Abstract
To investigate the roles of c-myc during hematopoietic proliferation induced by growth factors, we used factor-dependent human leukemic cell lines (MO7e and F36P) in which proliferation, cell cycle progression, and c-Myc expression were strictly regulated by granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-3 (IL-3). In these cell lines, both c-myc mRNA and c-Myc protein stability were not affected by GM-CSF and IL-3, suggesting a regulation of c-Myc protein at the translational level. However, rapamycin, an inhibitor of cap-dependent translation, did not block c-myc induction by GM-CSF and IL-3. Thus, we studied the cap-independent translation, the internal ribosome entry site (IRES), during c-Myc protein synthesis using dicistronic reporter gene plasmids and found that GM-CSF and IL-3 activated c-myc IRES to initiate translation. c-myc IRES activation, c-Myc protein expression, and cell cycle progression were all blocked by a phosphatidylinositol 3-kinase (PI3K) inhibitor, LY294002. In another factor-dependent cell line, UT7, we observed the cell cycle progression and up-regulation of c-Myc protein, c-myc mRNA, and c-myc IRES simultaneously, which were all inhibited by LY294002. Results indicate that hematopoietic growth factors induce cell cycle progression via IRES-mediated translation of c-myc though the PI3K pathway in human factor–dependent leukemic cells.
Introduction
The protooncogene c-myc plays pivotal roles in survival, proliferation, and differentiation of normal and leukemic hematopoietic cells.1-3 In these cells, the c-myc expression is regulated by hematopoietic growth factors (HGFs) including granulocyte-macrophage colony-stimulating factor (GM-CSF) and interleukin-3 (IL-3),4 which control proliferation and differentiation of progenitor cells of normal and leukemic origin.5 It has been reported that transduction of the aberrant c-myc gene into factor-dependent hematopoietic cell lines disturbed survival and G1 progression by HGFs.6-8 These data suggest that hematopoietic cell proliferation, particularly G1 cell-cycle progression, is closely associated with the appropriate regulation of c-myc expression by HGFs.
Although it is classically assumed that expression of c-Myc protein is determined by transcriptional control, it has recently been reported that c-myc gene is strongly subjected to posttranscriptional regulation such as control of translation.9 Most of the translation control occurs at the initiation phase, and this is the rate-limiting step of this process.10 There are 2 mechanisms used by eukaryotic cells to initiate translation, the cap-dependent scanning mechanism11 and internal ribosome entry,12,13 and it is reported that c-Myc protein synthesis can be initiated by both mechanisms.14
The cap-dependent scanning mechanism involves the recognition of the m7GpppN cap structure of the mRNA 5′-end by the eIF4E subunit of eIF4F translation initiation complex, and this is followed by binding of a 40S ribosomal subunit and scanning downstream to the initiation codon.15,16 Interestingly, c-myc mRNA has a long, highly structured 5′ untranslated region (UTR), which functions to suppress cap-dependent scanning,17,18 like other growth regulators.19 In spite of this, c-Myc protein can be translated in a cap-dependent manner by insulin or serum stimulation.11,20
Internal ribosome entry site (IRES) is known as one of the mechanisms that induces effective translation to mRNA with the 5′ encumbered UTR.21 IRES is the name given to a sequence that allows ribosomes to enter directly at an AUG codon rather than scanning from the capped 5′-end of the mRNA, and c-myc mRNA contains IRES within its 5′UTR.12,13 To date, c-myc IRES activity was observed only in situations where the cap-dependent translation machinery is inactive, for instance, in response to stress,22 apoptosis,23,24 and in cell-cycle G2-to-M transition.25 However, many important features of how IRES in c-myc mRNA mediates translation initiation are poorly understood. In addition, it is controversial whether growth factors can utilize an IRES mechanism for translation of their target genes that regulate cell proliferation.
In this study, we focused on the posttranscriptional control, particularly translational mechanism, of c-myc expression induced by the 2 HGFs, namely GM-CSF and IL-3. We found that both HGFs induced an increase in c-Myc protein via its enhanced translation rather than enhanced transcription or decreased degradation in human factor–dependent leukemic cell lines and that these 2 HGFs do this via the phosphatidylinositol 3-kinase (PI3K)–dependent pathway. This is the first observation that growth factor can regulate IRES-mediated translation to exert its proliferative activity.
Materials and methods
Cell culture and reagents
Human factor–dependent leukemic cell lines, MO7e,26 F36P,27 and UT7,28 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum and 10 ng/mL (MO7e) or 1 ng/mL (F36P, UT7) recombinant human GM-CSF (Immunex, Seattle, WA). F36P erythroleukemia cell line was obtained from RIKEN cell bank (Tsukuba, Japan). Recombinant human IL-3 was kindly provided by Kirin Brewery, Tokyo, Japan.
Centrifugal elutriation
Cells (1 × 108) were suspended in 10 mL phosphate-buffered saline (PBS) containing 0.5 mM EDTA (ethylenediaminetetraacetic acid) and 0.2% bovine serum albumin (Sigma, St Louis, MO) and were separated at 4°C by counterflow centrifugal elutriation (HITACHI CR20B2 centrifuge and SRR6Y rotor, Hitachi Koki, Tokyo, Japan) into each phase of the cell cycle.29 To evaluate the cell-cycle profile of each fraction, 5 × 105 cells were fixed by 80% ethanol, stained with propidium iodide, and were then analyzed by flow cytometry using a FACSCalibur (Nippon Becton Dickinson, Tokyo, Japan) as described.30
Antibodies and chemical inhibitors
Rabbit polyclonal antibodies recognizing Phospho-p44/42 (extracellular signal-regulated kinase 1/2 [Erk1/2]) (Thr202/Tyr204), Phospho-Akt (Ser473), Akt, Phospho-p70S6K (Thr421/Ser424), p70S6K, and 4E-BP1 were purchased from Cell Signaling Technology (Beverly, MA); rabbit polyclonal antibodies recognizing p44/42 (Erk1/2) and β-tubulin from Santa Cruz Biotechnology (Santa Cruz, CA); mouse monoclonal antibodies recognizing c-Myc protein for immunoblotting and immunoprecipitation from Becton Dickinson (San Jose, CA) and Santa Cruz Biotechnology, respectively; mouse monoclonal antibody recognizing retinoblastoma protein (Rb) from Becton Dickinson; and antimouse and antirabbit horseradish peroxidase (HRP)–conjugated secondary antibodies from Cell Signaling Technology. The PI3K inhibitor LY294002 and the MEK inhibitor PD98059 were purchased from Calbiochem (La Jolla, CA) and Biomol (Plymouth Meeting, PA), respectively.
Western blot analysis
The 1 × 106 cells were suspended in 100 μLof 1 × Laemmli sample buffer (50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl [pH 7.0], 4% SDS, 10% 2-mercaptoethanol, 1 mM EDTA, 10% glycerol, and 0.01% bromophenol blue) and boiled for 5 minutes. Total cell lysates (3 × 105 cells per lane) were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) on 7.5%, 10%, or 12.5% polyacrylamide gels, transferred to Immobilon polyvinylidene fluoride (PVDF) membrane (Millipore, Bedford, MA), and detected with HRP-conjugated secondary antibodies and immunofluorescence using the ECL system (Amersham, Piscataway, NJ).
RNase protection assay
Total RNA was isolated using the Total RNA Isolation Kit (Omega Bio-Tek, Doraville, GA) according to the manufacturer's recommendations. The presence of transcripts of the c-myc and internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was analyzed using Antisense Probe Templates (Ambion, Austin, TX). Biotin-labeled probes were produced by MAXIscript (Ambion) using Biotin-14-CTP (Invitrogen, Carlsbad, CA). Hybridization and RNase treatment were performed with the RPA III kit (Ambion) according to the manufacturer's recommendations. Protected transcripts were resolved by electrophoresis on denaturing 5% polyacrylamide gels and transferred to the nylon membrane by electroblotting and visualized by BrightStar BioDetect kit (Ambion).
Measurement of protein stability
Cells were prestarved by replacing the ordinary culture medium with PRMI 1640 medium without l-methionine for 15 minutes at 37°C. Then 1 × 106 cells were labeled in vivo with 400 μCi (14.8 MBq) of [35S]methionine for 30 minutes at 37°C. After labeling, cells were immediately washed twice with PBS and then incubated in RPMI 1640 medium supplemented with 10% fetal bovine serum for the indicated periods at 37°C. A total of 1 × 106 cells were lysed with 1 mL TNE buffer (50 mM Tris-HCl [pH 7.8], 1 mM EDTA, 1.0% Nonidet P-40, 150 mM NaCl, 0.5 mM dithiothreitol [DTT], 20 mM β-glycerophosphate, 1 mM Na2VO3, and 10 mM NaF) and incubated for 20 minutes at 4°C. Cell debris was removed by centrifugation at 10 000g for 5 minutes at 4°C. Extracts were precleared with protein G–Sepharose beads for 1 hour at 4°C. After incubation with anti–c-myc antibody (Santa Cruz Biotechnology) for 1 hour at 4°C, immunoglobulin complexes in the cell lysates were collected on protein G–Sepharose beads for 1 hour at 4°C. The beads were washed 4 times with TNE buffer. Protein complexes were recovered by boiling in 1 × Laemmli sample buffer and were analyzed by SDS-PAGE. Radioactivity in the gels was detected using a Phosphor Imager, BAS-1000 (Fuji, Tokyo, Japan).
Reporter gene assay
The dicistronic plasmids, pRMF (formerly pGL3utrR), phpRMF (formerly pGL3utrH), and pRF (formerly pGL3R), have been described previously13 (a kind gift from Dr Anne E. Willis, University of Leicester, United Kingdom). A total of 1 × 106 cells were transiently transfected with 5 μg dicistronic constructs using the Nucleofector technology31 (Amaxa, Cologne, Germany) in accordance with the manufacturer's recommendations. After incubation for 6 hours at 37°C, the cells were suspended with Passive Lysis Buffer (Promega). The activity of both firefly and Renilla luciferase was measured using a Dual-Luciferase Reporter Assay System (Promega). Light emission was quantified using a luminometer (Lumat LB 9507, Berthold Technologies, Wildbad, Germany). The activity of β-galactosidase in lysate prepared from cells transfected with pSV-β-gal (Promega) was measured using a β-galactosidase Enzyme Assay System (Promega) and was used for normalization of luciferase activity.
BrdU incorporation assay
Cells were pretreated with the inhibitors for 30 minutes at 37°C and were then incubated with GM-CSF or IL-3 in the presence of 10 μM bromodeoxyuridine (BrdU) for 24 hours at 37°C. After incubation, cells were fixed with 70% ethanol and the genomic DNA of the cells was denatured with 2N HCl for 30 minutes at room temperature. After the neutralization of acid by 0.1 M sodium tetraborate (pH 8.5), cells were resuspended in PBS containing 0.5% Tween 20 and incubated with fluorescein isothiocyanate (FITC)–conjugated anti-BrdU antibody (Becton Dickinson) for 30 minutes at room temperature. Fluorescence was analyzed by FACSCalibur (Becton Dickinson).
Results
The expression levels of c-Myc protein, but not its mRNA, were regulated by GM-CSF and IL-3 in factor-dependent cell lines MO7e and F36P
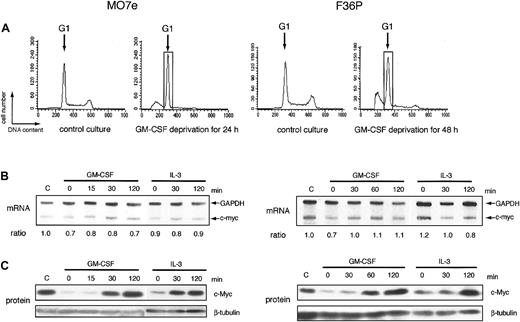
In many types of growth factor–dependent hematopoietic cell lines, the expression level of c-myc correlated tightly with growth factor–dependent progression of G1 phase of cell cycle.6-8 Human myeloblastic cell lines, MO7e and F36P, also can proliferate in response to GM-CSF and IL-3 and were arrested at the G1 phase by the factor deprivation (Figure 1A). Addition of GM-CSF or IL-3 to these G1-arrested cells released the cells from cell-cycle arrest and induced regrowth (data not shown). Under the same conditions, we determined the level of c-myc mRNA and c-Myc protein in these 2 cell lines. In this study, we purified G1 cells from the total factor-deprived cells using centrifugal elutriation, a method for purifying cells at each stage of the cycle,29 to eliminate the contamination of the apoptotic cells and the cells with the other phases of cell cycle. Purity of G1 cells increased from 68% to 92% in MO7e and from 56% to 91% in F36P after elutriation (data not shown).
mRNA and protein of c-myc during the induction of proliferation by GM-CSF and IL-3 in MO7e and F36P cells. (A) Cell cycle analysis of MO7e (left panel) and F36P (right panel) cells with or without GM-CSF. Propidium iodide staining and fluorescence analysis of cells were performed at the indicated culture condition (control culture and GM-CSF–deprived culture) to determine cell cycle distribution in each condition. The G1 cells indicated by the open box were separated by centrifugal elutriation for the following experiments. (B) After GM-CSF deprivation, MO7e (left panel) and F36P (right panel) cells were incubated with 10 ng/mL and 1 ng/mL GM-CSF or IL-3 for indicated periods at 37°C. Total cellular RNA was prepared and analyzed by RNase protection assay to determine intracellular level of c-myc mRNA. mRNA of GAPDH was used as control. The ratio was determined by densitometric analysis of the band and was calculated as compared with c-myc mRNA level of control cells (indicated as “C”). Lane-to-lane variation was corrected based upon mRNA level of GAPDH. (C) After GM-CSF deprivation, MO7e (left panel) and F36P (right panel) cells were incubated with 10 ng/mL and 1 ng/mL GM-CSF or IL-3 for the indicated periods at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted using specific antibodies for c-myc and β-tubulin. These show the representative results of a minimum of 3 independent experiments.
mRNA and protein of c-myc during the induction of proliferation by GM-CSF and IL-3 in MO7e and F36P cells. (A) Cell cycle analysis of MO7e (left panel) and F36P (right panel) cells with or without GM-CSF. Propidium iodide staining and fluorescence analysis of cells were performed at the indicated culture condition (control culture and GM-CSF–deprived culture) to determine cell cycle distribution in each condition. The G1 cells indicated by the open box were separated by centrifugal elutriation for the following experiments. (B) After GM-CSF deprivation, MO7e (left panel) and F36P (right panel) cells were incubated with 10 ng/mL and 1 ng/mL GM-CSF or IL-3 for indicated periods at 37°C. Total cellular RNA was prepared and analyzed by RNase protection assay to determine intracellular level of c-myc mRNA. mRNA of GAPDH was used as control. The ratio was determined by densitometric analysis of the band and was calculated as compared with c-myc mRNA level of control cells (indicated as “C”). Lane-to-lane variation was corrected based upon mRNA level of GAPDH. (C) After GM-CSF deprivation, MO7e (left panel) and F36P (right panel) cells were incubated with 10 ng/mL and 1 ng/mL GM-CSF or IL-3 for the indicated periods at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted using specific antibodies for c-myc and β-tubulin. These show the representative results of a minimum of 3 independent experiments.
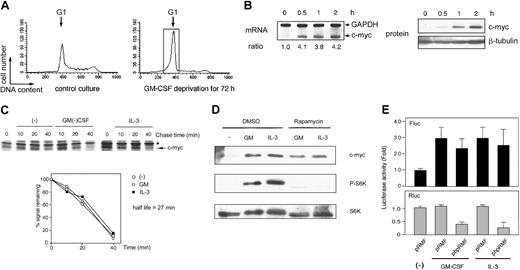
After GM-CSF deprivation for 24 or 48 hours, MO7e and F36P cells showed G1 cell-cycle arrest with a significant sub-G1 population (Figure 1A). Cells with G1 phase were further purified and isolated from G1-arrested MO7e and F36P cells using centrifugal elutriation and were then stimulated with GM-CSF or IL-3. As shown in Figure 1C, the c-Myc protein expression was reduced to an extremely low level in G1-arrested cells by GM-CSF withdrawal and began to increase as early as 30 minutes after the addition of GM-CSF or IL-3 and returned to the level of normal cultured cells at 2 hours. These findings indicate that GM-CSF and IL-3 regulated the level of c-Myc protein in these cell lines. Under the same condition, the levels of c-myc mRNA were almost constant (Figure 1B).
These data demonstrated that GM-CSF and IL-3 controlled the expression of c-Myc protein by the posttranscriptional mechanism in MO7e and F36P cells. In the following part of this paper, we performed a mechanistic study mainly using the 2 cell lines.
c-Myc protein stability was not modulated by GM-CSF and IL-3
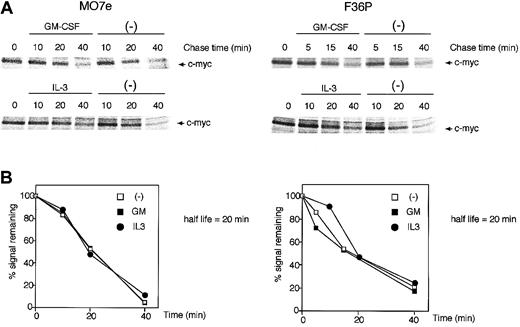
The c-Myc protein has a relatively short half-life (approximately 20 to 30 minutes)32 and is degraded by an ubiquitination-proteasome machinery.33,34 This degradation is dependent upon the phosphorylation level of the c-Myc protein, and this phosphorylation is known to be regulated by 2 distinct pathways: the classical mitogen-activated protein kinase (MAPK) (Ras/Raf/Erk) cascade and the PI3K pathway, which controls glycogen synthase kinase-3 (GSK-3) activity.35 Because GM-CSF and IL-3 also utilize these 2 pathways,36-38 which might result in acceleration or retardation of c-Myc degradation by the 2 HGFs, we evaluated whether GM-CSF and IL-3 affect the stability of c-Myc protein.
GM-CSF withdrawal-induced G1-arrested cells were purified by centrifugal elutriation and were then incubated in media without methionine and HGFs for 15 minutes. After being pulse-labeled by [35S]methionine for 30 minutes, the cells were cultured in the presence or absence of GM-CSF and IL-3. As shown in Figure 2, in both cell lines the half-life of the c-Myc protein was approximately 20 minutes, a value consistent with published data,32 and no difference was observed with or without GM-CSF and IL-3. These results clearly indicate that GM-CSF and IL-3 did not affect the stability of c-Myc protein.
c-Myc protein stability was unaltered by GM-CSF and IL-3. Determination of the half-life of the c-Myc protein in MO7e (left panels) and F36P (right panels) cells incubated with or without GM-CSF and IL-3. Pulse-labeling and immunoprecipitation were performed as described in “Materials and methods.” Cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 at 37°C. Immunoprecipitates were subjected to SDS-PAGE, and the amount of radiolabel incorporated into each band was determined using a Phosphor Imager. (A) Representative gels of pulse-labeling immunoprecipitations. (B) Phosphor Imager analysis of the gels shown in panel A. □ indicates cells incubated without HGFs; ▪, cells incubated with GM-CSF; •, cells incubated with IL-3. These show the representative results that were performed on a minimum of 3 independent experiments.
c-Myc protein stability was unaltered by GM-CSF and IL-3. Determination of the half-life of the c-Myc protein in MO7e (left panels) and F36P (right panels) cells incubated with or without GM-CSF and IL-3. Pulse-labeling and immunoprecipitation were performed as described in “Materials and methods.” Cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 at 37°C. Immunoprecipitates were subjected to SDS-PAGE, and the amount of radiolabel incorporated into each band was determined using a Phosphor Imager. (A) Representative gels of pulse-labeling immunoprecipitations. (B) Phosphor Imager analysis of the gels shown in panel A. □ indicates cells incubated without HGFs; ▪, cells incubated with GM-CSF; •, cells incubated with IL-3. These show the representative results that were performed on a minimum of 3 independent experiments.
GM-CSF/IL-3 signalings for c-Myc synthesis were not mediated by mTOR
Results shown in Figure 2 suggested that the increase of c-Myc protein was not due to decreased degradation but was rather controlled at the level of its synthesis, though the data in Figure 1 seem to rule out the transcriptional control, suggesting the translational control mechanism. To explore whether c-Myc synthesis induced by GM-CSF and IL-3 is executed in cap-dependent translation, we investigated the effects of cap-dependent translation inhibitor, rapamycin, on c-Myc induction. Rapamycin, after forming a complex with the immunophilin FKBP (FK506 binding protein), binds to a kinase named mammalian target of rapamycin (mTOR) and inhibits its kinase activity.39,40 mTOR controls the cap-dependent translation machinery via activation of the p70S6K protein kinase and via inhibition of the eIF4E inhibitor 4E-BP1.41,42 mTOR inhibition by rapamycin blocks the phosphorylation of p70S6K and 4E-BP1 and consequently inhibits cap-dependent translation.
As shown in Figure 3A, GM-CSF potently induced the phosphorylation of both p70S6K and 4E-BP1, which was almost completely blocked by rapamycin; 25 nM rapamycin was sufficient for this effect. Under these circumstances, c-Myc induction by GM-CSF was not affected by rapamycin (even by its higher concentrations). Similar findings were obtained for IL-3 (Figure 3B). These results demonstrated that although capable of activating mTOR-mediated cap-dependent translation, GM-CSF and IL-3 do not utilize this pathway at least for c-Myc protein synthesis.
Rapamycin did not inhibit the synthesis of c-Myc in MO7e and F36P cells. (A) Elutriated G1-arrested cells were pretreated with diluent (dimethyl sulfoxide [DMSO]) or 25, 250, and 2500 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF for 2 hours at 37°C. (B) Elutriated G1-arrested cells were pretreated with diluent (DMSO) or 250 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) IL-3 for 2 hours at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and immunoblotted. Western blotting for p70S6K was initially done using antiphosphorylated form-specific antibody followed by stripping and reblotting by antitotal p70S6K antibody. These show the representative results of 3 independent experiments.
Rapamycin did not inhibit the synthesis of c-Myc in MO7e and F36P cells. (A) Elutriated G1-arrested cells were pretreated with diluent (dimethyl sulfoxide [DMSO]) or 25, 250, and 2500 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF for 2 hours at 37°C. (B) Elutriated G1-arrested cells were pretreated with diluent (DMSO) or 250 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) IL-3 for 2 hours at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and immunoblotted. Western blotting for p70S6K was initially done using antiphosphorylated form-specific antibody followed by stripping and reblotting by antitotal p70S6K antibody. These show the representative results of 3 independent experiments.
c-myc translation was regulated by IRES-mediated mechanism
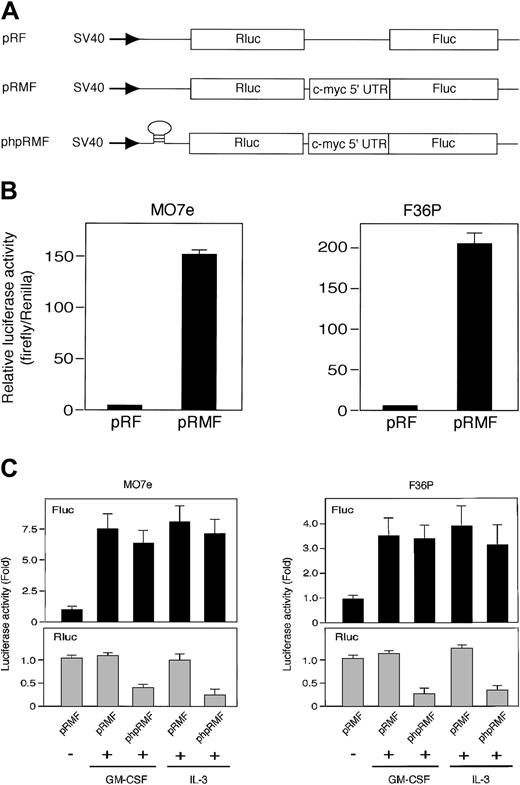
Rapamycin blocks mTOR-mediated cap-dependent translation but not cap-independent translation including IRES-mediated mechanism.43 Thus, the results described suggest that HGF-induced c-Myc protein synthesis in these cell lines occurs by an IRES-mediated mechanism. To test this, we tried to measure the IRES activities in cells using 2 types of dicistronic reporter gene plasmid (Figure 4A): pRMF, which contains the c-myc IRES between Renilla and firefly luciferase genes, and the control plasmid pRF, which contains a short linker sequence between 2 luciferases.13
The c-myc IRES was activated by GM-CSF and IL-3. (A) Schematic illustration shows dicistronic reporter gene constructs. The control plasmid pRF contains a short linker sequence between Renilla (Rluc) and firefly (Fluc) luciferase genes, and pRMF contains the c-myc IRES between 2 luciferase genes. phpRMF contains the c-myc 5′ untranslated region (UTR) between 2 luciferase genes and palindrome sequence upstream to Renilla luciferase gene. This palindrome sequence allows the formation of a stable hairpin in the RNA. Transcriptions of the 3 constructs were driven by SV40 promoter. (B) G1-arrested MO7e and F36P cells were transfected with pRF or pRMF by electroporation. After the incubation of cells with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF for 6 hours at 37°C, Renilla and firefly luciferase activities were determined, and IRES activities were expressed as ratios of firefly to Renilla luciferase. Horizontal axis represents the relative luciferase activity, which was calculated as relative to pRF, which was given a value of 1. (C) Elutriated G1-arrested cells were cotransfected with pSV-β-gal (used as control) and dicistronic plasmid (pRMF or phpRMF) and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 6 hours at 37°C. Both firefly (Fluc, upper panels) and Renilla (Rluc, lower panels) luciferase activities were normalized to β-galactosidase activity, and luciferase activity of stimulated cells was expressed as fold activation to that of unstimulated cells. Data presented are the means (± SEM) of duplicate samples from 3 independent experiments (B-C).
The c-myc IRES was activated by GM-CSF and IL-3. (A) Schematic illustration shows dicistronic reporter gene constructs. The control plasmid pRF contains a short linker sequence between Renilla (Rluc) and firefly (Fluc) luciferase genes, and pRMF contains the c-myc IRES between 2 luciferase genes. phpRMF contains the c-myc 5′ untranslated region (UTR) between 2 luciferase genes and palindrome sequence upstream to Renilla luciferase gene. This palindrome sequence allows the formation of a stable hairpin in the RNA. Transcriptions of the 3 constructs were driven by SV40 promoter. (B) G1-arrested MO7e and F36P cells were transfected with pRF or pRMF by electroporation. After the incubation of cells with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF for 6 hours at 37°C, Renilla and firefly luciferase activities were determined, and IRES activities were expressed as ratios of firefly to Renilla luciferase. Horizontal axis represents the relative luciferase activity, which was calculated as relative to pRF, which was given a value of 1. (C) Elutriated G1-arrested cells were cotransfected with pSV-β-gal (used as control) and dicistronic plasmid (pRMF or phpRMF) and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 6 hours at 37°C. Both firefly (Fluc, upper panels) and Renilla (Rluc, lower panels) luciferase activities were normalized to β-galactosidase activity, and luciferase activity of stimulated cells was expressed as fold activation to that of unstimulated cells. Data presented are the means (± SEM) of duplicate samples from 3 independent experiments (B-C).
We measured the activity of Renilla luciferase (Rluc) activity, which reflects the activity of cap-dependent translation with time after transfection. Reproducible value of the Rluc activity was detected at 6 hours after transfection in both MO7e and F36P cell lines. We also confirmed that elutriated G1-arrested cells were alive after 6 hours without HGF (data not shown). Thus, using this time point, dicistronic reporter gene assay was performed.
Elutriated G1-arrested cells transfected with each plasmid were incubated for 6 hours in the presence of GM-CSF, and the dual luciferase assay was performed as described in “Materials and methods.” The presence of c-myc IRES sequence increased the expression of firefly luciferase (Fluc) relative to Renilla luciferase (Rluc) by approximately 150-fold in MO7e cells and 200-fold in F36P cells (Figure 4B). These results indicated that c-myc IRES really functions as IRES in the presence of GM-CSF.
We then compared luciferase activity in the presence or absence of HGF. As shown in Figure 4C, Fluc activity in the presence of GM-CSF and IL-3 increased by 7.5-fold in MO7e cells and 3.5-fold in F36P cells as compared with Fluc activity in the absence of GM-CSF and IL-3. On the other hand, Rluc activity in the absence of GM-CSF and IL-3 was almost equivalent to that in the presence of GM-CSF and IL-3, indicating cap-dependent translation is active even without HGF. This finding is reasonable, because it has been reported that mTOR-independent cap-dependent mechanism works predominantly in mammalian cells.44
To further confirm the IRES function of c-myc 5′UTR and rule out cap-dependent translation of Fluc, we used another dicistronic plasmid, phpRMF, in which palindrome was inserted in front of Rluc gene (Figure 4A). This palindrome allows the formation of a stable RNA hairpin that inhibits the cap-dependent translation of Rluc. As shown in Figure 4C, increment of Fluc activity induced by GM-CSF and IL-3 was also observed in phpRMF, whereas Rluc activity was potently suppressed.
These results demonstrated that c-myc IRES functions well even when the cap-dependent translation was blocked by the hairpin structure and suggest that the increase of c-Myc protein by GM-CSF and IL-3 is controlled preferentially by IRES-dependent mechanism.
c-myc IRES was activated by PI3K-mediated pathway
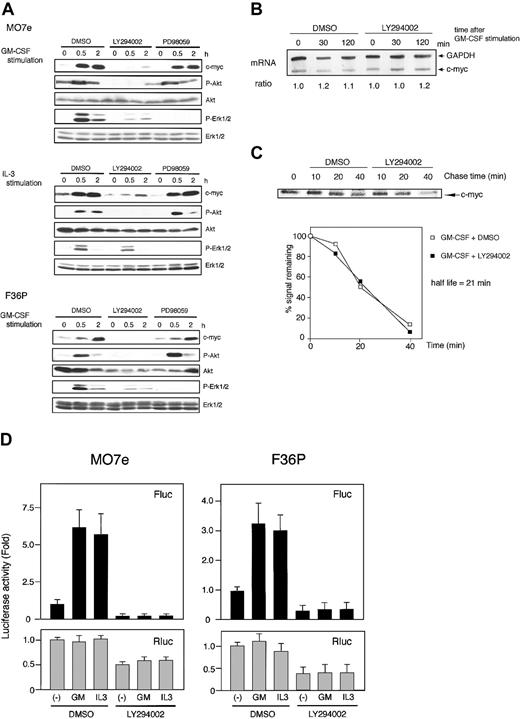
It is well known that GM-CSF and IL-3 utilize several signaling pathways (such as PI3K and Ras/Raf/Erk) for induction of their biologic action.4,36-38 To determine which pathway is critical for the activation of c-myc IRES, we analyzed c-Myc protein expression and c-myc IRES activation using a specific inhibitor for each pathway.
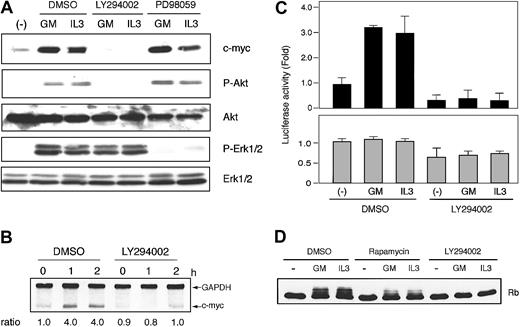
In both cell lines, LY294002, a specific inhibitor of PI3K, did block GM-CSF– and IL-3–induced protein kinase B (PKB)/Akt phosphorylation and potently suppressed GM-CSF– and IL-3–induced c-Myc expression (Figure 5A, data for IL-3–stimulated F36P cells are not shown). We then investigated the effect of LY294002 on c-myc mRNA expression and c-Myc protein stability and found that this inhibitor had no effect (Figure 5B-C).
Involvement of PI3K pathway during IRES-mediated c-Myc protein synthesis. (A) Elutriated G1-arrested cells were pretreated with diluent (DMSO), 50 μM LY294002, or 50 μM PD98059 for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and/or IL-3 for the indicated periods at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted. Western blotting for Akt and Erk1/2 was initially performed using antiphosphorylated form-specific antibodies followed by stripping and reblotting by antitotal Akt and Erk1/2 antibodies. These show the representative results of a minimum of 3 independent experiments. (B) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with 10 ng/mL GM-CSF for the indicated periods at 37°C. Analysis of c-myc mRNA was performed as described in the legend to Figure 1. (C) MO7e cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with 10 ng/mL GM-CSF at 37°C. Analysis of the half-life of the c-Myc protein was performed as in the legend to Figure 2, except for the presence of DMSO or 50 μM LY294002 during the incubation. (D) Elutriated G1-arrested cells transfected with pSV-β-gal and pRMF were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 6 hours at 37°C. Both Fluc and Rluc activities were normalized to β-galactosidase activity, and luciferase activity of stimulated cells was expressed as fold activation to that of unstimulated cells. Data presented are the means (± SEM) of 3 independent experiments.
Involvement of PI3K pathway during IRES-mediated c-Myc protein synthesis. (A) Elutriated G1-arrested cells were pretreated with diluent (DMSO), 50 μM LY294002, or 50 μM PD98059 for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and/or IL-3 for the indicated periods at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted. Western blotting for Akt and Erk1/2 was initially performed using antiphosphorylated form-specific antibodies followed by stripping and reblotting by antitotal Akt and Erk1/2 antibodies. These show the representative results of a minimum of 3 independent experiments. (B) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with 10 ng/mL GM-CSF for the indicated periods at 37°C. Analysis of c-myc mRNA was performed as described in the legend to Figure 1. (C) MO7e cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with 10 ng/mL GM-CSF at 37°C. Analysis of the half-life of the c-Myc protein was performed as in the legend to Figure 2, except for the presence of DMSO or 50 μM LY294002 during the incubation. (D) Elutriated G1-arrested cells transfected with pSV-β-gal and pRMF were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 6 hours at 37°C. Both Fluc and Rluc activities were normalized to β-galactosidase activity, and luciferase activity of stimulated cells was expressed as fold activation to that of unstimulated cells. Data presented are the means (± SEM) of 3 independent experiments.
To determine whether c-Myc suppression by LY294002 was caused by the inhibition of c-myc IRES-mediated translation, we measured c-myc IRES-driving luciferase activity. As shown in Figure 5D, GM-CSF– and IL-3–induced increase in Fluc activity was strongly suppressed in the presence of LY294002. This finding indicates that the mechanism of c-myc IRES activation is downstream of PI3K pathway. Interestingly, overall Rluc activity was also suppressed by approximately 50% in both cell lines by this inhibitor, a finding consistent with previous observations showing that PI3K pathway is involved in cap-dependent translation initiation.45
Inhibition of PI3K was reported to affect the Ras/Raf/Erk pathway.46 In this study, we also observed the slight attenuation of GM-CSF–activated phosphorylation of Erk1/2 by LY294002 (Figure 5A). Thus, we investigated the effect of Ras/Raf/Erk pathway on c-Myc induction using MEK inhibitor, PD98059. Treatment of cells by PD98059 suppressed Erk1/2 phosphorylation, during which the expression of c-myc was not altered (Figure 5A). This indicated that classic MAPK cascade was not involved in c-myc expression in these cell lines.
Inhibitory effect of LY294002 but not of rapamycin on Rb phosphorylation and cell-cycle progression
The results described together indicate that GM-CSF and IL-3 induce c-Myc protein synthesis via the PI3K-mediated (LY294002-sensitive) and cap-independent (rapamycin-insensitive) IRES pathway. Although the roles of c-myc during cell-cycle progression and cell proliferation have been established,1,3 a possible relationship between this pathway and downstream events remains to be determined. To address this question, we investigated the effects of LY294002 on the downstream events including retinoblastoma (Rb) protein phosphorylation,47 DNA synthesis, and cell cycle progression in our assay system using rapamycin as a control inhibitor.
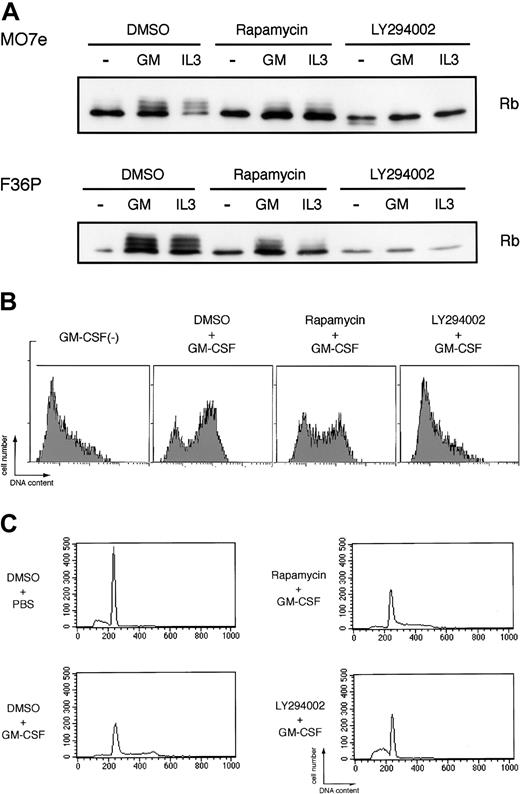
As shown in Figure 6A, elutriated G1-arrested cells incubated without HGF showed a single unphosphorylated band of Rb protein as determined by Western blotting, indicating early G1 phase of the cells. Addition of GM-CSF or IL-3 to the factor-deprived cells induced marked band shift (phosphorylation) of Rb (Figure 6A), reflecting cell-cycle progression from early G1 to mid or late G1 phase. LY294002 almost completely blocked Rb phosphorylation induced by GM-CSF and IL-3. In addition to Rb phosphorylation, we also confirmed that the addition of GM-CSF to the elutriated factor-deprived cells induced prominent DNA synthesis as determined by BrdU incorporation to the cells (Figure 6B) and cell-cycle progression from G1 phase to S and/or G2/M phases as determined by flow cytometric analysis (Figure 6C). LY294002 completely abolished both DNA synthesis and cell-cycle progression stimulated by GM-CSF (Figure 6B-C). On the other hand, rapamycin, in spite of its well-known broad effects on cap-dependent translation, exerted only minimal effects on Rb phosphorylation, BrdU incorporation, and cell-cycle progression induced by GM-CSF. Thus, PI3K-mediated IRES pathway seems to be specifically linked to the cell-cycle progression, and HGF-induced c-myc expression correlated well with the cell-cycle progression and proliferation.
Potent inhibitory effects of LY294002, but not rapamycin, on GM-CSF– and IL-3–induced Rb phosphorylation and cell cycle progression. (A) Elutriated G1-arrested cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 3 hours (MO7e) or 9 hours (F36P) at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted using specific antibody for Rb. (B) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 10 ng/mL GM-CSF in the presence of 10 μM BrdU for 24 hours at 37°C. Detection of incorporated BrdU was performed as described in “Materials and methods.” (C) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 10 ng/mL GM-CSF for 24 hours at 37°C. Analysis of cell cycle distribution was performed as in the legend to Figure 1.
Potent inhibitory effects of LY294002, but not rapamycin, on GM-CSF– and IL-3–induced Rb phosphorylation and cell cycle progression. (A) Elutriated G1-arrested cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with or without 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF and IL-3 for 3 hours (MO7e) or 9 hours (F36P) at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and were then immunoblotted using specific antibody for Rb. (B) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 10 ng/mL GM-CSF in the presence of 10 μM BrdU for 24 hours at 37°C. Detection of incorporated BrdU was performed as described in “Materials and methods.” (C) Elutriated G1-arrested MO7e cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 10 ng/mL GM-CSF for 24 hours at 37°C. Analysis of cell cycle distribution was performed as in the legend to Figure 1.
GM-CSF and IL-3 activate c-myc IRES in UT7 cells in which c-myc transcription is regulated by GM-CSF
Numerous studies have reported that the expression of c-myc is regulated at the level of transcription in various growth factor–dependent cell lines.48,49 To reveal whether HGF regulates c-myc IRES-dependent translation under the condition where its transcription is also regulated by HGF, we measured the IRES activity in another factor-dependent human leukemic cell line, UT7, in which the expression of both c-myc mRNA and protein was regulated by GM-CSF stimulation (Figure 7B). Also in this cell line, GM-CSF and IL-3 did not affect c-Myc protein stability (Figure 7C). In addition, c-Myc protein synthesis induced by these 2 HGFs was resistant to rapamycin (Figure 7D), indicating that HGF-induced c-Myc synthesis was not mediated by the cap-dependent translation also in this cell line. Finally, as shown in Figure 7E, the Fluc activity but not Rluc activity was increased approximately 3-fold after GM-CSF and IL-3 stimulation. Thus, IRES-dependent translation of c-myc appears to be able to work concomitantly with the up-regulation of c-myc mRNA in human hematopoietic cells stimulated by GM-CSF and IL-3.
GM-CSF and IL-3 induced c-myc IRES activity in another factor-dependent cell line UT7, in which c-myc mRNA was regulated by GM-CSF. (A) Cell cycle analysis of UT7 cells with or without 1 ng/mL GM-CSF. Determination of cell cycle distribution was performed as described in the legend to Figure 1. The G1 cells indicated by the open box were separated by centrifugal elutriation for the following experiments. (B) Elutriated G1-arrested UT7 cells were incubated with 1 ng/mL GM-CSF for the indicated periods at 37°C. Analyses of mRNA and protein were performed as described in the legend to Figure 1. (C) UT7 cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with or without 1 ng/mL GM-CSF or IL-3 at 37°C. Determination of the half-life of the c-Myc protein was performed as described in the legend to Figure 2. Asterisk indicates nonspecific band. (D) The effect of rapamycin on GM-CSF– and IL-3–induced c-Myc expression in UT7 cells. Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO) or 250 nM rapamycin for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for 2 hours at 37°C. Total cell lysates were subjected to Western blotting as in the legend to Figure 3. (E) Determination of GM-CSF– and IL-3–induced c-myc IRES activity in UT7. Elutriated G1-arrested UT7 cells were cotransfected with pSV-β-gal and dicistronic plasmid (pRMF or phpRMF) and were then incubated with or without 1 ng/mL GM-CSF or IL-3 for 6 hours at 37°C. Luciferase assay was performed as in the legend to Figure 4. Data presented are the means (± SEM) of duplicate samples from 3 independent experiments.
GM-CSF and IL-3 induced c-myc IRES activity in another factor-dependent cell line UT7, in which c-myc mRNA was regulated by GM-CSF. (A) Cell cycle analysis of UT7 cells with or without 1 ng/mL GM-CSF. Determination of cell cycle distribution was performed as described in the legend to Figure 1. The G1 cells indicated by the open box were separated by centrifugal elutriation for the following experiments. (B) Elutriated G1-arrested UT7 cells were incubated with 1 ng/mL GM-CSF for the indicated periods at 37°C. Analyses of mRNA and protein were performed as described in the legend to Figure 1. (C) UT7 cells were labeled with radioisotope for 30 minutes at 37°C and were then incubated for the indicated periods with or without 1 ng/mL GM-CSF or IL-3 at 37°C. Determination of the half-life of the c-Myc protein was performed as described in the legend to Figure 2. Asterisk indicates nonspecific band. (D) The effect of rapamycin on GM-CSF– and IL-3–induced c-Myc expression in UT7 cells. Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO) or 250 nM rapamycin for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for 2 hours at 37°C. Total cell lysates were subjected to Western blotting as in the legend to Figure 3. (E) Determination of GM-CSF– and IL-3–induced c-myc IRES activity in UT7. Elutriated G1-arrested UT7 cells were cotransfected with pSV-β-gal and dicistronic plasmid (pRMF or phpRMF) and were then incubated with or without 1 ng/mL GM-CSF or IL-3 for 6 hours at 37°C. Luciferase assay was performed as in the legend to Figure 4. Data presented are the means (± SEM) of duplicate samples from 3 independent experiments.
PI3K pathway was involved in both transcription and IRES translation of c-myc in UT7 cells
As shown in Figure 8A, PI3K inhibitor LY294002 almost completely blocked GM-CSF– and IL-3–induced c-Myc protein synthesis in UT7 cells whereas MEK inhibitor PD98059 had no effect. Interestingly, inhibitory effects of LY294002 on c-Myc induction were observed at both transcriptional and IRES-mediated translational levels (Figure 8B-C) in this cell line. In addition, LY294002 almost completely blocked Rb phosphorylation induced by GM-CSF and IL-3 in UT7 cells (Figure 8D). These findings suggest critical roles of PI3K in this cell line in which both c-myc transcription and IRES are regulated by GM-CSF and IL-3.
Potent inhibitory effects of LY294002 on GM-CSF– and IL-3–induced protein and mRNA synthesis of c-myc, c-myc IRES activity, and Rb phosphorylation in UT7 cells. (A) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO), 50 μM LY294002, or 50 μM PD98059 for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for 2 hours at 37°C. Total cell lysates were separated by SDS-PAGE and analyzed by immunoblotting as described in the legend to Figure 5. (B) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for the indicated periods at 37°C. Analysis of c-myc mRNA was performed as described in the legend to Figure 1. (C) Elutriated G1-arrested UT7 cells transfected with dicistronic luciferase plasmid (pRMF or phpRMF) and pSV-β-gal were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 1 ng/mL GM-CSF or IL-3 for 6 hours at 37°C. Luciferase assay was performed as described in the legend to Figure 4. Data presented are the means (± SEM) of 3 independent experiments. (D) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with 1 ng/mL GM-CSF or IL-3 for 9 hours at 37°C. Immunoblotting for Rb was performed as described in the legend to Figure 6.
Potent inhibitory effects of LY294002 on GM-CSF– and IL-3–induced protein and mRNA synthesis of c-myc, c-myc IRES activity, and Rb phosphorylation in UT7 cells. (A) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO), 50 μM LY294002, or 50 μM PD98059 for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for 2 hours at 37°C. Total cell lysates were separated by SDS-PAGE and analyzed by immunoblotting as described in the legend to Figure 5. (B) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then incubated with 1 ng/mL GM-CSF or IL-3 for the indicated periods at 37°C. Analysis of c-myc mRNA was performed as described in the legend to Figure 1. (C) Elutriated G1-arrested UT7 cells transfected with dicistronic luciferase plasmid (pRMF or phpRMF) and pSV-β-gal were pretreated with diluent (DMSO) or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with or without 1 ng/mL GM-CSF or IL-3 for 6 hours at 37°C. Luciferase assay was performed as described in the legend to Figure 4. Data presented are the means (± SEM) of 3 independent experiments. (D) Elutriated G1-arrested UT7 cells were pretreated with diluent (DMSO), 250 nM rapamycin, or 50 μM LY294002 for 30 minutes at 37°C and were then cultured with 1 ng/mL GM-CSF or IL-3 for 9 hours at 37°C. Immunoblotting for Rb was performed as described in the legend to Figure 6.
Discussion
In this study, we showed that 2 HGFs, GM-CSF and IL-3, induced c-Myc protein synthesis by the IRES-dependent cap-independent translation mechanism under the conditions in which cap-dependent translation was also active. This conclusion was established based upon the following 3 lines of evidence. First, c-myc mRNA expression and protein stability were not altered by HGF. Second, rapamycin did not affect c-Myc protein induction. Finally, an increase in c-myc IRES activity was observed after HGF stimulation. These findings together indicate that IRES of cellular mRNA has a significant role in growth regulation of the cells.
IRES elements were first discovered in picornavirus mRNAs, which are naturally uncapped but nonetheless efficiently translated.50,51 Subsequently, it was found that some cellular mRNAs that are normally capped could also be translated by an internal ribosome binding mechanism.15 When the first example of cellular mRNAs that contain IRES was found, it was not obvious what advantage there would be in having this alternative mechanism of translation initiation for capped mRNA.
To date, many studies reported a number of proteins that function during periods of cellular stress—for example, apoptosis,23,24,52 hypoxia,53,54 and amino acid starvation55 —are translated from mRNAs that contain an IRES, and these situations inhibit total cap-dependent translation. Thus, it is assumed that one role of cellular IRES is the protection against this form of suppression of translation. However, in this study, we have demonstrated that c-myc IRES was activated preferentially in situations where cells are growing and cap-dependent translation is active, indicating another type or role of cellular IRES.
Because canonical cap-dependent translation is constitutively active in viable cells, it is not suitable for gene-specific translation. In contrast, IRES-dependent translation is executed in the limited numbers of molecules with IRES structure in 5′UTR of its mRNA. In addition, it is conceivable that IRES-dependent translation would be strictly regulated by a specific mechanism because IRES requires each IRES-specific cellular RNA-binding proteins for its activation.15 Taken together, we consider that the role of IRES is to regulate specific gene expression required in some circumstances, including cell growth and under stress, regardless of activity of cap-dependent translation.
In contrast to the present results, c-myc can also be translated in an mTOR-mediated cap-dependent manner.14 Mendez et al have reported that c-myc induction by insulin was inhibited completely by rapamycin treatment.20 They used murine factor–dependent cell line 32D and stimulated the factor-deprived cells by insulin; thus, the circumstance of this experiment was similar to ours. On the other hand, in serum stimulation of Epstein-Barr virus–immortalized B-cell lines, the inhibitory effect of rapamycin on c-myc induction was reported to be only 50%.11
Previous studies have shown that mRNAs of elongation factors and ribosomal proteins44 and growth-regulated molecules,20 which contain encumbered 5′UTR,19 are translated by mTOR-mediated mechanisms, indicating that the mTOR pathway also controls the translation of growth-regulated genes by certain (unknown) mechanisms that render the translation of mRNA with cumbersome 5′UTR much more effectively. Thus, the expression of mRNAs with encumbered 5′UTR seems to be regulated at the translational level via dual mechanisms: mTOR-mediated cap-dependent and IRES-mediated cap-independent mechanisms. Which mechanism works might depend upon the types of cells and the mode of stimuli. This might explain why the translation of c-myc is regulated via distinct mechanisms in HGF-stimulated human hematopoietic cells versus insulin-stimulated murine hematopoietic cells. Fifty percent inhibition of c-myc induction by rapamycin in serum-stimulated B cells might result from heterogeneous substances in the serum, which may activate various mechanisms.
Little is known about the upstream signaling pathways to activate IRES-mediated translation machinery. In this study, we revealed that the PI3K pathway was involved in the activation of c-myc IRES. Interestingly, it is well known that the activation of PI3K induces mTOR activation,56,57 and we confirmed HGF-induced activation of Akt, a downstream target of PI3K. On the other hand, inhibition of mTOR by rapamycin did not inhibit c-myc induction by GM-CSF and IL-3 (Figures 3 and 7). Thus, HGF activates PI3K and thereby triggers downstream IRES-activating mechanisms other than mTOR. Another candidate of upstream signaling molecule of IRES may be p38 MAP kinase. p38 is reported to be involved in c-myc IRES activation during apoptosis by genotoxin.23 It is controversial whether GM-CSF and IL-3 exert some effects on p38,58 and we confirmed no effect of the 2 HGFs on p38 at least in MO7e and F36P cells (data not shown). The p38 pathway may be linked to the IRES-activating mechanism in limited situations such as apoptosis.
It is classically assumed that the intracellular level of c-Myc protein simply results from that of mRNA48,49 ; thus, transcriptional regulation has been focused on as the main mechanism by which HGF controls c-myc expression. In this study, we also observed the regulation of c-myc mRNA by GM-CSF and IL-3 in UT7 cells. In contrast, we observed the constitutive expression of c-myc mRNA in the other 2 cell lines (MO7e and F36P), suggesting dysregulation of c-myc expression in these malignant cells. In these 2 cell lines, however, we demonstrated the strict factor-dependent regulation of c-Myc protein via regulation of translation. Furthermore, the present results for UT7 cells in which these 2 HGFs regulate expression of c-myc not only at the transcriptional level but also at the translational level suggest that the translational regulation of proliferation-related protein might widely be executed in cells in which transcriptional regulation was also active. Thus, this is the first observation that HGF regulates the synthesis of c-Myc protein at 2 distinct levels to control hematopoietic cell proliferation.
In this study, we showed the relatively tight control of c-myc expression by the 2 HGFs, physiological regulators of hematopoietic cell proliferation.59 The c-myc proto-oncogene has widely been proposed as a key molecule for malignant transformation of cells, and it is well known that many kinds of leukemic cells overexpress c-myc.60,61 Furthermore, ectopic c-myc expression in HGF-dependent cell lines renders these cells factor independent.6-8 These findings together suggest that HGFs strictly regulate cell-cycle progression and proliferation via tight control of the c-myc expression in factor-dependent leukemic cell lines.
Whether GM-CSF and/or IL-3 controls c-Myc protein level via IRES-mediated translation also in vivo remains to be determined. Preliminary study in our laboratory did not provide sufficient evidence to show c-Myc IRES activation in normal human bone marrow cells stimulated by the 2 HGFs. These results do not rule out the presence of such mechanism in vivo, because experiments using primary cells have limitation. In this aspect, it should be noted that GM-CSF and IL-3 do not seem to be essential for normal myeloid cell proliferation because mice deficient for a receptor subunit shared by GM-CSF and IL-3 did not show apparent abnormality in myelopoiesis.62 In contrast, it has been well established that GM-CSF and IL-3 are potent growth factors for human leukemic cells. In primary human myeloid leukemia, the leukemic cells often produce GM-CSF and other cytokines by themselves, resulting in autonomous (autocrine or paracrine) proliferation of the leukemic blasts.63,64 In these cells, GM-CSF–induced growth signals might be constitutively active and would induce steady-state expression of c-Myc protein, which may at least in part explain c-myc overexpression in primary human leukemic cells. Thus, we would like to emphasize important roles of HGF and c-myc, particularly IRES-mediated c-myc regulation by HGF, during malignant transformation and/or maintenance of their proliferation. Physiological significance of these systems in normal human hematopoietic cells awaits future investigation.
Prepublished online as Blood First Edition Paper, July 10, 2003; DOI 10.1182/blood-2003-02-0567.
Supported by a Grant-in-Aid for the Second Term Comprehensive 10-year Strategy for Cancer Control from the Ministry of Health, Labour and Welfare, a Sasakawa Scientific Research Grant from the Japan Science Society, and a Young Researcher Grant from Mitsubishi Pharma Research Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Dr Anne E. Willis for the gift of plasmids (pRF, pRMF, and phpRMF).

![Figure 3. Rapamycin did not inhibit the synthesis of c-Myc in MO7e and F36P cells. (A) Elutriated G1-arrested cells were pretreated with diluent (dimethyl sulfoxide [DMSO]) or 25, 250, and 2500 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) GM-CSF for 2 hours at 37°C. (B) Elutriated G1-arrested cells were pretreated with diluent (DMSO) or 250 nM rapamycin for 30 minutes at 37°C and were then incubated with 10 ng/mL (MO7e) or 1 ng/mL (F36P) IL-3 for 2 hours at 37°C. Total cell lysates (3 × 105 cells per lane) were separated by SDS-PAGE and immunoblotted. Western blotting for p70S6K was initially done using antiphosphorylated form-specific antibody followed by stripping and reblotting by antitotal p70S6K antibody. These show the representative results of 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/9/10.1182_blood-2003-02-0567/6/m_h82135160003.jpeg?Expires=1769089245&Signature=QrQ9xQsp3TyH-iI6~iVeoWHJkenEzoCWlznYaZz1sZ2BX0HA0XBT1W9NbPvbAifwqjc97IfF3zvzgmqsURbAz7F3JJK58uQ2Kb2YxH0MQWfJssRPJYceT1NYBdP3z3gt4s1g29VXLhTleF294ly4SWSn5OqvvbQPgiOxw8fl2VK8TTRttMD5-jHRCyEO9hJveEDIYet1pvOPo1hUmwJhtVaH87Xnia~-rQ-ou6CaTiP9JPGzs7wvrW6hr0DQfjT4ikq-LTbYYGsVCZqMLeoRlrcF1MmhWjIUKL35vZeMZ7XOczpQkmaUsJ-E4onBMElZ80-kl4ywmfENpYB6vHX6bg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal