA fragment. A detailed restriction enzyme map of the region around the spi1 locus was produced (Figure 1A), facilitating subcloning and sequence analysis (GenBank accession no. AY293626-9). Comparison of genomic and cDNA sequences showed that the spi1 gene is distributed over 6 exons encompassing approximately 15 kb (Figure 1B), similar to other spi1 orthologs, the principal difference being the presence of an extra exon (exon 2), similar to the related spi-b genes.20 However, RT-PCR analysis revealed an alternatively spliced transcript that skips exon 2 and splices to an alternative exon 3 splice site (data not shown).
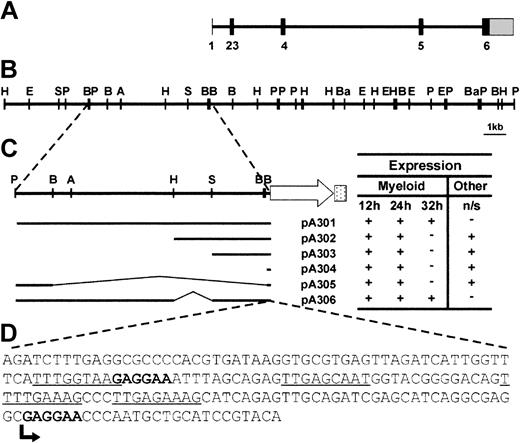
Analysis of the zebrafish spi1 locus and dissection of the promoter region. (A) Intron-exon structure of the spi1 gene. Solid bars represent translated exonic sequence, and the shaded bar shows the 3′ UTR. The 5′ UTR is too small to be visualized at this scale. (B) Linear depiction of the spi1 genomic locus. Letters indicate restriction enzyme sites (A = ApaI, B = BglII, Ba = BamHI, E = EcoRI, H = HindIII, P = PstI, S = SphI). (C) Activity of spi1 promoter fragments in transient expression assays. Embryos injected with the constructs indicated at the left were examined at 12, 24, and 32 hpf for patterns of EGFP expression that largely recapitulated endogenous spi1 expression pattern. This was scored as present (+) or absent (–). Nonspecific (n/s) expression at other sites (typically, the skeletal muscle and the eye) was similarly scored. All constructs were injected on at least 2 independent occasions, with 20% to 40% of injected embryos showing EGFP fluorescence in each case. Promoter-only injection controls gave no fluorescence. (D) Sequence analysis of the “core” spi1 promoter. Bases correspond to region of the spi1 promoter cloned upstream of EGFP in pA304, which produced early myeloid expression. Consensus Spi1 and c/ebpα sites are bolded and underlined, respectively, and the transcription start is indicated by an arrow.
Analysis of the zebrafish spi1 locus and dissection of the promoter region. (A) Intron-exon structure of the spi1 gene. Solid bars represent translated exonic sequence, and the shaded bar shows the 3′ UTR. The 5′ UTR is too small to be visualized at this scale. (B) Linear depiction of the spi1 genomic locus. Letters indicate restriction enzyme sites (A = ApaI, B = BglII, Ba = BamHI, E = EcoRI, H = HindIII, P = PstI, S = SphI). (C) Activity of spi1 promoter fragments in transient expression assays. Embryos injected with the constructs indicated at the left were examined at 12, 24, and 32 hpf for patterns of EGFP expression that largely recapitulated endogenous spi1 expression pattern. This was scored as present (+) or absent (–). Nonspecific (n/s) expression at other sites (typically, the skeletal muscle and the eye) was similarly scored. All constructs were injected on at least 2 independent occasions, with 20% to 40% of injected embryos showing EGFP fluorescence in each case. Promoter-only injection controls gave no fluorescence. (D) Sequence analysis of the “core” spi1 promoter. Bases correspond to region of the spi1 promoter cloned upstream of EGFP in pA304, which produced early myeloid expression. Consensus Spi1 and c/ebpα sites are bolded and underlined, respectively, and the transcription start is indicated by an arrow.
Characterization of the zebrafish spi1 promoter
Various fragments of genomic sequence upstream of the spi1 start codon were cloned in front of an EGFP reporter gene (Figure 1C). Linearized constructs were microinjected into one-cell zebrafish embryos and assayed for promoter activity by analyzing transient green fluorescence. All constructs tested reproduced the early spi1 expression profile,12 including rostral expression in the lateral plate mesoderm at 12 hpf, and over the yolk sac and in the posterior intermediate cell mass at 24 hpf (Figure 1C). However, only pA301 and pA306 produced fluorescent cells in circulation Figure 2A-B) and no nonspecific expression in other tissues (Figure 1C). This suggests a 181-bp core promoter capable of providing early myeloid-specific expression (Figure 1D), and other regions extending up to 5.3 kb upstream that are required for later expression in circulating cells as well as for suppressing nonspecific expression. A similar architecture of distant regulatory elements has also been identified in the murine Spi1 promoter.21 Analysis of the zebrafish core region revealed the presence of putative Spi1 binding sites, one beginning at bp –93 and another immediately after the transcription start site, with the latter arrangement conserved in both the murine22 and human23 Spi1 promoters. This suggests that Spi1 might autoregulate its expression in zebrafish as well.23 Sites for c/ebpα were also identified, as have been postulated for the murine promoter,24 although no sp1 or octamer binding sites were observed (Figure 1D). Analysis of the upstream promoter region failed to identify any conservation between the zebrafish and murine sequences (data not shown).
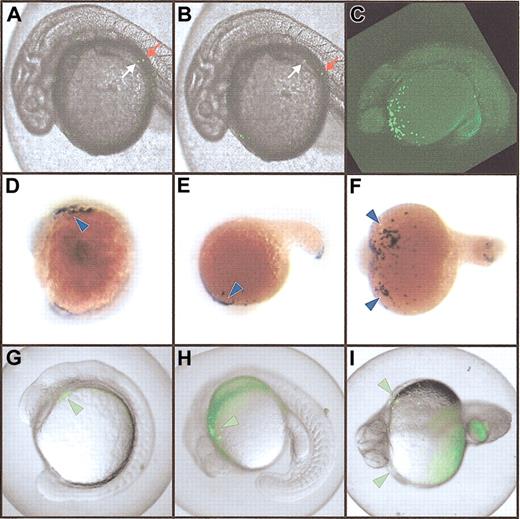
Expression of the spi1:EGFP transgene. (A-B) Transient expression in injected embryos. Two images taken 2 seconds apart, showing movement of fluorescent cells in circulation. The relative movement of 2 individual cells moving in contrary directions is indicated by white and red arrows. (C-I). Stable myeloid-specific expression. Panel C shows a vertical Z-series of images captured by confocal microscopy and projected to form a single image demonstrating EGFP-expressing cells over the yolk in green in an F2 embryo. Panels D-F show whole-mount in situ hybridization of embryos with an spi1 probe revealing expression in the rostral lateral plate mesoderm at around 14 hpf (D-F; blue arrowheads), and in cells over the yolk sac from around 18 hpf (E-F; arrowheads). Panels G-I show EGFP expression in pronouncedly fluorescent (flu++) F3 embryos (G-I; green arrowheads) recapitulating the in situ hybridization patterns shown. Original magnification, × 40.
Expression of the spi1:EGFP transgene. (A-B) Transient expression in injected embryos. Two images taken 2 seconds apart, showing movement of fluorescent cells in circulation. The relative movement of 2 individual cells moving in contrary directions is indicated by white and red arrows. (C-I). Stable myeloid-specific expression. Panel C shows a vertical Z-series of images captured by confocal microscopy and projected to form a single image demonstrating EGFP-expressing cells over the yolk in green in an F2 embryo. Panels D-F show whole-mount in situ hybridization of embryos with an spi1 probe revealing expression in the rostral lateral plate mesoderm at around 14 hpf (D-F; blue arrowheads), and in cells over the yolk sac from around 18 hpf (E-F; arrowheads). Panels G-I show EGFP expression in pronouncedly fluorescent (flu++) F3 embryos (G-I; green arrowheads) recapitulating the in situ hybridization patterns shown. Original magnification, × 40.
Generation of an spi1:EGFP transgenic zebrafish
Embryos were injected with the pA301 construct, and those scored as fluorescent (489 of 2077) were allowed to develop to adulthood. The 210 surviving adults were tested for transmission of the transgene into their progeny by bulk matings in 7 tanks containing 20 to 40 potential founder (F0) fish. Fluorescent (flu+) F1 embryos were recovered from one tank that displayed EGFP expression closely resembling that seen in the injected embryos. These were raised to adulthood. All (13 of 13) of the surviving F1 fish transmitted the transgene to their F2 progeny, suggesting 100% transmission of the transgene. When mated to wild-type fish, 48% (190 of 399) of the total progeny were scored as flu+. Crossing between pairs of F1 transgenics produced some F2 embryos with more pronounced fluorescence (flu++) (Figure 2C), with a proportion ratio of flu++/flu+/flu– of 0.22:0.51:0.27 (27, 64, and 34 fish, respectively. Both of these values are close to the expected Mendelian ratios, suggesting penetrance of nearly 100%. When any of these flu++ fish were crossed with wild-type fish, all F3 progeny were flu+ (Figure 2D-I). This suggests that the flu++ fish carry 2 copies of the transgene in the same chromosomal location, indicative of their derivation from the same F0 fish. This was confirmed by Southern blot analysis of genomic DNA (data not shown). This TG(spi1:EGFP)pA301 transgenic line will for the first time enable the direct visualization of myeloid cells in a living zebrafish embryo. This will augment our ongoing studies investigating myeloid development and its perturbation, forming the cornerstone of a mutagenesis screen for leukemia mutants in zebrafish, which will be identified by an expanded population of GFP+ cells. Such an approach has been further validated by the recent success in inducing T-cell leukemia in zebrafish by expression of a c-Myc–GFP fusion transgene.25
Prepublished online as Blood First Edition Paper, July 17, 2003; DOI 10.1182/blood-2003-03-0966.
Supported by a Sylvia and Charles Viertel Senior Medical Research Fellowship, an AMRAD Postdoctoral Award, a Clive and Vera Ramaciotti Equipment grant, and a National Health and Medical Research Council (NHMRC) project grant (no. 134510) (A.C.W); by a Deakin University International Research Scholarship, a Centre for Cellular and Molecular Biology (CCMB) Postgraduate Research Scholarship, and a Fredrika Bremer Stiffelsen Scholarship for Women in Research (S.M.N.O.); and by a Wellcome Trust Senior Research Fellowship in Medical Sciences in Australia (G.J.L.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors would like to thank Rowena Lewis for her assistance with sequence analyses.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal