Abstract
CD4 can be up-regulated on CD8+ T cells generating a CD4dimCD8bright phenotype. We previously demonstrated that the CD4dimCD8bright phenotype constitutes an activated phenotype of CD8+ T cells. We demonstrate here that the activated CD4dimCD8bright T cells are not undergoing apoptosis and do not produce significant intracellular levels of interferon γ (IFNγ), interleukin 2 (IL-2), or IL-10 but express elevated levels of intracellular IL-4 in comparison to CD8+CD4– and CD4+ T cells. In response to cytomegalovirus (CMV) peptide (pp65) priming, CD4dimCD8bright cells recognized CMV pp65 tetramer approximately 19-fold higher than CD4–CD8+ T cells, indicating that these cells are capable of antigen-specific recognition to a far greater extent than CD4–CD8+ T cells. CD4dimCD8bright T cells also express both CXCR4 and CCR5 but are susceptible to T-tropic and not M-tropic HIV infection. A soluble factor believed to be β-chemokine is responsible for the inhibition of M-tropic HIV infection in CD4dimCD8bright T cells. CD8+ T cells from HIV+ patients were capable of up-regulating CD4 on CD8+ T cells. We also provide evidence of the presence of peripheral blood CD4dimCD8bright T cells in HIV+ patients, albeit at low frequency. Collectively, these data suggest a role of CD4dimCD8bright T cells in both normal T-cell biology and HIV pathogenesis.
Introduction
Up-regulation of CD4 on CD8+ T cells is well established.1-4 Some of the physiologic stimulations that lead to CD4 up-regulation on CD8+ T cells include anti-CD3/CD28 costimulation,1,3 superantigen (staphylococcal enterotoxin B [SEB]),1,2 and interaction with antigen-presenting cells.4 Given the dim expression of CD4 on CD8-stimulated cells, these cells are referred to as CD4dimCD8bright T cells. The CD4 molecule on the CD8+ T cells is linked to p56lck,2 a critical kinase in CD4-mediated signal transduction, suggesting a biologic role of CD4 on CD8+ T cells. Recently, through extensive phenotypic characterization of CD4dimCD8bright T cells, we determined that they constitute an activated phenotype of CD8+ T cells.1 Specifically, the percentage of expression of CD25 (interleukin 2 [IL-2] α chain receptor), CD69 (early activation marker), CD38 (differentiation marker), CD95 (Fas receptor), CD28 (costimulatory molecule), and CD45RA+CD45RO+ (indicative of primed/transitional T cells) was significantly elevated on CD4dimCD8bright T cells in comparison to CD8+ T cells that did not up-regulate CD4 on their surface (CD8+CD4–).1 This finding suggests that CD4dim expression on CD8+ T cells may be an early marker of CD8+ T-cell activation.1
The lineage relationship between the CD8+ T-cell subtypes and the definition of the best phenotypic markers to differentiate between these subsets has been the topic of much debate.5-9 CD62L, CD45RA, and CCR7 surface markers have been used to differentiate between naive, effector memory, and central memory T cells.5,6 According to these markers, naive CD8+ T cells are CD45RA+CD62L+CCR7+. On antigen encounter, naive CD8+ T cells differentiate into central memory cells (CD45RA–CD62L+CCR7+), effector memory cells (CD45RA– CD62L–CCR7–), and terminally differentiated effector cells (CD45RA+CD62L–CCR7+). Alternatively, the combinations of CD45RA, CD27, and CD28 have also been used to differentiate between the various CD8+ T-cell subtypes. According to these markers, naive CD8+ T cells are defined as CD45RA+CD27+CD28+, memory cells as CD45RA–CD27+CD28+, and effector cells as CD45RA+ CD27–CD28–. The progression of naive CD8+ T cells to generate the effector or central memory population is also under intense investigation. Recently, Wherry et al7 proposed a linear progression model of CD8+ T-cell differentiation, whereby naive T cells give rise to effector T cells, effector memory, and central memory cells.7 Others have proposed that central and effector memory T cells arise from distinct lineages.8 Additional CD8+ T-cell lineage relationships may be conferred by CD4 up-regulation on CD8+ T cells, generating the CD4dimCD8bright T-cell phenotype.
Given that CD4 expression is critical in the pathogenesis of HIV infection, up-regulation of CD4 on CD8+ T cells may provide a mechanism for HIV infection of CD8+ T cells. Evidence of in vivo HIV infection of CD8+ T cells exists. Both peripheral blood CD8+ T cells2 and CD8+ T cells in the lung10 were identified to harbor HIV, as determined by HIV gag/pol DNA content, implicating CD8+ T cells as an additional reservoir for HIV. Limited data, however, exist on HIV productive infection of CD8+ T cells, as demonstrated by HIV p24 core antigen synthesis or by HIV gag/pol mRNA analysis. Discordant data have been reported for the susceptibility of CD4dimCD8bright T cells to infection by T-tropic and M-tropic isolates of HIV. Yang et al4 demonstrated that CD4dimCD8bright T cells are infected by CCR5-dependent strains and were resistant to CXCR4-dependent strains, whereas others demonstrated susceptibility of CD4dimCD8bright T cells to infection by CXCR4-dependent HIV strains.2,3 Given that CD4dimCD8bright cells may play a role in the immune response to HIV and that they may contribute to HIV pathogenesis in the CD8+ T-cell compartment, we evaluated cytokine production of these cells, evaluated evidence of their in vivo presence in HIV seropositive patients, and re-evaluated their in vitro susceptibility to T-tropic and M-tropic HIV infection. We also examined whether these CD4dimCD8bright cells are capable of antigen-specific recognition using cytomegalovirus (CMV) tetramer technology (reviewed in Constantin et al11 ).
Materials and methods
Isolation of CD8+ T cells
CD8+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) using immunoselection. Briefly, PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation from venous blood collected from healthy laboratory workers and HIV seropositive donors as previously described.12 Purified CD8+ T cells were isolated by negative immunoselection using magnetic beads (Dynal, Lake Success, NY). Depletion of CD4, CD14, CD16, CD19, and CD56 in these cultures was typically more than 99%. Isolated cells were suspended in RPMI 1640 medium (Biowhittaker, Walkersville, MD) supplemented with 10% human AB serum (Gemini Bio Products, Calabasas, CA), 1% penicillin/streptomycin (Sigma, St Louis, MO), and IL-2 (20 U/mL). Cultures were subsequently stimulated with 1 μg/mL SEB (Sigma) or 1 μg/mL soluble anti-CD3 and anti-CD28 antibodies (PharMingen, San Diego, CA) and maintained for 6 days, unless otherwise indicated.
Immunofluorescence surface staining and flow cytometric analysis
Cell surface staining of the lymphocyte subsets was performed at various time points following stimulation. Briefly, cells were incubated at room temperature for 20 minutes with the appropriate antibody panel conjugated to fluorescein isothiocyanate (FITC), phycoerythrin (PE), peridinin chlorophyll protein (PerCP)/Cy-Chorme (CyC), or allophycocyanin (APC). Following immunofluorescent labeling (if whole blood was used, the red blood cells were first lysed with 1 × FACSLyse [Becton Dickinson (BD), San Jose, CA] for 10 minutes), the cells were washed with 1 × phosphate-buffered saline (PBS) and fixed in 1% formaldehyde. All surface antibodies were purchased from either Becton Dickinson or PharMingen. Three- or 4-color flow cytometric analysis was preformed using a FACSCalibur Flow Cytometer using CELLQuest software (BD). Dual expression of CD4 and CD8 was determined by gating on CD3+CD8bright lymphocytes.
Fluorochrome-conjugated, peptide-loaded major histocompatibility class I tetramer complex detection of antigen-specific recognizing cells
Four donors with the haplotype HLA-A*0201, who are CMV immunoglobulin G (IgG) positive, indicating a past history of infection, were used in these studies. CD8+ T cells, depleted of monocyte/macrophages (CD14), CD4+ T cells, and B cells (CD19) were isolated from the 4 HLA-A*0201/CMV+ donors and were stimulated with SEB (1 μg/mL), primed with a CMV peptide (10 μg/mL), or left untreated. The CMV peptide, a 9-mer with the amino acid sequence NLVPMVATV, is based on the lower matrix protein pp65. Approximately 95% of the protein mass of “dense bodies” in CMV-infected cells is made up of matrix protein pp65, which is also an immunodominant epitope recognized by CD8+ T cells. The pp65 peptide was synthesized by Sigma Genosys (The Woodlands, TX) and was more than 95% pure as determined by reverse-phase high-performance liquid chromatography (HPLC) and mass spectral analysis. The tetramer, consisting of HLA-A*0201 with either CMV or Epstein-Barr virus (EBV) peptide within its groove and conjugated to streptavidin-phycoerythrin, was generated as previously described.13 The tetramers were a generous gift from John Altman (Department of Microbiology and Immunology, Emory University, Atlanta, GA). Six days after stimulation, the cells were stained for CD3:CD4:CD8: and CMV or EBV tetramer. Both isotype controls and CD8 compensation tubes were used as negative controls to set the gates. Stained samples were analyzed using FACSCalibur Flow Cytometer using CELLQuest software.
Detection of intracellular cytokine and β-chemokine secretion
CD8+ T cells were stimulated for 6 days as previously described. Following stimulation, cells were collected, treated with 10 μg/mL Brefeldin A (BFA; Sigma) for 1 to 3 hours, and intracellular cytokine (IL-2, interferon γ [IFNγ], IL-10, IL-4) or β-chemokine (macrophage inflammatory protein 1α [MIP-1α], regulated on activation, normal T-cell expressed and secreted [RANTES]) expression was detected using the Caltag Fix and Perm kit, according to the manufacturer's instructions (Caltag Laboratories, Burlingame, CA). Briefly, 2 to 5 × 105 cells were incubated with 100 μL fixation medium (Caltag Laboratories) for 15 minutes at room temperature. After fixation, cells were washed and resuspended in 100 μL permeabilization medium (Caltag Laboratories) in addition to antibodies for β-chemokine detection (RANTES-PE or MIP-1α–PE) or antibodies for cytokine detection (IL-2–PE or INF-γ–PE) along with CD4-FITC or APC and CD8-PerCP for 15 minutes at room temperature, in the dark. Cells were then washed and resuspended in 1% formaldehyde. Three- or 4-color flow cytometric analysis was preformed using a FACSCalibur Flow Cytometer using CELLQuest software (BD).
CXCR4 and CCR5 quantitation
Voltages for the flow cytometer were set according to Shero Rainbow Calibration Particles (Sherotech, Libertyville, IL). Samples were then acquired via flow cytometry. PE quantitation was determined by using QuantiBRIGHT Beads (BD), according to the manufacturer's instructions. The number of CXCR4 and CCR5 molecules per cell was determined by subtracting the number of molecules expressed on the appropriate isotype control.
Tunel assay
SEB-stimulated CD8+ T cells were first surface-stained for both CD4-PE and CD8-PerCP and fixed for at least 2 hours in 1% paraformaldehyde, as previously described, prior to performing the Tunel assay. Cells were permeabilized with 1 × fluorescence-activated cell sorter (FACS) Permeabilizing Solution (BD) for 2 minutes and vigorously washed. Low molecular weight DNA fragments (mononucleosomes and oligonucleosomes) as well as single-strand breaks (nicks) in high molecular weight DNA found in the cells were labeled with terminal deoxynucleotidyl transferase (TdT) (In Situ Cell Death Detection Kit-Fluorescein; Roche, Indianapolis, IN). FITC labels incorporated into nucleotide polymers were detected and quantitated by flow cytometry gating on CD4dimCD8bright T cells and CD8+CD4– T cells, respectively. As a positive control, cells were treated with DNase to induce nicks and fragments in the DNA prior to the addition of TdT. As a negative control, TdT was not added to the samples.
HIV-1 infection of CD4dimCD8bright T cells and intracellular p24 detection
Various primary isolates of HIV-1 (302073, 302140, 302056, 302072, and 302144), determined by ghost cell analysis to use CCR5 coreceptor, and a laboratory-adapted strain of HIV-1 (IIIB, CXCR4 using) were used to infect activated CD8+ T cells that had been stimulated for 6 days to induce CD4 expression. These viruses were obtained through the National Institutes of Health (NIH) AIDS Research and Reference Program, Division of AIDS, NIAID, NIH (Rockville, MD). Virus stocks were added to the cell pellets at 2 ng HIV p24/1 × 106 cells in a total volume of 1 mL. Cells were then incubated at 37°C under 5% CO2 for 2 hours. Following incubation with HIV, cells were washed twice with 1 × PBS and resuspended at a final concentration of 1 × 106 cells/mL using RPMI media supplemented with 10% human AB serum and 20 U/mL IL-2. Following 7 days in culture, HIV p24 core antigen was measured using a flow-based assay for the detection of intracellular p24.14 Briefly, 7 days after infection, 2 to 5 × 105 cells were incubated with 100 μL fixation medium (Fix and Perm kit, Caltag Laboratories) for 15 minutes at room temperature. After fixation, the cells were washed and resuspended in 100 μL permeabilization medium (Caltag Laboratories) in addition to intracellular KC57-FITC (Coulter, Miami, FL), CD4-PE, and CD8-PerCP antibodies for 15 minutes at room temperature in the dark. The KC57 antibody identifies HIV-1 core antigens (p55, p39, p33, and p24). Cells were then washed and resuspended in 1% formaldehyde. Three-color flow cytometric analysis was preformed using a FACSCalibur Flow Cytometer using CELLQuest software. HIV-1 core antigen production (p24) was also measured by lysing cell-free supernatants with 1% Triton X-100 and measuring HIV p24 antigen using conventional enzyme-linked immunosorbent assay (ELISA; assay kit purchased from the AIDS vaccine program, Fredrick, MD).
Supernatant blocking and neutralization assays
Supernatants from stimulated CD8+ T-cell cultures induced to express CD4dimCD8bright were harvested at day 6 and used immediately or frozen at –80°C until later use. PBMCs were isolated as previously described and stimulated for 3 days with 4 μg/mL phytohemagglutinin (PHA; Sigma). PBMCs were then harvested, infected with HIV, and resuspended at 2 × 106 cells/mL. Harvested supernatants were left untreated or either treated with β-chemokine blocking antibody (Ab) (anti-RANTES, anti–MIP-1α, and anti–MIP-1β) or control at 10 μg/mL (each) for 2 hours, at room temperature. Harvested supernatant from each culture was added to infected PBMCs that were then cultured in a 96-well plate at a final concentration of 2 × 105 cells/well. After 7 days of culture, supernatants were harvested, and p24 values were measured by ELISA (NIH AIDS vaccine program, Fredrick, MD).
HIV-seropositive cohort identification
CD8+ T cells from HIV+ patients in early (CD4 count > 500 cells/μL; n = 6), moderate (CD4 count between 200 and 500 cells/μL; n = 5), and advanced (CD4 count < 100 cells/μL; n = 5) stages of HIV disease were isolated by negative immunoselection as described earlier, and their ability to up-regulate CD4 in response to anti-CD3/CD28 or SEB stimulation was monitored by surface expression of CD4dimCD8bright T cells. Criteria for early HIV disease stage consisted of patients that had never experienced a decline in CD4+ cell count lower than 500 cells/μL. Similarly, patients in moderate HIV disease had not experienced a decline in CD4 count less than 200 cells/μL. All patients were seen at the Mark Weiss Infectious Disease Clinic at Rush-Presbyterian-St Luke's Medical Center (Chicago, IL), and research was conducted in accordance with institutional and U.S. government guidelines on human research.
Approval was obtained from Rush-Presbyterian-St Luke's Medical Center institutional review board for these studies. Informed consent was provided according to the Declaration of Helsinki.
Results
CD4dimCD8bright T cells do not appear to originate from the gut
Our previous data demonstrate that CD4dimCD8bright T cells are an activated phenotype of CD8+ T cells, as determined by elevated levels of a number of activation and functional markers (CD95, CD25, CD38, CD69, CD28, and CD45RA+CD45RO+) in comparison to their CD8+CD4– counterparts.1 To evaluate whether the CD8+ T cells that up-regulated CD4 on their surface are peripheral CD8+ T cells that originally resided in the gut, we examined the phenotype of the T-cell receptor (TCR) in CD4dimCD8bright T cells, whether it is αβ or γδ, and the CD8+ molecule, whether it is αα or αβ. γδ TCR and αα CD8 molecules are indicative of gut-derived lymphocytes. Highly purified CD8+ T cells were isolated by negative immunoselection as described previously1 and stimulated for 6 days with 1 μg/mL SEB to induce CD4dimCD8bright expression (Figure 1A). Six days after stimulation, the phenotype of the TCR and CD8 molecules was evaluated by flow cytometric analysis. The TCR was determined to be TCRαβ (Figure 1B) and the CD8 molecule was αβ (Figure 1C), indicating that these cells are not gut derived. CD1a expression, a biomarker for premature thymocytes, was also negative on CD4dimCD8bright T cells (data not shown). Similar results were obtained after CD3/CD28 costimulation to generate the CD4dimCD8bright T-cell phenotype (data not shown).
TCR and CD8 molecule expression on CD4dimCD8bright T cells. CD4dimCD8bright cells were generated by SEB stimulation of highly purified CD8+ T cells from healthy donors. (A) Representative level of CD4dimCD8bright expression, as indicated by the boxed gate in quadrant 2, is shown. (B) SEB-stimulated CD8+ T cells were stained on day 6 for surface expression of TCRαβ. The open histogram represents the isotype control and the filled histogram represents TCRαβ cells. Expression of CD8αβ is shown in panel C. Data are representative of 3 healthy donors.
TCR and CD8 molecule expression on CD4dimCD8bright T cells. CD4dimCD8bright cells were generated by SEB stimulation of highly purified CD8+ T cells from healthy donors. (A) Representative level of CD4dimCD8bright expression, as indicated by the boxed gate in quadrant 2, is shown. (B) SEB-stimulated CD8+ T cells were stained on day 6 for surface expression of TCRαβ. The open histogram represents the isotype control and the filled histogram represents TCRαβ cells. Expression of CD8αβ is shown in panel C. Data are representative of 3 healthy donors.
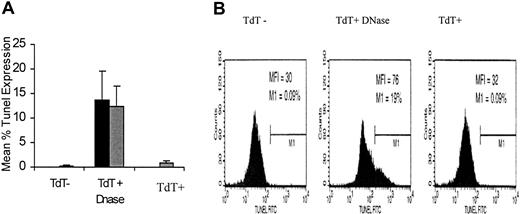
Given that we have previously shown that these cells are more activated than CD8+ T cells that do not up-regulate CD4 on their surface, it was critical to determine whether CD4dimCD8bright T cells were undergoing apoptosis. Apoptosis of CD4dimCD8bright T cells was evaluated using the Tunel assay. Spontaneous apoptosis of CD4dimCD8bright T cells was comparable to CD8+CD4– cells and much less than that of DNase-induced DNA fragmentation in both cultures of CD8+CD4– and CD4dimCD8bright T cells (Figure 2).
Apoptotic profile of CD4dimCD8bright T cells. CD8+ T cells were stimulated with SEB, and the Tunel assay was performed using DNase-treated cultures as a positive control and TdT-negative cultures as a negative control. Percentage of Tunel+ cells was determined for CD4dimCD8bright T cells (▦) and CD8+CD4– cells (▪) exposed to the same stimulation conditions as those generating the CD4dimCD8bright phenotype. Data represent the mean percentage of tunnel expression of 3 healthy donors, and the error bars represent ± SD. (B) Flow cytometric representation of experiment shown in panel A.
Apoptotic profile of CD4dimCD8bright T cells. CD8+ T cells were stimulated with SEB, and the Tunel assay was performed using DNase-treated cultures as a positive control and TdT-negative cultures as a negative control. Percentage of Tunel+ cells was determined for CD4dimCD8bright T cells (▦) and CD8+CD4– cells (▪) exposed to the same stimulation conditions as those generating the CD4dimCD8bright phenotype. Data represent the mean percentage of tunnel expression of 3 healthy donors, and the error bars represent ± SD. (B) Flow cytometric representation of experiment shown in panel A.
CD4dimCD8bright T cells preferentially produce IL-4
The CD4 molecule that is expressed on CD4dimCD8bright T cells is associated with p56lck, a protein tyrosine kinase, suggesting that these cells may be capable of signaling through the CD4 molecule.15 To determine whether CD4dimCD8bright cells differ in their cytokine expression and potentially their effector function, type 1 (IL-2, IFNγ) and type 2 (IL-10, IL-4) intracellular cytokines in CD4dimCD8bright T cells were measured using a flow-based assay after SEB or anti-CD3/CD28 costimulation of purified CD8+ T cells. CD4dimCD8bright T cells expressed elevated levels of intracellular IL-4 after SEB and anti-CD3/CD28 costimulation in comparison to CD8+CD4– T cells, under the same stimulation conditions. Specifically, 87% of CD4dimCD8bright cells after SEB stimulation and 54% after anti-CD3/CD28 costimulation were positive for intracellular IL-4, whereas IL-4 levels in CD8+CD4– cells were at approximately 40% and 25%, respectively (Figure 3A-C). These IL-4 levels detected in CD4dimCD8bright T cells are equivalent to a 2-fold increase in IL-4 expression compared with CD8+CD4– T cells and are also higher than intracellular IL-4 levels detected in stimulated CD4+ T cells (Figure 3). CD4dimCD8bright, CD8+CD4–, and CD4+ T cells were all negative for intracellular IL-10 and IL-2, whereas intracellular INF-γ production in CD4dimCD8bright T cells was minimal (2.7%) (Figure 3). We also examined the level of perforin expression by intracellular perforin staining. Similar levels of intracellular perforin expression were also detected in CD4dimCD8bright T cells than in their CD4– or CD4–CD8+ T cell counterparts (Figure 3D).
CD4dimCD8bright T cells produce elevated levels of intracellular IL-4. (A-B) Type 1 (IL-2, IFNγ) and type 2 (IL-4) intracellular cytokines were evaluated by gating on CD4dimCD8bright (▪) and CD8+CD4– (□) cells after SEB stimulation (A) or anti-CD3/CD28 (B). Data represent the mean percentage of cytokine expression ± SD of 3 healthy donors. (C) Intracellular cytokine staining of CD4dimCD8bright and CD8+CD4– T cells generated after SEB stimulation, as well as SEB-stimulated CD4+ T cells, is shown. (D) CD8+ T cells were stimulated with SEB or anti-CD3/CD28, and intracellular perforin expression was evaluated by flow cytometry at day 6, when the CD4dimCD8bright phenotype is generated. Data represent the mean percentage of expression of perforin ± SD of 3 donors
CD4dimCD8bright T cells produce elevated levels of intracellular IL-4. (A-B) Type 1 (IL-2, IFNγ) and type 2 (IL-4) intracellular cytokines were evaluated by gating on CD4dimCD8bright (▪) and CD8+CD4– (□) cells after SEB stimulation (A) or anti-CD3/CD28 (B). Data represent the mean percentage of cytokine expression ± SD of 3 healthy donors. (C) Intracellular cytokine staining of CD4dimCD8bright and CD8+CD4– T cells generated after SEB stimulation, as well as SEB-stimulated CD4+ T cells, is shown. (D) CD8+ T cells were stimulated with SEB or anti-CD3/CD28, and intracellular perforin expression was evaluated by flow cytometry at day 6, when the CD4dimCD8bright phenotype is generated. Data represent the mean percentage of expression of perforin ± SD of 3 donors
CD4dimCD8bright T cells recognize an antigen-specific target
To determine whether CD4dimCD8bright T cells are capable of recognizing an antigen-specific target, we monitored antigen recognition using CMV tetramer staining technology. CMV tetramer, a complex of 4 HLA-A*0201 class I major histocompatibility complex (MHC) molecules bound to the CMV pp65 peptide and conjugated with PE was used in this study. Four healthy donors that are CMV IgG+ and have the haplotype HLA-A*0201 were identified. CD8+ T cells from these donors were isolated by negative immunoselection. The isolated CD8+ T-cell population was more than 98% CD4– (Figure 4Ai). Purified CD8+ T cells were cultured for 6 days in the presence of CMV pp65 (10 μg/mL), SEB (1 μg/mL), or left untreated. Fresh purified CD8+ T cells were also used in these experiments and denoted by day 0. At day 6, viable T cells (Figure 4Aii) that are CD3+CD8+ T cells were gated on, and this population was further discriminated as CD4dimCD8bright or CD4–CD8+ T cells (Figure 4 Aiii). Subsequently, the percentage of CMV tetramer+ cells within the CD4dimCD8bright or CD4–CD8+ T-cell subpopulations was determined (Figure 4B). At day 0, total CMV+ tetramer recognition was between 1% and 3% (Figure 4C). However, after CMV pp65 priming for 6 days, the median percentage of CD4dimCD8bright T cells recognizing the CMV tetramer increased by 19-fold, as compared with day 0 (fresh) samples (Figure 4B-C). CMV alone, however, did not alter the percentage of expression of these double-positive cells (data not shown). However, SEB, as previously demonstrated, enhanced the total percentage of expression of CD4dimCD8bright T cells without antigen exposure by approximately 40% (Figure 4Aiii). These cells did not demonstrate enhancement in CMV antigen recognition (Figure 4B-C). These striking data indicate that CD4dimCD8bright T cells are capable of antigen-specific recognition to a far greater extent than their single positive (CD4–CD8+) T-cell counterparts. The data also suggest that CD4dimCD8bright T cells may be capable of a potent antigen-specific cytotoxic T lymphocyte (CTL) response.
CD4dimCD8bright cells recognize an antigen-specific target more than CD4–CD8+ T cells.(A) CD8+ T cells were isolated by negative immunoselection (i), and the cells were either untreated or treated with CMV pp65 and SEB for 6 days. Freshly purified CD8+ T cells from the same donors are denoted as day 0. On day 6 after treatment, viable T cells were gated (ii) and further analyzed for CD4–CD8+ or CD4dimCD8bright expression as shown after SEB stimulation (iii). (B) From the CD4–CD8+ or CD4dimCD8bright T-cell population gates shown in panel A, percentages of CMV tetramer-positive cells were determined. Data in panel B are representative of 4 CMV IgG+/HLA-A*0201+ donors, and values are presented as median fold increase from day 0 (fresh) samples. CMV pp65/nonspecific tetramer refers to priming with CMV pp65 but staining with an EBV-tetramer (negative control). Asterisk denotes significant value (P = .03) using the Friedman Q Test. (C) Representative flow diagram from 1 donor is shown.
CD4dimCD8bright cells recognize an antigen-specific target more than CD4–CD8+ T cells.(A) CD8+ T cells were isolated by negative immunoselection (i), and the cells were either untreated or treated with CMV pp65 and SEB for 6 days. Freshly purified CD8+ T cells from the same donors are denoted as day 0. On day 6 after treatment, viable T cells were gated (ii) and further analyzed for CD4–CD8+ or CD4dimCD8bright expression as shown after SEB stimulation (iii). (B) From the CD4–CD8+ or CD4dimCD8bright T-cell population gates shown in panel A, percentages of CMV tetramer-positive cells were determined. Data in panel B are representative of 4 CMV IgG+/HLA-A*0201+ donors, and values are presented as median fold increase from day 0 (fresh) samples. CMV pp65/nonspecific tetramer refers to priming with CMV pp65 but staining with an EBV-tetramer (negative control). Asterisk denotes significant value (P = .03) using the Friedman Q Test. (C) Representative flow diagram from 1 donor is shown.
CD4dimCD8bright T cells express both CXCR4 and CCR5 on their surface
Given that no data exist on the number of CXCR4 and CCR5 chemokine coreceptors per cell on CD4dimCD8bright in comparison to CD8+CD4– and CD4+ T cells, we quantified the expression of CXCR4 and CCR5 on these cells using SEB and anti-CD3/CD28 costimulation of purified CD8+ T cells (to generate CD4dimCD8bright and CD8+CD4– phenotypes) and total PBMCs (to gate on CD4+ T cells). As seen in Figure 5, CCR5 expression on CD4dimCD8bright, CD8+CD4–, and CD4+ T cells was comparable, regardless of SEB or anti-CD3/CD28 mode of stimulation. However, after SEB stimulation, CXCR4 expression on CD4dimCD8bright and CD4+ T cells was lower than on CD8+CD4– cells (Figure 5A), whereas after anti-CD3/CD28 costimulation, CXCR4 expression on CD4dimCD8bright and CD8+CD4– cells was lower than on CD4+ T cells (Figure 5B). CXCR4 levels on CD4dimCD8bright T cells were also equivalent with either SEB or anti-CD3/CD28 costimulation (Figure 5A-B). Specifically, the number of CXCR4 molecules on CD4dimCD8bright T cells following SEB and anti-CD3/CD28 costimulation is 3249 and 2903 molecules/cell, respectively. CD8+CD4– T cells expressed 7726 and 4145 molecules/cell after SEB and anti-CD3/CD28 costimulation, respectively. CD4+ T cells expressed 4903 and 7446 CXCR4 molecules/cell following SEB and anti-CD3/CD28 costimulation, respectively. These data indicate that CD4dimCD8bright T cells express both CCR5 and CXCR4 on their surface at similar levels, regardless of the mode of stimulation.
CXCR4 and CCR5 molecules per cell on CD4dimCD8bright T cells. Quantibright beads were used to quantify the number of CXCR4 and CCR5 molecules on (A) SEB- and (B) anti-CD3/CD28–stimulated CD8+ T cells to generate CD4dimCD8bright (▪) and CD8+CD4– (□) phenotypes. In addition, SEB- and anti-CD3/CD28–stimulated CD4+ T cells from PBMC cultures (▦) were also quantitated for CXCR4 and CCR5 expression. Data represent the mean number of molecules per cell of at least 8 donors ± SD. Asterisk denotes statistical significance (P = .002) between SEB-treated CD8+CD4– and CD4dimCD8bright, as determined by the Wilcoxon signed rank test. (C) A representative histogram of the data pertaining to CD4dimCD8bright T cells is shown, with CXCR4 and CCR5 MI referring to the geo mean fluorescent intensity on CD4+ and CD8+CD4– cells shown in the data below the respective histogram.
CXCR4 and CCR5 molecules per cell on CD4dimCD8bright T cells. Quantibright beads were used to quantify the number of CXCR4 and CCR5 molecules on (A) SEB- and (B) anti-CD3/CD28–stimulated CD8+ T cells to generate CD4dimCD8bright (▪) and CD8+CD4– (□) phenotypes. In addition, SEB- and anti-CD3/CD28–stimulated CD4+ T cells from PBMC cultures (▦) were also quantitated for CXCR4 and CCR5 expression. Data represent the mean number of molecules per cell of at least 8 donors ± SD. Asterisk denotes statistical significance (P = .002) between SEB-treated CD8+CD4– and CD4dimCD8bright, as determined by the Wilcoxon signed rank test. (C) A representative histogram of the data pertaining to CD4dimCD8bright T cells is shown, with CXCR4 and CCR5 MI referring to the geo mean fluorescent intensity on CD4+ and CD8+CD4– cells shown in the data below the respective histogram.
Infection of CD4dimCD8bright T cells in vitro
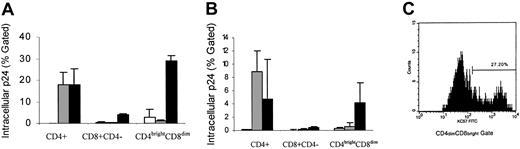
To evaluate the susceptibility of CD4dimCD8bright T cells to T-tropic and M-tropic HIV infection, CD8+ T cells were stimulated for 6 days with either SEB or anti-CD3/CD28. Six days after stimulation, CD8+ T cells were infected with a CCR5-dependent primary isolate (302056) or a laboratory-adapted CXCR4-dependent strain (IIIB) of HIV-1. HIV infection was evaluated by intracellular p24 production using a flow-based assay. Intracellular HIV p24 is indicative of productive infection and was detected in approximately 29% of SEB and 4% of anti-CD3/CD28 induced CD4dimCD8bright T cells infected with IIIB (Figure 6A-C). Although less than 4% HIV IIIB p24 was detected in the CD8+CD4– population, this level of infection may represent infection that originated in CD4dimCD8bright T cells that led to the down-regulation of the CD4 molecule, especially because we observed an equivalent increase in the CD8+CD4– population and a decline in the CD4dimCD8bright T cell population after infection. HIV M-tropic intracellular p24 was not detected in CD4dimCD8bright T cells, with low levels (< 1%) detected in this subpopulation being similar to background levels measured from uninfected cultures (Figure 6).
T-tropic infection of CD4dimCD8bright T cells. (A) SEB- or (B) anti-CD3/CD28–stimulated CD8+ T cells were infected with HIV-1 IIIB (CXCR4 isolate; ▪) or 302056 (CCR5 isolate; ▦). Expression of HIV was monitored by intracellular staining of p24. Positively selected CD4+ T cells were also infected, and intracellular expression of p24 was determined as a positive control. □ indicates uninfected cells. Values represent the mean intracellular p24 expression on respective gated cells from at least 2 donors performed in triplicates ± SD. (C) A representative histogram of p24 intracellular staining.
T-tropic infection of CD4dimCD8bright T cells. (A) SEB- or (B) anti-CD3/CD28–stimulated CD8+ T cells were infected with HIV-1 IIIB (CXCR4 isolate; ▪) or 302056 (CCR5 isolate; ▦). Expression of HIV was monitored by intracellular staining of p24. Positively selected CD4+ T cells were also infected, and intracellular expression of p24 was determined as a positive control. □ indicates uninfected cells. Values represent the mean intracellular p24 expression on respective gated cells from at least 2 donors performed in triplicates ± SD. (C) A representative histogram of p24 intracellular staining.
Inhibition of HIV M-tropic replication in CD4dimCD8bright T cells is mediated by a soluble factor(s)
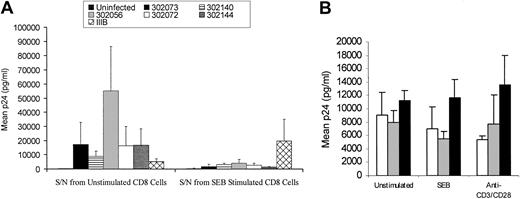
To examine the refractoriness of HIV M-tropic infection in CD4dimCD8bright T cells with our stimulation conditions (SEB and anti-CD3/CD28), we evaluated whether a soluble factor may be mediating the inhibition of HIV M-tropic productive infection. Supernatant from SEB-stimulated CD8+ T cells following 6 days of stimulation was added to PHA-stimulated PBMCs that were infected with a number of primary isolates of HIV (302073, 302140, 302056, 302072, 302144) or IIIB. Supernatants from SEB-stimulated cultures potently inhibited M-tropic viruses but had no affect on T-tropic HIV (Figure 7A). The level of SEB supernatant-mediated M-tropic HIV inhibition was between 3- and 12-fold, depending on the primary isolate used, in comparison to cultures treated with unstimulated supernatant. Similar results were obtained using supernatant from anti-CD3/CD28–costimulated CD8+ T cells (data not shown).
Soluble factor(s) mediates resistance of CD4dimCD8bright T cells to M-tropic HIV infection. (A) Supernatant from SEB-stimulated CD8+ T cells or unstimulated CD8+ T cells was added to primary isolate (302073, 302140, 302056, 302072, 302144) or IIIB-infected PBMC cultures. HIV replication was determined by extracellular production of HIV p24, evaluated by ELISA on day 7 after infection. Values represent mean p24 (picogram per milliliter) production from 4 donors. A comparison of each virus treatment between unstimulated supernatant and SEB-stimulated supernatant demonstrated that all individual p24 values decreased with SEB-stimulated supernatant (Wilcoxon signed rank = 0.068). Although not significant at the .5 level, this is the smallest P value attainable for this sample size. Because the expected behavior of supernatant inhibition by primary virus strains was similar (decreased), all the data were combined and a 2-way repeated measures analysis of variance (ANOVA) on the log counts showed there was a main effect of SEB-stimulated supernatant addition (F1,3 = 25.929, P = .015). (B) Supernatant from SEB-stimulated CD8+ T cells or unstimulated CD8+ T cells was treated with neutralizing antibodies to β-chemokines then added to primary isolate (302073)–infected PBMC cultures. HIV p24 expression was evaluated by ELISA 7 days after infection and treatments. □ indicates no antibody; ▦, control antibody; and ▪, anti–β-chemokine antibody.
Soluble factor(s) mediates resistance of CD4dimCD8bright T cells to M-tropic HIV infection. (A) Supernatant from SEB-stimulated CD8+ T cells or unstimulated CD8+ T cells was added to primary isolate (302073, 302140, 302056, 302072, 302144) or IIIB-infected PBMC cultures. HIV replication was determined by extracellular production of HIV p24, evaluated by ELISA on day 7 after infection. Values represent mean p24 (picogram per milliliter) production from 4 donors. A comparison of each virus treatment between unstimulated supernatant and SEB-stimulated supernatant demonstrated that all individual p24 values decreased with SEB-stimulated supernatant (Wilcoxon signed rank = 0.068). Although not significant at the .5 level, this is the smallest P value attainable for this sample size. Because the expected behavior of supernatant inhibition by primary virus strains was similar (decreased), all the data were combined and a 2-way repeated measures analysis of variance (ANOVA) on the log counts showed there was a main effect of SEB-stimulated supernatant addition (F1,3 = 25.929, P = .015). (B) Supernatant from SEB-stimulated CD8+ T cells or unstimulated CD8+ T cells was treated with neutralizing antibodies to β-chemokines then added to primary isolate (302073)–infected PBMC cultures. HIV p24 expression was evaluated by ELISA 7 days after infection and treatments. □ indicates no antibody; ▦, control antibody; and ▪, anti–β-chemokine antibody.
Given that the supernatant from activated CD8+ T cells inhibited M-tropic viruses but had no effect on T-tropic isolates, we evaluated whether β-chemokines (MIP-1α, MIP-1β, and/or RANTES) were mediating this anti–M-tropic HIV effect. Adding anti–β-chemokine cocktail antibodies directly to the SEB- or anti-CD3/CD28–stimulated CD8+ T-cell cultures was not feasible because the duration of the in vitro generation of the CD4dimCD8bright phenotype is 6 days, and released β-chemokines may be able to down-regulate/block the CCR5 receptor prior to HIV M-tropic infection on day 6. Alternatively, we pretreated the supernatant harvested from SEB-stimulated CD8+ T-cell cultures induced to express the CD4dimCD8bright phenotype with anti–β-chemokine antibodies, and we then added the treated supernatant to PBMCs infected with the primary isolate 302073. As previously noted, supernatants from SEB-stimulated, CD4dimCD8bright-induced cultures inhibited HIV replication of strain 302073 by 6-fold (Figure 7A, from approximately 8000 pg/mL to 1000 pg/mL). Neutralization of β-chemokines from the supernatant reversed this inhibitory effect as demonstrated by an increase in p24 levels after anti–β-chemokine antibody neutralization (Figure 7B). This finding suggests that the inhibition of M-tropic infection seen in CD4dimCD8bright T cells may be due to β-chemokine secretion. CD4dimCD8bright T cells produced RANTES and MIP-1α, as determined by intracellular chemokine detection, similar to levels that were detected in CD8+CD4– cells (data not shown).
Generation of CD4dimCD8bright phenotype is not affected by the stage of HIV disease
To determine whether the ability to induce CD4dimCD8bright T cells was affected by the stage of HIV disease, HIV seropositive individuals from early (CD4 count > 500 cells/μL), moderate (CD4 count 200-500 cells/μL), and advanced (CD4 count < 100 cells/μL) disease were evaluated for their in vitro ability to up-regulate CD4 on CD8+ T cells. CD8+ T cells from HIV-seropositive individuals, independent of the stage of HIV disease, were still able to generate the CD4dimCD8bright phenotype as effectively as those from healthy donors (Figure 8).
Effect of stage of HIV disease on generation of CD4dimCD8bright T-cell phenotype. CD8+ T cells from HIV+ patients in early (n = 5), moderate (n = 5), and advanced (n = 6) stages of HIV disease were stimulated with SEB or anti-CD3/CD28 costimulation, as previously described. Percentage of expression of CD4dimCD8bright was determined by gating on CD3+CD8+ T cells (A). Values represent median percentage of expression of CD4dimCD8bright T cells, and a representative diagram from 1 donor each in early, moderate, and advanced HIV disease stage is shown in panel B.
Effect of stage of HIV disease on generation of CD4dimCD8bright T-cell phenotype. CD8+ T cells from HIV+ patients in early (n = 5), moderate (n = 5), and advanced (n = 6) stages of HIV disease were stimulated with SEB or anti-CD3/CD28 costimulation, as previously described. Percentage of expression of CD4dimCD8bright was determined by gating on CD3+CD8+ T cells (A). Values represent median percentage of expression of CD4dimCD8bright T cells, and a representative diagram from 1 donor each in early, moderate, and advanced HIV disease stage is shown in panel B.
Evidence of CD4dimCD8bright T cells in vivo in HIV-seropositive patients
To determine whether CD4dimCD8bright T cells could be detected in vivo in HIV-seropositive patients, a retrospective study of more than 150 HIV-seropositive individuals was performed to determine the percentage of double-positive T-cell expression. The CD4 and CD8 dual expression in most of the HIV+ patients ranged from 0.4% to 3.4%. However, levels of CD4dimCD8bright T cells more than 5% were detected among 6 HIV+ individuals (Figure 9). The frequency of this elevated in vivo CD4dimCD8bright T cells ranged from 5% to 16% of CD3+ T cells. Patient 1 was retroviral naive, and CD4+CD8+ expression was at 9%, which was maintained through 24 weeks of antiretroviral therapy. In some individuals, not only were CD4dimCD8bright T cells detected but also CD8dim expression on CD4+ T cells (CD4brightCD8dim) was detected. No correlation with viral load, CD4 count, or clinical status could be inferred because of the limited number of patients demonstrating this elevated level of CD4dimCD8bright expression.
Detection of in vivo elevated levels of CD4dimCD8bright T cells in HIV+ patients. Increased CD4dimCD8bright T cells were detected among 6 HIV+ patients (A-F), and the percent of dual CD4 and CD8 expression is indicated in the upper right quadrant. Patient 1 represents an antiretroviral-naive patient who was monitored at baseline, week 12, and week 24 after treatment. The percent expression of CD4dimCD8bright T cells in patient 6 is shown by gating on total CD3+ T lymphocytes or on CD3+CD8+ lymphocytes.
Detection of in vivo elevated levels of CD4dimCD8bright T cells in HIV+ patients. Increased CD4dimCD8bright T cells were detected among 6 HIV+ patients (A-F), and the percent of dual CD4 and CD8 expression is indicated in the upper right quadrant. Patient 1 represents an antiretroviral-naive patient who was monitored at baseline, week 12, and week 24 after treatment. The percent expression of CD4dimCD8bright T cells in patient 6 is shown by gating on total CD3+ T lymphocytes or on CD3+CD8+ lymphocytes.
Discussion
Dual expression of CD4 and CD8 molecules on T cells exists at a frequency of about 3% to 5% in the periphery of healthy adults.16-19 These cells were believed to be prematurely released from the thymus. With the phenotypic evaluation of CD4dimCD8bright T cells, we determined that these cells were negative for CD1a. Given that CD4dimCD8bright cells are generated from highly purified CD8+ T cells, it is unlikely that these cells are prematurely released from the thymus but are rather differentiated cells that are capable of up-regulating CD4 on their surface. We also determined whether the mature peripheral CD8+ T cells that can up-regulate CD4 on their surface are predominately CD8+ T cells that originated from the gut. Intestinal CD8+ T cells are characterized by TCRγδ and CD8αα expression. We showed that these cells were more than 99% CD8αβ and TCRαβ, indicating that the responsive CD8+ T cells to CD4 up-regulation are not originally from the gut but are rather thymically derived. Despite our previous report that CD4dimCD8bright T cells are more activated than their CD8+CD4– counterpart, we show here that these cells are not undergoing apoptosis at a rate different from CD8+CD4– cells.
To understand the possible functional relevance of CD4 expression on CD8+ T cells, we evaluated intracellular type 1 and type 2 cytokine expression in CD4dimCD8bright T cells. We demonstrate that CD4dimCD8bright T cells predominately produce IL-4 at levels that were higher than CD8+CD4– and CD4+ T cells under the same stimulation conditions. IL-4 is reported to activate B cells and to induce IgE and IgG1 class switching.20 Elevated IL-4 production by CD4dimCD8bright T cells may play a role in B-cell immunoglobulin class switching. IL-4 is also a key cytokine in inducing the expression of CD8 on CD4+ T cells, generating the reported CD4brightCD8dim phenotype.21-23 Given that the CD4 molecule on CD8+ T cells is linked to p56lck, it is possible that it may confer helperlike function to the CD4dimCD8bright T cells. Predominately IL-4 production by these cells suggests that the CD4dimCD8bright T cells may have an increased ability to provide type-2 helper function. Recently, CD4dim expression has been shown to be biologically relevant.24 The natural ligand for CD4 is IL-16, a chemotactic factor, and CD4dimCD8bright T cells were shown to be able to migrate in response to IL-16. This chemotactic effect was inhibited by blocking CD4 expression on CD8+ T cells.
To evaluate whether CD4dimCD8bright T cells are capable of antigen-specific recognition, we performed CMV pp65 tetramer staining experiments. We demonstrated that after CMV pp65 priming, it is the CD4dimCD8bright T-cell population and not the CD4–CD8+ T-cell population that is capable of a potent antigen-specific recognition. This CMV-specific recognition was up to 19-fold higher among CD4dimCD8bright T cells than CD4–CD8+ T cells. Although we have demonstrated elevated IL-4 cytokine expression within the CD4dimCD8bright T cells, determining the cytokine profile, especially for IFNγ expression, after CMV pp65 priming among the CMV tetramer+ CD4dimCD8bright T-cell population will be critical in evaluating the functional role of these double-positive cells. Enhanced tetramer recognition, nonetheless, indicates that CD4dimCD8bright T cells can potentially function as cytolytic T cells.
Two models exist describing the lineage differentiation of effector and central memory T cells. The first model is based on a separate lineage of these cells, originating from the naive T-cell population.8 More recently, a progressive linear regression model was proposed whereby naive T cells give rise to effector memory which then gives rise to central memory T cells.7 Our data demonstrating CMV-specific recognition in response to the recall antigen (CMV) among the CD4dimCD8bright cells and not the CD4–CD8+ cells are especially intriguing, considering these models of CD8+ T cell differentiation.
If CD8+ memory T cells are defined by 2 criteria, (1) antigen-independent homeostatic turnover of the population and (2) rapid response to a recall antigen, then CD4dimCD8bright cells fit these criteria and can be classified as memory T cells. Further studies are warranted to verify whether these cells constitute the central memory or effector memory CD8+ T-cell populations and to determine the role of CD4 expression in this lineage progression. These studies will include both characterization of tetramer+ CD4dimCD8bright and CD4–CD8+ T cells at the level of key surrogate markers of central versus effector memory CD8+ T cells (CD62L, CCR7, CD27), cytokine production of tetramer+ cells (IFNγ, tumor necrosis factor α [TNFα], IL-2), and, if experimentally possible, direct assays of cytotoxicity.
Accumulating data point to HIV infection of CD8+ T cells.2,4,25-28 Recently, 2 HIV primary isolates were reported to proficiently replicate in CD8+ T cells, including CD8+ HeLa cells.28 The origin of these isolates may have been in CD4dimCD8bright T cells that were infected and subsequently down-regulated their CD4 surface expression, especially because the cloning process of CD8+ T cells was based on human T-cell leukemia virus (HTLV) cell immortalization, and HTLV infection leads to the transient expression of CD4 on CD8+ T cells.29 Discordant data exist in the literature on HIV infectivity of CD4dimCD8bright T cells. One group reported the susceptibility of CD4dimCD8bright to M-tropic and not T-tropic isolates of HIV,4 others demonstrated that these cells were susceptible to infection by T-tropic isolates.2,3 We demonstrate that CD4dimCD8bright T cells are susceptible to productive T-tropic strain infection but are refractory to M-tropic HIV infection. We believe that the mode of stimulation to generate CD4dimCD8bright may affect the outcome of HIV infection, whether using immobilized CD3/CD28,3 soluble anti-CD3/CD28, SEB,2 or anti-CD3/CD28 cross-linked on CD32- and B7.1-transfected L cells.4 The β-chemokine levels were reported to be affected by the mode of stimulation, resulting in variable HIV infection outcomes.30 Although laboratory-adapted CXCR4 strains can use lower levels of CD4 in comparison to primary CXCR4 stains,31 it is unlikely that differences between the ability of primary and lab-adapted strains to use low CD4 levels is playing a role in the inhibition, given that supernatant from SEB-treated CD8+ T cells inhibited HIV infection of primary isolates in PHA-stimulated PBMCs. This soluble factor is most probably a member of the β-chemokine family. Therefore, the microenvironment plays a key role in modulating HIV infection of CD4dimCD8bright T cells, and the discordant data in the literature may be accounted for by mode of CD8+ T cell stimulation.
Given that we demonstrated the generation of CD4dimCD8bright T cells in response to CD8+ T-cell activation1 and that HIV disease causes hyperactivation of the immune system,32-34 we predicated that we would be able to detect CD4dimCD8bright T cells at a high frequency in HIV infection. However, despite extensive screening of lymphocytes from more than 150 HIV+ patients, we found this population to be elevated in only a few patients (n = 6), corresponding to about 4% of the HIV+ cohort we examined. Given that the duration of this population in vivo is not known and that post-HIV infection CD4 may be down-regulated on the surface of these cells, it may explain our reported low frequency of this population in the periphery of HIV+ patients. Additionally, if CD4dimCD8bright T cells are the central memory T cells, they are expected to be found at higher frequency in the lymph nodes.7 Nonetheless, these few elevated examples are critical to illustrate that these populations do exist in vivo, albeit at a lower frequency. Another report has also confirmed the presence of these CD4dimCD8bright T cells in a small (n = 8) cohort of HIV+ patients.26 By gating on CD71, an activation marker, and CD8+ T cells (CD8+CD71+), Imlach et al26 reported that approximately 57% of CD8+CD71+ cells were also CD4+. The elevated frequency (5%-16%) that we reported of the presence of these cells is based on total T lymphocytes and not on gating on a CD8+ activation subset. On the basis of our previous in vitro observation, CD4dimCD8bright T cells constitute an activated phenotype of CD8+ T cells.1 Therefore, gating on a CD8+ activation subset would enrich for the presence of these double-positive cells. Specifically, on the basis of our previous in vitro studies, by gating on any of the CD8+ activation markers (CD38, CD45RA+CD45RO+, CD69, CD25, and CD95), we observed that most of the cells were also CD4+. Demonstration of in vivo infection of these cells highlights the role that they may play in HIV-mediated dysregulation in the CD8+ T-cell compartment.26
Double-positive (CD4+CD8+) T cells have been reported in a number of clinical conditions such as HTLV-1 infection,29 chronic T-lymphoid leukemia,35 Sjögren syndrome,36 myasthenia gravis,17-19,37 multiple sclerosis,38 idiopathic thrombocytopenic purpura,39 and Behçet syndrome.40 These cells may play a role not only in HIV pathogenesis but also may constitute a normal response to CD8+ T-cell activation and, hence, may also play a role in normal T-cell biology.
Prepublished online as Blood First Edition Paper, June 5, 2003; DOI 10.1182/blood-2002-07-1972.
A.Z. and Y.B.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
This work is submitted in partial fulfillment of the requirements for the degree of Doctor of Philosophy for A.Z. and Y.B.S. at Rush University (Chicago, IL). We thank John Altman (Department of Microbiology and Immunology, Emory University, Atlanta, GA) for providing the CMV and EBV tetramers and for helpful discussions. We thank Allison Truckenbrod, MPAS, PA-C for coordinating patient samples through the Mark Weiss Infectious Disease Clinic at Rush-Presbyterian-St Luke's Medical Center, Chicago, IL. We thank Drs Shande Chen (Department of Preventative Medicine, Rush Medical College, Chicago, IL) and David Wright (Westat, San Luis Obispo, CA) for performing the statistical analyses.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal