Abstract
Translocations and somatic mutations are common genetic alterations of the BCL-6 gene on chromosome 3q27 in B-cell lymphoma, with implications for lymphomagenesis. The 2 events may have linked origins and can influence juxtaposed loci. To evaluate this further, we compared mutations occurring within the major mutation cluster region of the translocated and untranslocated BCL-6 alleles in 7 t(3;14)(q27;14q32) lymphomas. In 6 of 7 cases, the translocated allele revealed significantly higher mutations (mean, 5.8 × 10–2 bp–1) than did the untranslocated allele (mean, 5.3 × 10–3 bp–1; P < .01). The increase mapped to der(14q32), which retains the BCL-6 promoter and is transcriptionally active, as revealed by fusion transcripts and ongoing somatic mutations, absent in the der(3q27) region. These results indicate that enhanced mutational activity at the translocated allele may be a consequence of loss of cis regulatory elements or gain of IgH enhancer elements. Junctional sequences indicate translocation origins from earlier BCL-6 mutations and switch recombinase events.
Introduction
The BCL-6 gene on chromosome 3q27 encodes a potent transcriptional repressor that regulates normal germinal center (GC) formation and function1 and is suggested as an important factor in lymphomagenesis.2
Rearrangements of this locus are common events in diffuse large B-cell lymphoma (DLBCL)3,4 and include chromosomal translocations mapping within the first noncoding BCL-6 exon and the first intron.4,5 A functional consequence is that regulatory and coding sequences on partner chromosomes are juxtaposed, generating a potential for aberrant gene expression on both rearranged partners.6,7 A second type of genetic alteration targeting this locus involves somatic mutations, which map to a region of approximately 2 kilobase (kb) downstream of the initiation site, encompassing the major mutation cluster (MMC).8,9 These can potentially dysregulate BCL-6 expression.10,11
There is some evidence to suggest that the 2 somatic events, that is, somatic mutations and translocations, may be related. A 120–base pair (bp) breakpoint hypercluster region located within the MMC12 displays a high BCL-6 mutational rate in lymphoma, with up to 35% of identifiable somatic mutations occurring in this region.13 Additional mechanisms may be involved. In t(3;14)(q27;q32) lymphomas, translocations usually involve the switch regions (SH) of the IgH locus, suggesting a role for the switch recombinase.12 This region also contains enhancer elements located 3′ of the Cμ gene in IgH that affect somatic mutations in V genes.14
To assess the role of surrounding genetic elements on somatic mutation, we investigated the effects of the t(3;14) translocation on BCL-6 mutations by contrasting the untranslocated and translocated alleles in individual lymphoma cases. Junctional sequences were also analyzed to identify mechanisms likely to play a role in translocation origins.
Study design
Patient material
We analyzed 7 lymphomas: 5 DLBCLs with more than 80% tumor cells and 2 follicular lymphomas (FLs) with more than 60% tumor cells, classified by World Health Organization criteria,15 each bearing the t(3;14) (3q27;14q32) translocation detected by conventional cytogenetic methods, with BCL-6 gene rearrangement confirmed by Southern blot analysis.2
Molecular analysis of BCL-6
To amplify the IgH/BCL-6 junctions, primers were designed according to the molecular anatomy of the t(3;14)(q27;q32) translocation described recently.12 To amplify the untranslocated allele, primers were designed to span the breakpoints (Figure 1). The polymerase chain reactions (PCRs) were performed using established conditions,16 and each product was cloned for sequence analysis. Identical amounts of DNA template ensured a comparable Taq polymerase rate error for both alleles, confirmed by repeated PCR analysis. In 4 of 4 cases with available RNA, fusion transcripts were detected by reverse transcriptase–PCR (RT-PCR) as previously described.7,17
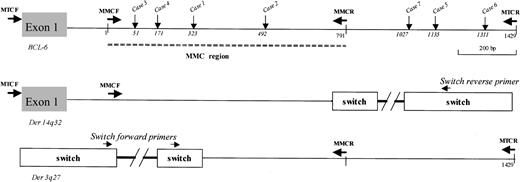
Schematic representation of the first BCL-6 intron and derivative chromosomes after t(3;14) translocation. Breakpoints on der(3q27) are indicated (vertical arrows) for each case and located according to the MMC germ line sequence (GenBank accession no. AF191831). MTC or MMC forward (F) and reverse (R) primers, previously designed,9,16 were used to amplify the junction site and the translocated allele in combination with Switch μ or Switch γ primers (Sμ 01F 5′-ggcaatgagatggctttagctg, Sμ 02F 5′-tgggctgagcaggttgtac, Sμ 01R 5′-tgccatgtccttccagttcatcc, SγF 5′-gggcagaatggtcataat, SγR 5′-atgttctcccctgttccctgag). To exclusively amplify the untranslocated allele, MTC or MMC primers spanning the breakpoints were used under PCR conditions identical to those used for amplification of the BCL6-IgH junctions.
Schematic representation of the first BCL-6 intron and derivative chromosomes after t(3;14) translocation. Breakpoints on der(3q27) are indicated (vertical arrows) for each case and located according to the MMC germ line sequence (GenBank accession no. AF191831). MTC or MMC forward (F) and reverse (R) primers, previously designed,9,16 were used to amplify the junction site and the translocated allele in combination with Switch μ or Switch γ primers (Sμ 01F 5′-ggcaatgagatggctttagctg, Sμ 02F 5′-tgggctgagcaggttgtac, Sμ 01R 5′-tgccatgtccttccagttcatcc, SγF 5′-gggcagaatggtcataat, SγR 5′-atgttctcccctgttccctgag). To exclusively amplify the untranslocated allele, MTC or MMC primers spanning the breakpoints were used under PCR conditions identical to those used for amplification of the BCL6-IgH junctions.
Statistical analysis
The frequency of mutations occurring between the translocated and the untranslocated allele were compared using the χ2 test or the Fisher exact test.
Results and discussion
In 7 of 7 cases, both the translocated and untranslocated alleles were amplified. Of the 7 t(3;14) translocations, reciprocal breakpoint regions were identified in 6 cases (Figures 1, 2), with der(14) not being found in 1 case (Table 1).
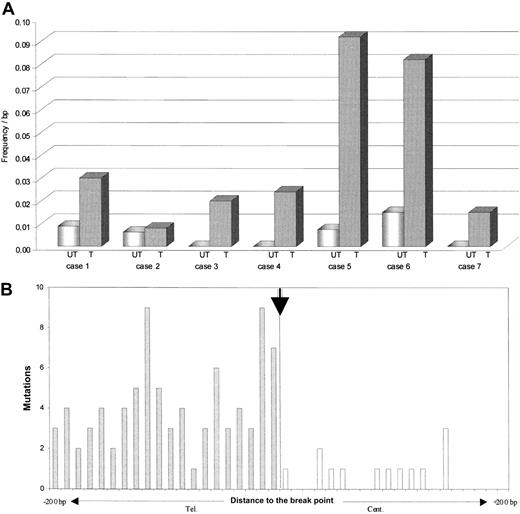
Distribution of the BCL-6 somatic mutations in both alleles of t(3;14) lymphoma. (A) Frequency of mutations in the BCL-6 major mutation cluster region observed within the translocated (T) and the untranslocated (UT) allele. (B) Mutations in BCL-6 first intron relative to junction sites in 6 lymphomas with t(3;14) translocation. The figure is a composite and shows mutations in BCL-6 in der(14q32) (shaded bars) and in der(3q27) (open bars). Mutational frequency in the BCL-6 locus permitted a direct comparison of rates for all derivative chromosomes.
Distribution of the BCL-6 somatic mutations in both alleles of t(3;14) lymphoma. (A) Frequency of mutations in the BCL-6 major mutation cluster region observed within the translocated (T) and the untranslocated (UT) allele. (B) Mutations in BCL-6 first intron relative to junction sites in 6 lymphomas with t(3;14) translocation. The figure is a composite and shows mutations in BCL-6 in der(14q32) (shaded bars) and in der(3q27) (open bars). Mutational frequency in the BCL-6 locus permitted a direct comparison of rates for all derivative chromosomes.
Clonal mutations observed in the major mutation cluster region of the first BCL-6 intron in 7 cases of lymphoma with (3;14) translocation
Case no. and diagnosis . | Mutations located within the MMC or 3′ of the MMC . | Intraclonal heterogeneity . | No. of sequenced clones . |
|---|---|---|---|
| 1. DLBCL | |||
| Untranslocated allele | 119C>G; 123C>T; 265G>A; 378G>T; 423C>T; 519C>T; 709A>G | Yes | 6 |
| Translocated allele | |||
| der(3q27) | 345G>C; 347G>A; 349-350insG; 350T>G; 359A>G; 365G>C; 423C>G; 429C>G; 441G>A; 443G>A; 478T>G; 685C>T; 697G>A; 737delT; 741T>A; 780G>A | No | 9 |
| der(14q32) | Unknown* | ||
| 2. Grade 3 FL | Yes | 9 | |
| Untranslocated allele | 18T>C; 95T>A; 542T>C; 548C>T; 782A>T | ||
| Translocated allele | |||
| der(3q27) | No clonal mutation | No | 6 |
| der(14q32) | 18T>C; 375G>A; 416G>A; 423C>G | Yes | 7 |
| 3. DLBCL | |||
| Untranslocated allele | No clonal mutation | No | 18† |
| Translocated allele | |||
| der(3q27) | 51C>G; 87T>A; 90C>G; 100A>G; 107A>G; 136A>G; 142A>G; 170C>T; 196A>T; 252C>T; (387-393)del | No | 9 |
| der(14q32) | 6G>A; 7C>A; 30T>C; 36T>C; 38T>A | Yes | 6 |
| 4. DLBCL | |||
| Untranslocated allele | No clonal mutation | No | 18† |
| Translocated allele | |||
| der(3q27) | 520delT | No | 6 |
| der(14q32) | 8T>G; 16A>T; 23T>C; 44T>G; 45C>A; 71A>C; 76T>C; 88C>A; 89T>A; 97T>A; 105T>G; 107T>A; 110T>G; 123C>G; 143T>C; 145C>T; 149A>C; 151-152insT, 153T>G | No | 6 |
| 5. DLBCL | |||
| Untranslocated allele | 202A>T; 226A>G; 267T>G; 269T>A; 344A>T; 434G>T | No | 4 |
| Translocated allele | |||
| der(3q27)‡ | 1249G>C; 1260G>C; 1284C>T; 1285C>T | No | 4 |
| der(14q32) | 23T>C; 24A>G; 32A>G; 43T>C; 44T>G; 47G>C; 57A>G; 71A>G; 73G>C; 75T>A; 84G>C; 87T>A; 90A>G; 100A>G; 104A>C; 108A>C; 116A>G; 121A>C; 122G>C; 140A>G; 141A>G; 176delA; 178G>T; 181delA; 183delA; 184G>T; 188G>A; 209T>G; 211G>C; 222G>A; 234G>C; 257T>G; 298A>C; 309G>A; 312T>A; 328G>A; 335G>C; 347G>C; 349T>G; 350T>G; 356G>C; 360T>C; 377G>A; 390A>C; 393C>G; 394C>T; 402G>A; 421C>G; 431G>A; (433-442)del; 459C>G; 466T>G; 477G>A; 500T>G; 501T>C; 505A>C; 509G>A; 510C>G; 519C>T; 526G>A; 608G>A; 620G>C 623G>A; 632T>G; 659C>G; 692C>G; 694G>A; 732T>C; 734delG; 738T>C; 742G>A; 769G>A; 775A>G; 779A>G; 782A>T | No | 9 |
| 6. DLBCL | |||
| Untranslocated allele | 46C>G; 84G>A; 142A>G; 193G>A; 211G>A; 502G>A; 520delT; 573T>C; 599T>C; 677T>A; 717A>G; 737T>G; 750T>G; 752T>A | Yes | 8 |
| Translocated allele | |||
| der(3q27)‡ | 1314G>C | No | 5 |
| der(14q32) | 25C>T; 26C>T; 84G>A; 100A>G; 106A>G; 108A>C; 119C>T; 121A>C; 122G>T; 139T>C; 173C>G; 191A>T; 195A>C; 197T>C; 200T>C; 217T>C; 261T>C; 275T>A; 279T>C; 328G>C; 329C>T; 334A>C; 337A>G; 345G>A; 350-351linsG; 400G>C; 423C>G; 424T>C; 425C>G; 445A>G; 478T>A; 479G>A; 486C>T; 487C>A; 488C>T; 499C>G; 503C>G; 507C>G; 508T>A; 509G>C; 512T>A; 524T>G; 541T>G; 545C>G; 555T>A; 561C>T; 574C>A; 579C>T; 614A>G; 617A>G; 650C>G; 671T>G; 677T>A; 679C>G; 697C>T; 718T>G; 720C>G; 727A>G; 728C>T; 735G>A; 741T>C; 761 61T>A; 763A>C; 766C>G | No | 15 |
| 7. Grade 1 FL | |||
| Untranslocated allele | No clonal mutation | No | 13† |
| Translocated allele | |||
| der(3q27) | No clonal mutation | No | 4 |
| der(14q32) | 73G>C; 177G>C; 339G>C; 397G>C; 441G>A; 445A>C; 475C>T; 504A>T; 549C>T; 613A>T; 633C>T; 735G>A; 736G>A | Yes | 4 |
Case no. and diagnosis . | Mutations located within the MMC or 3′ of the MMC . | Intraclonal heterogeneity . | No. of sequenced clones . |
|---|---|---|---|
| 1. DLBCL | |||
| Untranslocated allele | 119C>G; 123C>T; 265G>A; 378G>T; 423C>T; 519C>T; 709A>G | Yes | 6 |
| Translocated allele | |||
| der(3q27) | 345G>C; 347G>A; 349-350insG; 350T>G; 359A>G; 365G>C; 423C>G; 429C>G; 441G>A; 443G>A; 478T>G; 685C>T; 697G>A; 737delT; 741T>A; 780G>A | No | 9 |
| der(14q32) | Unknown* | ||
| 2. Grade 3 FL | Yes | 9 | |
| Untranslocated allele | 18T>C; 95T>A; 542T>C; 548C>T; 782A>T | ||
| Translocated allele | |||
| der(3q27) | No clonal mutation | No | 6 |
| der(14q32) | 18T>C; 375G>A; 416G>A; 423C>G | Yes | 7 |
| 3. DLBCL | |||
| Untranslocated allele | No clonal mutation | No | 18† |
| Translocated allele | |||
| der(3q27) | 51C>G; 87T>A; 90C>G; 100A>G; 107A>G; 136A>G; 142A>G; 170C>T; 196A>T; 252C>T; (387-393)del | No | 9 |
| der(14q32) | 6G>A; 7C>A; 30T>C; 36T>C; 38T>A | Yes | 6 |
| 4. DLBCL | |||
| Untranslocated allele | No clonal mutation | No | 18† |
| Translocated allele | |||
| der(3q27) | 520delT | No | 6 |
| der(14q32) | 8T>G; 16A>T; 23T>C; 44T>G; 45C>A; 71A>C; 76T>C; 88C>A; 89T>A; 97T>A; 105T>G; 107T>A; 110T>G; 123C>G; 143T>C; 145C>T; 149A>C; 151-152insT, 153T>G | No | 6 |
| 5. DLBCL | |||
| Untranslocated allele | 202A>T; 226A>G; 267T>G; 269T>A; 344A>T; 434G>T | No | 4 |
| Translocated allele | |||
| der(3q27)‡ | 1249G>C; 1260G>C; 1284C>T; 1285C>T | No | 4 |
| der(14q32) | 23T>C; 24A>G; 32A>G; 43T>C; 44T>G; 47G>C; 57A>G; 71A>G; 73G>C; 75T>A; 84G>C; 87T>A; 90A>G; 100A>G; 104A>C; 108A>C; 116A>G; 121A>C; 122G>C; 140A>G; 141A>G; 176delA; 178G>T; 181delA; 183delA; 184G>T; 188G>A; 209T>G; 211G>C; 222G>A; 234G>C; 257T>G; 298A>C; 309G>A; 312T>A; 328G>A; 335G>C; 347G>C; 349T>G; 350T>G; 356G>C; 360T>C; 377G>A; 390A>C; 393C>G; 394C>T; 402G>A; 421C>G; 431G>A; (433-442)del; 459C>G; 466T>G; 477G>A; 500T>G; 501T>C; 505A>C; 509G>A; 510C>G; 519C>T; 526G>A; 608G>A; 620G>C 623G>A; 632T>G; 659C>G; 692C>G; 694G>A; 732T>C; 734delG; 738T>C; 742G>A; 769G>A; 775A>G; 779A>G; 782A>T | No | 9 |
| 6. DLBCL | |||
| Untranslocated allele | 46C>G; 84G>A; 142A>G; 193G>A; 211G>A; 502G>A; 520delT; 573T>C; 599T>C; 677T>A; 717A>G; 737T>G; 750T>G; 752T>A | Yes | 8 |
| Translocated allele | |||
| der(3q27)‡ | 1314G>C | No | 5 |
| der(14q32) | 25C>T; 26C>T; 84G>A; 100A>G; 106A>G; 108A>C; 119C>T; 121A>C; 122G>T; 139T>C; 173C>G; 191A>T; 195A>C; 197T>C; 200T>C; 217T>C; 261T>C; 275T>A; 279T>C; 328G>C; 329C>T; 334A>C; 337A>G; 345G>A; 350-351linsG; 400G>C; 423C>G; 424T>C; 425C>G; 445A>G; 478T>A; 479G>A; 486C>T; 487C>A; 488C>T; 499C>G; 503C>G; 507C>G; 508T>A; 509G>C; 512T>A; 524T>G; 541T>G; 545C>G; 555T>A; 561C>T; 574C>A; 579C>T; 614A>G; 617A>G; 650C>G; 671T>G; 677T>A; 679C>G; 697C>T; 718T>G; 720C>G; 727A>G; 728C>T; 735G>A; 741T>C; 761 61T>A; 763A>C; 766C>G | No | 15 |
| 7. Grade 1 FL | |||
| Untranslocated allele | No clonal mutation | No | 13† |
| Translocated allele | |||
| der(3q27) | No clonal mutation | No | 4 |
| der(14q32) | 73G>C; 177G>C; 339G>C; 397G>C; 441G>A; 445A>C; 475C>T; 504A>T; 549C>T; 613A>T; 633C>T; 735G>A; 736G>A | Yes | 4 |
To analyze the MMC, sequences were aligned with the reported BCL-6 germ line gene9 (GenBank accession nos. AF 191831 and Z79581). The first nucleotide of the amplified BCL-6 first intron corresponds to the first nucleotide of the MMCF primer, defining position +1. In each case, multiple clones were obtained from at least 2 independent PCRs using different BCL-6 primers. Clonal mutations are observed in at least 2 independent clones. Taq error rate (1 × 10-3 bp-1) was also calculated by amplification and cloning of BCL-6 using an identical amount of monocyte DNA template not targeted for somatic mutation. MMC indicates major mutation cluster region; DLBCL, diffuse large B-cell lymphoma; FL, follicular lymphoma. Polymorphisms are shown in boldface type; biallelic mutations are underlined. del indicates deletion.
In case 1, no PCR product was obtained using different BCL-6 forward primers and switch reverse primers.
In absence of clonal mutations, 1 or 2 mutations per clone may have resulted from Taq error or may represent unrelated cells.
In these cases, mutations were located 3′ of the MMC.
BCL-6 mutations were analyzed in each region. Polymorphic mutations (397G>C, 502G>A, or 520delT) allowed intra-allelic discrimination. Clonal mutations were observed. Mutations included point mutations and short additions and deletions (5 of 7 cases), biallelic mutations, and identical, recurrent mutations between cases (Table 1).
Notably, a significant imbalance in the BCL-6 mutational load was observed between the translocated (mean, 5.8 × 10–2 bp–1) and the untranslocated (mean, 1.5 × 10–3 bp–1) allele in 6 of 7 cases (P < .01; Table 1 and Figure 2A). Mutations in BCL-6 can occur in the absence or presence of chromosomal rearrangements,8 but the influence of such events on mutational activity has not been delineated. These data show that in BCL-6, a dramatic increase in mutational rate can occur in t(3;14) lymphoma, which is preferentially targeted to the translocated allele. The data demonstrate clear differences in the level of BCL-6 mutations between the translocated and untranslocated alleles. This suggests that mechanistic constraints that regulate the level of mutational activity at the BCL-6 locus can be overcome in this case by involving IgH regulator elements. Whether this mutational activity results in higher levels of BCL-6 mutations as compared with lymphoma cases without the t(3;14) translocation lies outside the scope of this study, and large numbers of cases will clearly be required to reach statistical validity, for example, in correlating prognostic significance of BCL-6 mutations in lymphoma.18
By comparing BCL-6 mutations in both the derivative arms of the translocated allele, we obtained further insights into preferential targeting at this allele. Mutational activity in the translocated BCL-6 allele was notably in excess in der(14q32) compared with der(3q27), with 74% of mutations associating with der(14q32) (Table 1 and Figure 2B). The BCL-6 promoter is clearly transcriptionally active on der(14q32), as revealed by the presence of BCL-6–IgH fusion transcripts (2 of 4 cases; data not shown), and it is clear that transcription is a necessary prerequisite for somatic mutation.19 This suggests that enhancer elements downstream of the Cμ gene in IgH or loss of putative silencer elements in 3q27 affect the BCL-6 promoter and result in the high mutation rate observed in der(14q32). It reveals potential synergistic interaction between regulatory factors that control hypermutation at 2 different loci, BCL-6 and V genes.
Furthermore, somatic mutations in BCL-6 in der(14q32) were apparently ongoing, as revealed by intraclonal heterogeneity at this site, but this was not true in der(3q27) in the same cases (Table 1 and Figure 2B). This argues for the acquisition of mutations after the chromosomal translocation event. Continuing mutational activity was also observed in the untranslocated allele, indicating biallelic targeting. The relationship between mutations in BCL-6 and V genes is not yet clear, but ongoing mutations are a feature of V genes in DLBCL or FL.20 Translocation of der(14q32) to other mutationally susceptible loci can potentially have an impact on mutations in partner genes, as in t(8;14) Burkitt lymphoma21 or in t(14;18) FL.22
The translocated breakpoints were identified. The MMC was directly involved at the junction site in 4 of 7 cases (cases 1-4), with 3 of 7 cases mapping downstream of MMC (Figure 1). Translocation mapped to Sμ in 6 cases and in 1 case was located in Sγ (Figure 3). In cases translocated to Sμ, a GAGCT or GGGCT pentameric repeat motif was identified at the breakpoint. Deletions in Sμ were identified in 6 of 6 cases, consistent with known features of isotype switch.23 BCL-6 sequences at the der(3q27) and der(14q32) junctional regions showed short deletions or duplications (cases 2, 4, 5, and 6) or precise double-stranded DNA breaks (Figure 3). Overall, in 5 of 7 cases the breakpoint was within or in close proximity to RGYW/WRCY motifs in BCL-6 (Figure 3).
Nucleotide sequence analysis of the breakpoint junctions of 6 lymphomas with t(3;14) translocation. Sequence identities with the BCL-6 germ line gene (GenBank accession nos. AF191831 and Z79581) are indicated by the vertical lines; asterisks indicate location of mutations. The SH–BCL-6 junctions were analyzed by alignment to the closest switch (SH) sequence (GenBank accession nos. X56795, D78345, AL122127). RGYW and WRCY motifs are boxed; pentameric repeat motifs of the Sμ region are underlined. In case 3, partial homology with the BCL-6 germ line and the absence of homology with the Switch μ germ line sequence did not allow an exact location of the breakpoint. These sequences have been submitted to the GenBank database (GenBank accession nos. AJ557805 to AJ557817).
Nucleotide sequence analysis of the breakpoint junctions of 6 lymphomas with t(3;14) translocation. Sequence identities with the BCL-6 germ line gene (GenBank accession nos. AF191831 and Z79581) are indicated by the vertical lines; asterisks indicate location of mutations. The SH–BCL-6 junctions were analyzed by alignment to the closest switch (SH) sequence (GenBank accession nos. X56795, D78345, AL122127). RGYW and WRCY motifs are boxed; pentameric repeat motifs of the Sμ region are underlined. In case 3, partial homology with the BCL-6 germ line and the absence of homology with the Switch μ germ line sequence did not allow an exact location of the breakpoint. These sequences have been submitted to the GenBank database (GenBank accession nos. AJ557805 to AJ557817).
Although switch events are involved in the translocated allele, we failed to find any homology with switch recognition motifs at the BCL-6 junction site on 3q27. This suggests involvement of additional proteins in addition to the switch recombinase. BCL-6 sequences at the junctional regions showed short deletions or duplications or precise double-stranded DNA breaks. Short-template insertions, sometimes with single substitutions, suggest a short-patch error-prone DNA synthesis. These features are consistent with a role for the hypermutation mechanism. A common enzyme, activation-induced cytidine deaminase, is required for V gene somatic mutation as well as switch recombination.24 Whether this key enzyme is implicated here in t(3;14) lymphoma is as yet not known. Our data show that at least 2 separate mechanisms, somatic mutation and isotype switch, underlie this frequent and important translocation in lymphoma.
Prepublished online as Blood First Edition Paper, May 29, 2003; DOI 10.1182/blood-2002-12-3630.
Supported by the Leukaemia Research Fund (United Kingdom) and the Multiple Myeloma Research Foundation (United States). F.J. was the recipient of a grant from La Fondation De France.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal