Abstract
Severe factor V (FV) deficiency is a rare bleeding disorder, whose genetic bases have been characterized only in a limited number of cases. We investigated 6 unrelated patients with extremely reduced plasma FV levels, associated with a bleeding tendency ranging from moderately severe to severe. Clinical manifestations were substantially concordant with the previously established spectrum of hemorrhagic symptoms of the disease. Molecular analysis of FV gene identified 9 different mutations, 7 hitherto unknown, and 2 previously reported (Arg712ter and Tyr1702Cys). Four of 6 analyzed patients were compound heterozygotes, indicating the high allelic heterogeneity of this disease. Among novel mutations, 5 led to premature termination codons, because of nonsense (Arg1002ter, Arg1606ter, and Trp1854ter), or frameshift mutations (5127-5128insA and 6122-6123insAACAG). The remaining 2 were missense mutations (Cys472Gly and Val1813Met), located in FV A2 and A3 domains. Their effect on FV expression was studied by transient transfection experiments, demonstrating that the presence of each mutation impaired FV secretion. These data increase the number of severe FV deficiency–causing mutations by about 50%. The high number of “private” mutations identified in FV-deficient families indicates that full mutational screening of FV gene is still required for molecular diagnosis.
Introduction
Coagulation factor V (FV) takes part in the coagulation cascade as an essential nonenzymatic cofactor of the prothrombinase complex, which catalyses the conversion of prothrombin to thrombin.1,2 Human FV is a 330-kDa glycoprotein synthesized by hepatocytes and megakaryocytes; about 75% of FV circulates in the blood as a precursor molecule, whereas the remaining 25% is stored into the platelet α-granules.3 FV shows high structural and functional homology with factor VIII (FVIII).4 The 2 proteins share the same A1-A2-B-A3-C1-C2 architecture. The 3 A domains of FV and FVIII share about 30% amino acid identity with each other and with the triplicated A domains of ceruloplasmin, the major plasma copper-transport protein.5 The large B domain, which has no known homology with any other protein, is proteolytically removed during activation of both FV and FVIII. The 2 C domains are tandem modules of about 150 amino acids and belong to the major subfamily of discoidin domains.6 The 3-dimensional structures of the 3 A domains of FV and FVIII have been predicted on the basis of the crystal structure of ceruloplasmin.7-9 The X-ray structures of the C2 domain of FV and FVIII have been resolved,10,11 whereas the C1-domain structures were predicted by homology modeling.12
After secretion, FV circulates as single polypeptide expressing low procoagulant activity. Conversion of FV into its active form (FVa) is promoted by thrombin and/or activated factor X proteolytic cleavages at 3 arginine residues (Arg709, Arg1018, and Arg1545).13,14 FVa is composed of a 105-kDa heavy chain at the N-terminus and of a 74- or 71-kDa light chain at the C-terminus, linked by a single Ca++ ion. Cleavage by activated protein C (APC) at 3 arginine residues (Arg306, Arg506, and Arg679) located in the FV heavy chain results in the inactivation of the procoagulant factor.15
Severe FV deficiency is an autosomal recessive bleeding disorder, characterized by very low FV coagulant and antigen levels, associated with bleeding symptoms ranging from mild to severe.16,17 Its incidence is about 1 in 1 million. The study of the molecular basis of severe FV deficiency started in 1998 with the identification of the first causative mutation.18 After that, 15 additional point mutations were described in FV-deficient patients, all located in FV gene (F5).19-31 Most mutations give rise to null alleles, either because of nonsense or frameshift mutations. Of 6 missense mutations identified so far, only 1 has been characterized by in vitro expression studies, which demonstrated an impairment of FV secretion associated with intracellular degradation of the mutant protein.28
In this report, 6 patients with severe FV deficiency were studied. All patients were symptomatic, suffering from a bleeding diathesis of variable severity. F5 analysis revealed 7 novel mutations (3 nonsense, 2 frameshift, and 2 missense) as well as 2 previously described genetic defects. The causal role of the newly identified missense mutations (Cys472Gly and Val1813Met), occurring in different domains of FV, was investigated by transient transfection experiments in COS-1 cells with plasmids expressing the mutant FV molecules, followed by FV antigen measurements both in cell lysates and in conditioned media.
Patients, materials, and methods
Patients
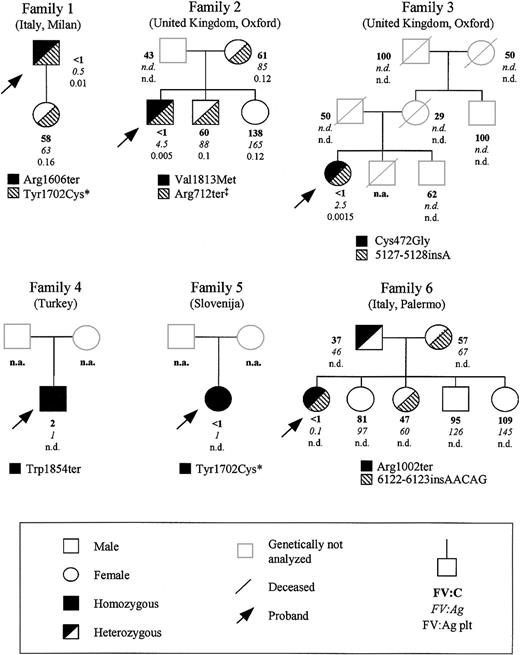
Six unrelated patients from different European countries affected by severe FV deficiency were studied; their family pedigrees are reported in Figure 1. Approval for these studies was obtained from the institutional review board of the University of Milan.
Family pedigrees of the 6 FV-deficient probands analyzed in this study, showing FV levels and segregation of the identified mutations. Plasma coagulant (percentage), immunoreactive (percentage), and intraplatelet (percentage per 109 platelets) FV levels are indicated in this order below each symbol. The normal ranges for the 3 assays were 58% to 140%, 64% to 139%, and 0.1% to 0.3%/109 platelets, respectively. * and ‡ mark previously reported mutations20,32,33 ; n.d., not done; and n.a., not available.
Family pedigrees of the 6 FV-deficient probands analyzed in this study, showing FV levels and segregation of the identified mutations. Plasma coagulant (percentage), immunoreactive (percentage), and intraplatelet (percentage per 109 platelets) FV levels are indicated in this order below each symbol. The normal ranges for the 3 assays were 58% to 140%, 64% to 139%, and 0.1% to 0.3%/109 platelets, respectively. * and ‡ mark previously reported mutations20,32,33 ; n.d., not done; and n.a., not available.
Patient 1, a 56-year-old man from Italy, first came to medical attention at the age of 3 years, because of prolonged gum bleeding. Subsequently, he presented with recurrent muscle hematomas and hemarthroses that occurred spontaneously or after minor trauma. He received prophylactic treatment with fresh-frozen plasma before dental extractions. No history of bleeding is reported in the family.
Patient 2 is a 50-year-old man from the United Kingdom. During childhood he suffered mainly from epistaxis and easy bruising. At the age of 11 years, he was treated with plasma infusion after dental extraction; subsequently, he was prophylactically treated with fresh-frozen plasma before each operative procedure (dental extractions and anal polyp excision). In the past few years, his bleeding pattern changed to spontaneous hemarthroses and posttraumatic hematomas (often requiring transfusion of fresh-frozen plasma). No clinical bleeding problems in his father and brother were apparent, whereas the proband's mother suffered from mild menorrhagia and his sister experienced both bleeding after a dental extraction and menorrhagia.
Patient 3 is a 46-year-old woman from the United Kingdom. At the age of 9 months she developed bleeding of the soft tissues of the mouth. In the following years she frequently suffered from severe epistaxis (resolved by fresh-frozen plasma infusion), hematomas, melena, and, after puberty, menorrhagia and hemarthroses. Severe bleeding episodes occurred after a dental extraction and as a consequence of a minor trauma, which caused spleen rupture and required splenectomy. At the age of 40 years, the proband successfully underwent ovariectomy under prophylactic treatment with fresh-frozen plasma. No bleeding tendency was reported for the other family members, except for bruising in the proband's mother.
Patient 4 is a 16-year-old boy from Turkey. During childhood and puberty, he had recurrent nasal hemorrhages, suffered from severe spontaneous bleedings from the oral cavity, and developed postinjection and posttraumatic hematomas, requiring fresh-frozen plasma transfusions.
Patient 5 is from Slovenia, and we have only blood and plasma samples available. No specific clinical data, except for a bleeding tendency suggestive of a coagulation disorder, could be obtained.
Patient 6 is a 24-year-old woman from Southern Italy. She first came to clinical attention at the age of 3 years for a large frontal hematoma caused by an accidental trauma. On this occasion, diagnosis of severe FV deficiency was made. Since then, the patient suffered from repeated traumatic hemorrhages resolved with the administration of fresh-frozen plasma. After puberty, severe menorrhagia required multiple treatments with plasma, and in 7 episodes red blood cell transfusion was necessary to control anemia. At the age of 20 years, she underwent a monolateral ovariectomy (right) because of the presence of a hemorrhagic anovulatory follicle. After an attempt to control menorrhagia with oral contraceptives, a life-threatening hemorrhage in the pouch of Douglas suggested the opportunity to remove the left ovary. Ever since, the hemorrhagic symptoms eased as did the requirement for plasma infusions. No bleeding tendency was reported for the proband's relatives.
Blood collection and genomic DNA extraction
All subjects signed an informed consent according to the Declaration of Helsinki before blood withdrawal. Peripheral venous blood was collected in 1:10 volume of 0.11 M trisodium citrate, pH 7.3. Plasma was obtained by centrifugation at 2000g for 10 minutes, and aliquots were stored at –80°C until use. Genomic DNA was extracted from whole blood using the Nucleon BACC1 Kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
Coagulation tests
FV coagulant activity was measured in plasma by a functional assay based on the degree of normalization of the prothrombin time of a commercial immunodepleted FV-deficient plasma sample (Hemoliance; IL Instrumentation Laboratory, Milan, Italy). FV antigen levels in plasma and in platelet lysates were measured by an enzyme-immuno assay (EIA), based on a sheep antihuman polyclonal antibody (Affinity Biologicals, Hamilton, ON, Canada).19 In both assays, FV levels were expressed as a percentage of control plasma pooled from 40 healthy individuals, set as 100%. The sensitivity of the functional and immunologic tests was 1% and 0.01%, respectively. Normal ranges were 58% to 140% (FV coagulant activity, FV:C), 64% to 139% (FV antigen in plasma, FV:Ag), and 0.1% to 0.3%/109 platelets (FV antigen in platelets, FV:Ag plt).
Polymerase chain reaction (PCR) amplifications and DNA sequencing
PCRs were performed on 200 ng genomic DNA in a standard volume of 25 μL. The reaction mixtures contained 1 × reaction buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 8.3, 50 mM KCl, 0.01% gelatin), 1.5 mM MgCl2, 0.4 μM of each primer (Life Technologies, Inchinnan, Paisley, United Kingdom), 200 μM deoxynucleoside triphosphates, and 1.25 U REDTaq DNA polymerase (Sigma, St Louis, MO). PCRs were carried out on a PTC-100 thermal cycler (MJ-Research, Watertown, MA), under standard conditions. DNA sequencing was performed on both strands directly on ammonium acetate–precipitated PCR products, using the BigDye Terminator Kit and an automated ABI-3100 DNA sequencer (Applied Biosystems, Foster City, CA). Primers used for both PCR amplifications and sequencing reactions were designed on the basis of the sequence of F5 (GenBank accession no. Z99572), and their sequences are available on request. Factura and Sequence Navigator software packages (Applied Biosystems) were used for mutation detection.
Site-directed mutagenesis
The pMT2/FV mammalian expression plasmid, containing the full-length FV complementary DNA (cDNA), was kindly provided by Dr R. J. Kaufman (Howard Hughes Medical Institute, University of Michigan Medical School, Ann Arbor). The 2 identified missense mutations were independently introduced in pMT2/FV by the QuikChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA), according to the manufacturer's instruction. Each mutant plasmid (pMT2/FV-Cys472Gly and pMT2/FV-Val1813Met) was checked by sequencing the whole FV cDNA insert as well as 100 to 200 base pairs (bp) of flanking DNA on both sides of the cloning site. Large-scale plasmid preparations were obtained using the EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany).
Cell cultures and transfections
African green monkey kidney COS-1 cells were cultured at 37°C in a 5% CO2 humidified atmosphere in Dulbecco modified Eagle medium (DMEM) supplemented with fetal calf serum (10%), glutamine (1%), and antibiotics (penicillin and streptomycin, 100 IU/mL and 100 μg/mL, respectively). Cells were transfected with either wild-type or each mutant plasmid using the LIPOFECTAMINE 2000 reagent (Invitrogen, Carlsbad, CA) in 6-well plates, as described.28
FV antigen measurements in conditioned media and cell lysates
FV antigen levels were measured 72 hours after transfection in conditioned media and cell lysates by EIA; standard curves were constructed with reference plasma diluted 1:100 to 1:6400 in Tris-buffered saline (50 mM Tris, 150 mM NaCl), pH 7.5. Conditioned media were collected in prechilled tubes containing a protease inhibitor (Complete; Roche, Basel, Switzerland) and stored at –80°C, after removal of cell debris by centrifugation. To obtain cell lysates, cells were washed 3 times with prechilled phosphate-buffered saline (PBS) and lysed for 1 hour on ice with lysis buffer containing 1 × PBS, 1% Triton X-100, and 1 × Complete. Cell lysates were centrifuged to remove cell debris.
Results
Laboratory measurements
All the 6 probands showed markedly reduced plasma levels of FV (Figure 1), which were unmeasurable by functional assay in all except proband 4, who had a value of 2%. Antigen levels ranged from 0.1% (proband 6) to 4.5% (proband 2). In 3 families (ie, family 1, 2, and 3), intraplatelet FV antigen levels could also be measured in some individuals. In the probands from these families, platelet levels of immunoreactive FV were also very low (about 1%-5% of the mean normal value). Among the probands' relatives, all individuals with approximately half normal FV levels were genetically heterozygotes (next section).
Mutational screening
In all 6 investigated probands, mutation screening was performed by sequencing PCR fragments amplified from genomic DNA spanning the whole F5 coding region, including exon-intron boundaries and about 300 bp of the promoter region. Besides 2 previously described mutations (ie, Arg712ter32 and Tyr1702Cys20,33 ), 7 novel causative genetic alterations were identified. Their segregation in the probands' pedigrees and their position within FV gene and protein are reported in Figures 1 and 2, respectively. Their numbering from here onward is according to Jenny et al,34 GenBank accession number M16967; amino acid positions are indicated omitting the signal peptide.
Mutations responsible for FV deficiency identified in the analyzed patients. (A) Position of the identified mutations on FV gene and protein. In the schematic representation of F5, exons are represented by boxes, introns are drawn (not to scale) as lines, and mutations are named on the basis of the DNA alteration involved (numbering according to Jenny et al34 ). In the lower part of the panel, FV protein is shown with its domain structure. Mutations are named on the basis of the predicted effects at the amino acid level (numbering refers to the mature protein); * and ‡ mark previously reported mutations.20,32,33 (B) Predicted effects of the 2 frameshift mutations. The 5127-5128insA mutation introduces a frameshift with an aberrant sequence of 7 amino acids (bolded) followed by a PTC at codon 1659 of the mature protein. The 6122-6123insAACAG mutation results in a frameshift with an aberrant sequence of 6 amino acids and a PTC at position 1989. For both mutations, the inserted nucleotides are underlined.
Mutations responsible for FV deficiency identified in the analyzed patients. (A) Position of the identified mutations on FV gene and protein. In the schematic representation of F5, exons are represented by boxes, introns are drawn (not to scale) as lines, and mutations are named on the basis of the DNA alteration involved (numbering according to Jenny et al34 ). In the lower part of the panel, FV protein is shown with its domain structure. Mutations are named on the basis of the predicted effects at the amino acid level (numbering refers to the mature protein); * and ‡ mark previously reported mutations.20,32,33 (B) Predicted effects of the 2 frameshift mutations. The 5127-5128insA mutation introduces a frameshift with an aberrant sequence of 7 amino acids (bolded) followed by a PTC at codon 1659 of the mature protein. The 6122-6123insAACAG mutation results in a frameshift with an aberrant sequence of 6 amino acids and a PTC at position 1989. For both mutations, the inserted nucleotides are underlined.
Patient 1. Sequencing of F5 identified a novel nonsense mutation and a previously reported missense mutation (Tyr1702Cys),20,33 both present in the heterozygous state. The novel nonsense mutation (Arg1606ter) results from a C>T transition in exon 14, corresponding to cDNA position 4990. The only available family member (ie, the proband's daughter) was heterozygous for the Tyr1702Cys mutation and homozygous wild type for the Arg1606ter mutation, suggesting that the 2 mutations are located in trans in patient 1.
Patient 2. Patient 2 is a compound heterozygote. One mutation was a novel G>A transition occurring in exon 17 at cDNA position 5611 and causing the substitution of Val1813 with a methionine (Val1813Met). The other mutation was the previously described nonsense Arg712ter mutation.32 The proband's mother and brother were heterozygous for the Arg712ter mutation, whereas the propositus' sister was normal for both mutations. The proband's father, who had reduced FV functional levels, was not available for genetic analysis.
Patient 3. Genetic analysis showed that the patient is heterozygous for 2 novel mutations. The first one is a 1-bp insertion (5127-5128insA) occurring in exon 15. This mutation results in a frameshift from amino acid 1652 followed by 7 aberrant residues and a premature termination codon (PTC) at position 1659 (Figure 2B). The second mutation is a T>G transversion in exon 10 (cDNA position 1588), causing the substitution of Cys472 with a glycine (Cys472Gly). Cys472, located within FV A2 domain and involved in a disulfide bridge with Cys498,35 is a buried residue that is part of a β-strand belonging to the first β-barrel motif of the A2 domain (Figure 3).
Homology models of FV A2 and A3 domains showing the position of the 2 identified missense mutations. Ribbon diagrams of human FV A2 (left panel) and A3 (right panel) domain homology models were produced using SwissPDB 3.7 software and the coordinates under Protein Data Bank entry 1FV4.9 In each panel, the N- and C-terminal β-barrels and the amino(N) and carboxy-termini (C) of the domain are indicated. Each A domain is oriented to best display the sites of mutated residues (Cys472 and Val1813). The position of Cys498, involved in a disulfide bridge with Cys472 in the wild-type protein, is also indicated.
Homology models of FV A2 and A3 domains showing the position of the 2 identified missense mutations. Ribbon diagrams of human FV A2 (left panel) and A3 (right panel) domain homology models were produced using SwissPDB 3.7 software and the coordinates under Protein Data Bank entry 1FV4.9 In each panel, the N- and C-terminal β-barrels and the amino(N) and carboxy-termini (C) of the domain are indicated. Each A domain is oriented to best display the sites of mutated residues (Cys472 and Val1813). The position of Cys498, involved in a disulfide bridge with Cys472 in the wild-type protein, is also indicated.
Patient 4. The F5 of this patient showed a novel homozygous nonsense mutation in exon 18 at cDNA position 5736, where a G>A transition changed the Trp1854 residue into a stop codon. This patient is the only available family member.
Patient 5. Mutation screening of F5 revealed the already described missense mutation (Tyr1702Cys) in the homozygous state.20,33 No other family members were available.
Patient 6. Direct sequencing of F5 revealed patient 6 to be a compound heterozygote for 2 novel mutations: a nonsense mutation located in exon 13 and a 5-bp insertion occurring in exon 21. The nonsense mutation consists of a C>T transition at cDNA position 3178 that introduces a PTC at residue 1002 (Arg1002ter) within the B domain of the protein. The 5-bp insertion (6122-6123insAACAG), which represents a perfect duplication of nucleotides 6118-6122, causes a frameshift from amino acid 1983 followed by a PTC at position 1989 preceded by 6 aberrant residues (Figure 2B).
The proband's father was heterozygous for the Arg1002ter mutation, whereas the mother and the only other analyzed relative with reduced FV levels were both heterozygous for the 6122-6123insAACAG mutation. The remaining siblings, with normal FV levels, were all genotypically homozygous wild type.
Polymorphisms identified in F5
Sequencing of F5 in all analyzed probands revealed several additional nucleotide variations. All of them were previously reported polymorphisms (Dr H. L. Vos, personal electronic communication, April 17, 2003), except for a C>T transition at nucleotide 3834 in exon 13, which was found in the heterozygous state in patient 1 and in the homozygous state in patient 4. This nucleotide variation results in a synonymous codon (ACC/ACT) and is likely to represent a new F5 polymorphism.
Expression of wild-type and mutant recombinant FV in COS-1 cells
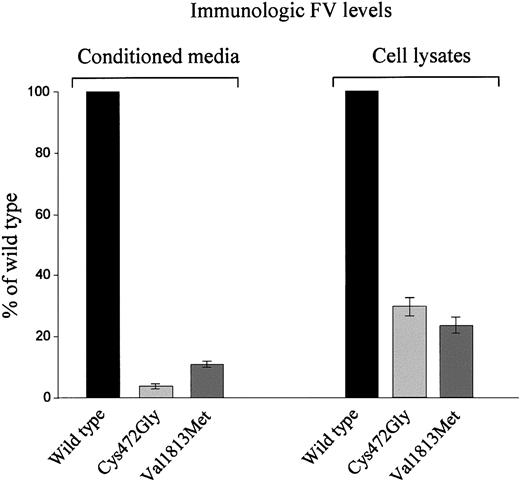
To study the effects of the newly identified missense mutations on FV secretion, each mutant FV molecule was expressed, by transient transfection experiments, in COS-1 cells, which do not express endogenous FV. To this purpose, the pMT2/FV expression plasmid, containing the full-length FV cDNA underwent 2 independent site-directed mutagenesis experiments, to produce pMT2/FV-Cys472Gly and pMT2/FV-Val1813Met expression plasmids. Each mutant protein was analyzed in 3 independent transfection experiments, each performed in duplicate. Cells were independently transfected either with the wild-type or each mutant plasmid. Transfection results are shown in Figure 4. In media conditioned by cells expressing the wild-type FV, antigen levels ranged from 100 to 200 ng/mL, whereas in the corresponding lysates levels of immunoreactive FV were between 60 and 100 ng/mL. Both mutations caused a marked reduction (96% and 89% for Cys472Gly and Val1813Met, respectively) of FV antigen level in the corresponding conditioned media. The same situation was found intracellularly, even though in this case antigen levels of mutant proteins were reduced to a lesser extent (70%-80%).
Transient expression of FV missense mutants in COS-1 cells. Plasmids containing wild-type (pMT2/FV) or mutant (pMT2/FV-Cys472Gly and pMT2/FV-Val1813Met) FV cDNA were transiently transfected in COS-1 cells by using LIPOFECTAMINE 2000 reagent. Antigen levels of recombinant FV were measured both in conditioned media and in the corresponding cell lysates by an EIA assay 72 hours after transfection. Bars represent means ± SD of 3 independent experiments, each performed in duplicate. The mean values of wild-type FV are set as 100%; SDs of wild-type FV measurements never exceeded 10%.
Transient expression of FV missense mutants in COS-1 cells. Plasmids containing wild-type (pMT2/FV) or mutant (pMT2/FV-Cys472Gly and pMT2/FV-Val1813Met) FV cDNA were transiently transfected in COS-1 cells by using LIPOFECTAMINE 2000 reagent. Antigen levels of recombinant FV were measured both in conditioned media and in the corresponding cell lysates by an EIA assay 72 hours after transfection. Bars represent means ± SD of 3 independent experiments, each performed in duplicate. The mean values of wild-type FV are set as 100%; SDs of wild-type FV measurements never exceeded 10%.
Discussion
Severe FV deficiency is a rare inherited bleeding condition whose clinical manifestations are usually relatively mild, even though severe intracranial and gastrointestinal hemorrhages and potentially handicapping hematomas and hemarthroses are observed in a number of patients.33 We studied 6 unrelated European families with severe FV deficiency. All patients had unmeasurable functional plasma FV levels (< 1%), the only exception being represented by the proband of family 4, who had values slightly exceeding the lower limit of the assay (Figure 1). Clinical symptoms of the analyzed probands were in general good accordance with the pattern of bleeding symptoms observed in the largest patient cohort so far analyzed (45 Iranian patients).36
Sequencing of F5 in all analyzed patients identified a total of 9 point mutations. Among them, 7 were described for the first time, and included 5 mutations causing premature truncation of FV (nonsense or frameshift) and 2 missense mutations. Four of 6 probands were compound heterozygotes (Figure 1), confirming the high level of allelic heterogeneity underlying the disease.28
The truncating mutations may cause severe FV deficiency either by affecting protein stability, competence for secretion, or both, or by promoting the degradation of the corresponding mRNA because of the nonsense-mediated mRNA decay pathway.37 Among previously described F5 mutations, introducing PTCs, in at least 4 cases the mutant mRNA level was highly reduced compared with the wild-type one.19,21,32,38,39 Quantitation of mutant and wild-type transcripts in a heterozygous patient to look for a reduced rate of transcription or an increased mRNA degradation of the mutant mRNA was unfeasible, because no biologic samples (platelets or mononuclear cells) suitable for RNA analysis could be obtained from the probands or their heterozygous relatives (families 1, 2, 3, 4, and 6). However, should the PTC-carrying mRNAs escape nonsense-mediated decay, the resulting truncated FV proteins are likely to be retained and degraded intracellularly by the quality control system of secretory proteins.
The 2 novel missense mutations were located in FV A2 (Cys472Gly) and A3 (Val1813Met) domains. The involvement of the identified amino acid substitutions in type I FV deficiency was first postulated on the basis of computer-assisted structural predictions (Figure 3). In particular, Cys472 is involved in a disulfide bridge with Cys498, forming a small α-loop of 26 amino acids. This structure is conserved not only among the 3 A domains of FV but also in the corresponding regions of FVIII and ceruloplasmin.35 In addition, the amino acid sequences of the α-loops of the 3 proteins are extremely conserved (identity up to 78% between FV and FVIII α-loop of the A1 domain).35 The replacement of Cys472 with a glycine disrupts this highly conserved disulfide-bonded organized structure and leaves an additional free thiol group at position 498 within the A2 domain. As a result, the loss of the A2 domain α-loop may affect the overall tertiary structure of the domain, causing the retention of the mutant protein. Moreover, the presence of an exposed thiol group by itself may act as an “intracellular retention element,” through the interaction with endoplasmic reticulum resident proteins.40
As far as Val1813Met mutation is concerned, this relatively conservative amino acid substitution involves a residue that is invariant among mammals' FVs (Homo sapiens, Sus scrofa, Bos taurus, and Mus musculus) but is not conserved in FVIII or ceruloplasmin. Within FV A3 domain, Val1813 participates in the formation of a 6-residue β-strand belonging to the C-terminal β-barrel (Figure 3). Inspection of the 3-dimensional model of activated FV bound to APC (PDB accession no. 1FV4)9 revealed that this β-strand is located at the interface between the light and the heavy chains. This β-strand includes His1815 and His1817 that, together with His85 (domain A1), coordinate a copper ion that has been proposed to contribute to the stabilization of both the A3 domain and the A1/A3 interface.9 Because the currently accepted structural model of FV entails the light and heavy chains being associated and oriented in a very similar way in the procofactor and in FVa,41 the substitution of Val1813 with a methionine is likely to have a strong effect on protein tertiary structure at the A1/A3 domain interface.
To confirm the causal role of the 2 identified missense mutations, the corresponding mutant FV proteins were in vitro expressed. For both mutations, a defective expression in transiently transfected COS-1 cells that parallels the extremely low FV plasma levels observed in affected patients was found (Figure 4). Antigen levels of the 2 mutant FV proteins were reduced up to 89% and 70% in conditioned media and cell lysates, respectively. These data, besides confirming the causative role of the identified missense mutations, suggest that the secretion defect might be associated with a rapid intracellular degradation, as reported for the only missense mutation leading to severe FV deficiency so far characterized by in vitro expression.28 However, the possible existence of differences in translational efficiency, mRNA stability, posttranslational modifications, and stability of secreted FV cannot be completely ruled out.
Among the 9 genetic defects identified, 2 mutations (Tyr1702Cys and Arg712ter), found in families 1, 2, and 5, had already been identified. Tyr1702Cys represents a recurrent mutation among FV-deficient patients, at least in the Italian population.20 This mutation, which determines the exposure of a novel cysteine residue, has been proposed to cause protein instability by disrupting the A3 domain scaffold and by interfering with the correct disulfide bond formation.20 The Arg712ter mutation was previously reported to account for a case of pseudo-homozygous APC resistance in a thrombophilic heterozygous patient.32 This mutation is here described for the first time associated with severe FV deficiency.
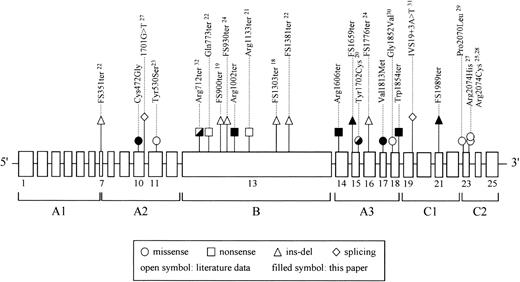
Altogether, the F5 mutations reported in this paper increase by approximately 50% the number of mutations underlying severe or moderately severe FV deficiency (Figure 5). Taking into account all mutations, most of them (16 of 24) are truncating mutations; they are scattered over the whole gene, except for exons (1-6) coding for the N-terminal A1 domain, where no mutations have been identified so far. The remaining 9 missense mutations reported so far are almost equally distributed among FV A2, A3, and C2 domains. Among them, 3 mutations were characterized by expression studies (this paper and Duga et al28 ), and all resulted in a secretion defect. Similarly to the FVIII gene (F8), whose mutational spectrum has been extensively studied, no missense mutations were found within the large FV B domain (encoded by exon 13 in F5 and exon 14 in F8). This probably reflects its “tolerance” to variations, as demonstrated by the fact that it is highly polymorphic, poorly conserved among species, and without sequence similarity to any other protein.
Mutational spectrum of severe and moderately severe FV deficiency. All known causative mutations for type I FV deficiency are projected on the exon-intron structure of F5. Exons (approximately to scale) are represented by boxes, introns (not to scale) by lines. The FV domain structure is also indicated. All genetic defects, except for splicing mutations, are indicated on the basis of their predicted effect at the protein level.
Mutational spectrum of severe and moderately severe FV deficiency. All known causative mutations for type I FV deficiency are projected on the exon-intron structure of F5. Exons (approximately to scale) are represented by boxes, introns (not to scale) by lines. The FV domain structure is also indicated. All genetic defects, except for splicing mutations, are indicated on the basis of their predicted effect at the protein level.
Looking at the whole spectrum of FV deficiency–causing mutations, no truncating mutations have been so far identified in FV-deficient patients at the homozygous state in A1 and A2 domains, except for a recently reported splicing mutation located in the last nucleotide of exon 10.25 Nevertheless, even though those researchers convincingly demonstrated the effect of the mutation on pre-mRNA processing, resulting in a PTC at codon 498, it cannot be excluded that trace amounts of wild-type transcript could be produced, as suggested by the residual FV immunologic levels measured in the proband's plasma (8%). FV knock-out mice have been shown to die either during embryonic development or at birth of fatal hemorrhages.42 Conversely, partial rescue of the lethal FV–/– phenotype was obtained by very low levels of FV expression in transgenic mice, carrying a FV minigene, suggesting that trace expression of FV could significantly improve hemostasis.43 The absence of “severe” mutations in the FV heavy chain, together with the low residual FV activity measured in most human FV-deficient patients, leaves the question whether in humans complete FV deficiency is compatible with life. Solving this question requires the genetic characterization of a larger number of FV-deficient patients. Knowledge of the mutations underlying this rare coagulation disorder is a prerequisite to perform prenatal diagnosis in at-risk families as well as a precious tool to highlight important residues and/or protein regions involved in FV folding and secretion.
Prepublished online as Blood First Edition Paper, June 19, 2003; DOI 10.1182/blood-2003-03-0922.
Supported by MURST (Ministero dell'Università e della Ricerca Scientifica e Tecnologica) grant no. 2001057917 and IRCCS Maggiore Hospital, Milan, Italy; and partially founded by a grant from Fondazione Italo Monzino (F.P. and P.M.M.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank all family members for their participation in this study. We thank Dr Randal J. Kaufman for kindly providing the pMT2/FV expression plasmid. We wish to acknowledge Dr P. L. F. Giangrande (Oxford Haemophilia Centre, The Churchill, Oxford, United Kingdom), Prof G. Mancuso (Department of Paediatrics, Haemophilia Centre, Ospedale dei bambini G. Di Cristina, and University of Palermo, Italy), Dr M. Benedik-Dolnicar (Division of Paediatrics, Unit of Oncology and Haematology, University Medical Centre, Ljubljana, Slovenia), and Dr K. Kavakli (Ege University Hospital, Departments of Paediatrics and Paediatric Hematology, Ege Hemophilia Center, Bornova, Izmir, Turkey) for the clinical identification of the probands, family history, and blood collection, as well as Dr Rossella Bader and Maria Teresa Bajetta (Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Maggiore Hospital, Milan, Italy) for FV antigen quantitation. Finally, we wish to thank Dr H. L. Ves (Department of Hematology, Leiden University Medical Center, the Netherlands) for critical commentary on this paper.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal