Abstract
The development of dendritic cells (DCs) from hematopoietic progenitors is not well understood. Using a spleen-derived long-term culture (LTC) system, it has been possible to continuously generate DCs from progenitors maintained in culture. The nonadherent LTC-DC population is composed of 2 major subsets. These are the small LTC-DC or DC precursors and their progeny, the large LTC-DCs that phenotypically resemble immature DCs. In this study, subtracted cDNA libraries were generated containing sequences differentially expressed in small or large LTC-DCs. Differential screening was then used on plated library clones to select genes expressed in either the small or the large cell population. Real-time polymerase chain reaction (PCR) has been used to verify the selection procedure for several genes of particular interest. Known genes isolated from subtracted libraries were related to stages in DC development and supported previous findings regarding the function of small and large LTC-DCs. Large LTC-DCs expressed a number of immunologically important genes encoding CD86, CCR1, osteopontin, and lysozyme. Small LTC-DCs resembled progenitor DCs expressing genes related to the organization of the cytoskeleton, the regulation of antigen processing, and a number of mitochondrial and ribosomal proteins. Novel transcripts were isolated from small and large LTC-DC–subtracted libraries that could encode novel proteins important in DC development. This study describes changes in gene expression related to the development of CD11c+CD11b+ major histocompatibility complex 2 low (MHC2lo) CD8α– DCs from precursors in a stroma-dependent culture system in the absence of exogenous cytokines.
Introduction
Dendritic cells (DCs) are potent antigen-presenting cells responsible for the activation of naive T cells and the generation of a primary immune response.1,2 They are also able to interact with a number of other immune cells, including B cells,3 and they play an important role in immune tolerance.4 Progeny DCs have been derived from a number of different progenitor cells, including hematopoietic stem cells and lineage-restricted progenitors in the bone marrow,5 peripheral blood monocytes,6 and thymic precursors.7 However, the exact pathway of DC development from progenitors is not well characterized. Immature DCs are located in the periphery, and they function in the uptake and processing of antigen.8 They express little major histocompatibility complex 2 (MHC 2) on their surfaces, but abundant intracellular MHC 2 is present within specialized MHC 2-rich endocytic compartments as part of an efficient antigen-processing system.9 Inflammatory signals stimulate DC migration and trigger irreversible phenotypic and functional changes. Activated DCs lose attributes associated with antigen capture and processing, and they function instead to present antigen to T cells in the secondary lymphoid organs.8 They up-regulate MHC 2 peptide complexes, along with MHC 1 and costimulatory molecules, to form an immunologic synapse for activating naive T cells.10 The study of DC function and development has been further complicated by the existence of a number of DC subsets. Murine spleen DCs can be divided into at least 3 functionally distinct subsets on the basis of CD4 and CD8α expression.11 The most recent evidence suggests that they do not represent DCs of separate lineage5 but that they may reflect different stages of maturation or may regulate different arms of the immune response.12,13
During this study we generated DCs in a spleen-derived long-term culture (LTC) system in which hematopoiesis is supported by a stromal layer in the absence of exogenous cytokines.14 Two distinct cell subsets are present within the nonadherent cell population of LTCs. Small LTC-DCs represent committed DC precursors. Their low expression of DC markers and their weak functional capacity identify them as at an earlier stage of development than large LTC-DCs.15 These factors preclude, however, that small LTC-DCs are early hematopoietic progenitor cells, though they are heterogeneous and could contain progenitors representing various stages in development. Small LTC-DCs have the capacity to proliferate, grow, develop dendrites, and up-regulate DC markers to resemble large LTC-DCs when sorted and cultured for 12 to 20 days on irradiated stroma.15 However, these DC precursors are not capable of long-term self-renewal and are strictly dependent on stromal cell contact for development.16 Large LTC-DCs represent a homogeneous population of immature DCs because they are highly endocytic, express low levels of surface MHC 2, and lack CD40 expression. However, they do express the costimulatory molecules CD80 and CD86.15 LTC represents an in vitro model highly suitable for the study of DC development from committed precursors. In cytokine-supported colony assays, LTCs generate DCs and no other cell type.17
Gene expression analysis offers the opportunity to identify genes that determine DC lineage and function and to study the changes that underlie their development from progenitors. Studies reported so far have examined gene expression changes in DCs generated from monocytes by cytokine exposure, or they have compared DC gene expression before and after activation with stimuli such as lipopolysaccharide.18-21 Furthermore, the identification of novel DC-associated genes can lead to the characterization of novel proteins that contribute to the unique function of DCs.22 To date there is no reported study of gene expression related to DC development from precursors in the absence of cytokines or other forms of activation. In the current study, 2 subtracted cDNA libraries were generated to identify genes specifically expressed in small LTC-DCs and not large LTC-DCs (small subtracted cDNA) and in large LTC-DCs but not small LTC-DCs (large subtracted cDNA). It was hypothesized that a comparison of the gene expression of small and large LTC-DCs would identify genes related to DC development and function. The LTC system represents an ideal system for such a study because gene expression is measured in distinct cell populations maintained within the same culture, eliminating background gene expression related to common cell functions. The purpose of this study was to verify the LTC system as a producer of precursor and progeny myeloid-like DCs by generating a small but highly specific gene expression profile. The identification of new genes was also anticipated.
Materials and methods
Establishment and maintenance of LTCs
B10.A(2R) strain mice were bred at the John Curtin School of Medical Research (Canberra, Australia) under specific pathogen-free conditions. LTCs were established from whole spleen of 6- to 8-week-old female mice and were maintained in supplemented Dulbecco modified Eagle medium (sDMEM) as described previously.23 Nonadherent LTC-DCs were collected from culture flasks without disturbance to the stroma and were filtered through a 200-gauge nylon filter to remove cell aggregates.
Analysis of cell surface marker expression
The indirect 2-color antibody staining protocol used has been described previously.15 Primary antibodies used for fluorescence-associated cell sorter (FACS) analysis included affinity-purified monoclonal antibodies specific for CD8α (53-6.7; phycoerythrin (PE)–conjugated rat immunoglobulin [G2a] IgG2a), CD11c (HL3; biotinylated hamster IgG), CD11b (M1/70; biotinylated rat IgG2b), CD86 (GL1; biotinylated rat IgG2a), and MHC 2 (AF6-120.1; fluorescein isothiocyanate (FITC)–conjugated mouse IgG2a), all purchased from PharMingen (San Diego, CA), and CD205 (NLDC-145; rat IgG2a), purchased from Serotec (Oxford, United Kingdom). Culture supernatant collected from hybridoma cells was used as a source of antibody specific for the DC marker 33D1 (33D1; rat IgG2b) and the macrophage marker F4/80 (F4/80; rat IgG2b). Fluorescent conjugates included PE-conjugated avidin from PharMingen, FITC-conjugated antirat immunoglobulin (goat F(ab′)2) from Southern Biotechnology (Birmingham, AL), and FITC-conjugated antihamster immunoglobulin (goat IgG) from Kierkegaard and Perry Laboratories (Gaithersburg, MD). Quadrants based on background staining attributed to secondary conjugates only were used to distinguish positively and negatively staining cells.
Sorting LTC-DCs into small and large cell subsets
After collection from LTCs, nonadherent cells were resuspended in Hanks balanced salt solution (HBSS)/5% fetal calf serum (FCS). Cells were sorted into small and large LTC-DC subsets using a FACStar Plus cell sorter (Becton Dickinson, Franklin Lakes, NJ). These 2 subsets were gated on the basis of forward scatter and side scatter. Sorted cells were deposited into sDMEM/25% FCS to maintain viability. Cell viability was checked by trypan blue staining. Cells were transferred to cryotubes and were washed in ice-cold PBS. Cell pellets were stored in liquid nitrogen until required.
Preparation of cDNA for subtraction
Total RNA was isolated from cells using the TRIzol Reagent (Gibco BRL, Grand Island, NY) according to the manufacturer's instructions. The quality and yield of total RNA were assessed using agarose gel electrophoresis and ultraviolet (UV) spectrophotometry. cDNA was synthesized and amplified using the Smart PCR cDNA Synthesis Kit (Clontech Laboratories, Palo Alto, CA) following the manufacturer's instructions. Amplified cDNA was then prepared for subtraction using both the SMART PCR cDNA Synthesis Kit and the PCR-Select cDNA Subtraction Kit (Clontech Laboratories) following the manufacturer's instructions. Briefly, cDNA was digested with RsaI and purified. Different adaptors were ligated to the ends of separate populations of purified target cDNA.
Generation of subtracted cDNA libraries
Subtractive hybridization and suppressive polymerase chain reaction (PCR) were performed using the PCR-Select cDNA Subtraction Kit according to the manufacturer's instructions. Briefly, first and second hybridizations were carried out, and the resultant hybridization products were used as the PCR template in 2 rounds of suppressive PCR using primers supplied by the manufacturer. This process is described in detail by Diatchenko et al.24 Subtracted PCR products were visualized by agarose gel electrophoresis. Subtraction efficiency was monitored by PCR to compare the abundance of the housekeeping gene GAPDH in cDNA samples before and after subtraction using primers supplied by the manufacturer (Clontech Laboratories). Aliquots were removed from the PCR reaction at 18, 23, 28, and 33 cycles and were analyzed using agarose gel electrophoresis.
Subtracted PCR products were cloned using the CloneAmp pAMP10 System (Life Technologies, Gibco BRL, Rockville, MD) following the manufacturer's instructions. In brief, first-round suppressive PCR products were reamplified using primers identical to those used during suppressive PCR except that they possessed deoxy-UMP residues at their 5′ end (5′-CUACUACUACUATCGAGCGGCCGCCCGGGCAGGT-3′ and 5′-CUACUACUACUAAGCGTGGTCGCGGCCGAGGT-3′). These PCR products were purified (Nucleospin Extraction Kit; Clontech Laboratories) and cloned into the pAMP10 vector, and libraries of potentially subtracted cDNAs were established. Approximately 500 subtracted clones were randomly selected from libraries for further analysis.
Differential screening of subtracted cDNA clones
A screening step was performed to identify unique cDNA clones and to detect cDNA clones common to small and large LTC-DC populations. Differential screening of clones was performed using the PCR-Select Differential Screening Kit (Clontech Laboratories) according to the manufacturer's instructions. In brief, alkaline-denatured PCR-amplified clone inserts were dotted onto Hybond N+ membranes (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom) and were hybridized separately with 4 different 32P-labeled probes prepared from small LTC-DC subtracted cDNA, small LTC-DC cDNA (not subtracted), large LTC-DC subtracted cDNA, or large LTC-DC cDNA (not subtracted). Membranes were washed and exposed to autoradiography film (Kodak, Rochester, NY). Exposed films were examined visually and by densitometry.
Analysis of subtracted cDNA clones
The nucleotide sequence of selected differentially expressed clones was determined using the Big Dye Terminator Reaction Mix (Applied Biosystems, Foster City, CA) and automated DNA sequencing (model 377; Applied Biosystems). Analysis was carried out using Applied Biosystems Sequencing Software (version 3.0). Sequence results were submitted to BLAST searches of various online databases to elucidate the identity of clones. These included the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) nr (nonredundant GenBank, EMBL, DDBJ, and PDB), EST (nonredundant GenBank, EMBL, and DDBJ EST divisions), and conserved domain (Pfam and Smart) databases and the Ensembl (http://www.ensembl.org) mouse genome server.
Known mouse genes identical to subtracted clones were the first choice for annotation. For subtracted clones with nonidentical similarity to known genes, prefix terms including similar to and homologue to were used to indicate that the sequence description was derived from nonidentical mouse and nonmouse mammal, respectively. Subtracted sequences with no significant similarity to known sequences were classified as unknown.
Real-time PCR
Total RNA (300 ng) was treated with DNase I (Promega, Madison, WI), and SuperScript II (Invitrogen, Carlsbad, CA) and oligo(dT) were used for reverse transcription according to the manufacturers' instructions. Real-time PCR reactions were performed in a 25-μL volume containing diluted cDNA, Sybr Green PCR Master Mix (Applied Biosystems), and 2.5 μM each gene-specific primer. An ABI SDS7700 analyzer (Applied Biosystems) was used at 50°C for 2 minutes and 95°C for 10 minutes, followed by 40 cycles at 95°C for 15 seconds and 60°C for 1 minute. Cycle threshold (Ct) values were exported onto Excel worksheets for analysis. Test cDNA results were normalized to GAPDH measured on the same plate. After cycling, the specificity of amplification was validated by the generation of a melting curve through slow denaturation of the PCR products and then by gel electrophoresis. Fold differences in gene expression between small and large LTC-DC samples were determined using the 2–ΔΔCt method.25 This procedure involved measurement of a gene of interest relative to GAPDH in each of the small and large cell populations. Primers were designed from the sequence of the subtracted clones of interest rather than from sequences of homologous genes; they are available on request.
Results
LTC-DCs display a myeloid-like DC phenotype
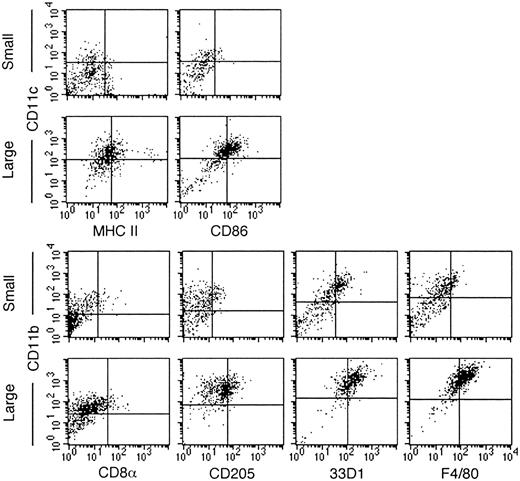
Recent studies have indicated that murine splenic DCs can be divided into a number of subsets on the basis of cell surface marker expression.11 Lymphoid-like DCs express CD8α and CD205, whereas myeloid-like DCs are characterized by the expression of CD11b, 33D1, and F4/80, and they may or may not express CD4.11,26 Two-color staining using specific monoclonal antibodies was undertaken to determine whether nonadherent LTC-DCs resembled a previously described murine DC subset. As previously shown,15 the large LTC-DC subset was a homogeneous population of cells; approximately 66% expressed detectable levels of CD11c. These CD11c+ cells also expressed CD86 and low levels of MHC 2 (Figure 1). For many markers, including CD86 and MHC 2, quadrants based on background staining separate the large cell population. This indicates a lower level of marker expression over the population, rather than the presence of clear positive and negative subpopulations. Combined data from many experiments and different LTCs show consistency in marker expression on small and large LTC-DCs (Table 1). CD11c+ large LTC-DCs also express CD11b15 (and data not shown); hence, 66% of large cells resemble CD11c+CD11b+ myeloid-like DCs. Consistent with this designation, large cells uniformly express F4/80 (69.0%) and 33D1 (74.0%) but only low levels of CD205 (37.1%) and CD8α (2.7%).
LTC-DCs resemble CD8α– myeloid-like DCs. Cells were labeled with antibodies specific for DC markers using an indirect 2-color staining technique. Cells were separated into small and large LTC-DC subsets using postacquisitional gating based on forward scatter and side scatter. Quadrants distinguishing positively and negatively staining cells were placed to represent background staining with only labeled conjugate. Data are representative of many separate experiments. The quadrants for CD8α are set at a lower intensity because acquisition was performed using lower FL1 and FL2 voltages than were used for other markers.
LTC-DCs resemble CD8α– myeloid-like DCs. Cells were labeled with antibodies specific for DC markers using an indirect 2-color staining technique. Cells were separated into small and large LTC-DC subsets using postacquisitional gating based on forward scatter and side scatter. Quadrants distinguishing positively and negatively staining cells were placed to represent background staining with only labeled conjugate. Data are representative of many separate experiments. The quadrants for CD8α are set at a lower intensity because acquisition was performed using lower FL1 and FL2 voltages than were used for other markers.
Percentage of positively staining cells
Surface Marker . | No. of Experiments . | % cell staining . | . | |
|---|---|---|---|---|
| . | . | Small LTC-DC . | Large LTC-DC . | |
| CD11c | 9 | 26.8 ± 4.2 | 66.5 ± 3.9 | |
| MHC II | 7 | 11.0 ± 2.2 | 20.7 ± 1.6 | |
| CD86 | 4 | 10.6 ± 1.4 | 46.6 ± 0.6 | |
| CD11b | 8 | 51.7 ± 0.6 | 91.9 ± 1.4 | |
| 33D1 | 2 | 44.9 ± 5.3 | 74.0 ± 0.7 | |
| F4/80 | 4 | 15.6 ± 3.4 | 69.0 ± 2.8 | |
| CD8α | 4 | 2.8 ± 1.6 | 2.7 ± 1.6 | |
| CD205 | 4 | 16.9 ± 2.9 | 37.1 ± 4.4 | |
Surface Marker . | No. of Experiments . | % cell staining . | . | |
|---|---|---|---|---|
| . | . | Small LTC-DC . | Large LTC-DC . | |
| CD11c | 9 | 26.8 ± 4.2 | 66.5 ± 3.9 | |
| MHC II | 7 | 11.0 ± 2.2 | 20.7 ± 1.6 | |
| CD86 | 4 | 10.6 ± 1.4 | 46.6 ± 0.6 | |
| CD11b | 8 | 51.7 ± 0.6 | 91.9 ± 1.4 | |
| 33D1 | 2 | 44.9 ± 5.3 | 74.0 ± 0.7 | |
| F4/80 | 4 | 15.6 ± 3.4 | 69.0 ± 2.8 | |
| CD8α | 4 | 2.8 ± 1.6 | 2.7 ± 1.6 | |
| CD205 | 4 | 16.9 ± 2.9 | 37.1 ± 4.4 | |
Data were calculated by subtracting background from specific antibody staining and represent means ± SEs across multiple experiments.
Again, for small LTC-DCs, quadrant lines do not delineate clear subpopulations of cells (Figure 1). Some small cells express low levels of CD86 (10.6%) and MHC 2 (11.0%). Approximately 50% of small LTC-DCs express CD11b, and half of these are also CD11c+ (26.8%).15 CD11b+ small cells also express 33D1 (44.9%), F4/80 (15.6%), and CD205 (16.8%). Only 2.6% of small cells express low levels of CD8α. Lineage-related markers could not distinguish the small and large subsets of cells. Because LTC-DCs have been shown previously to lack expression of CD4,27 the LTC system generates cells that resemble CD8α–CD4– myeloid-like DCs.
Generation of subtracted cDNA libraries derived from small or large LTC-DCs
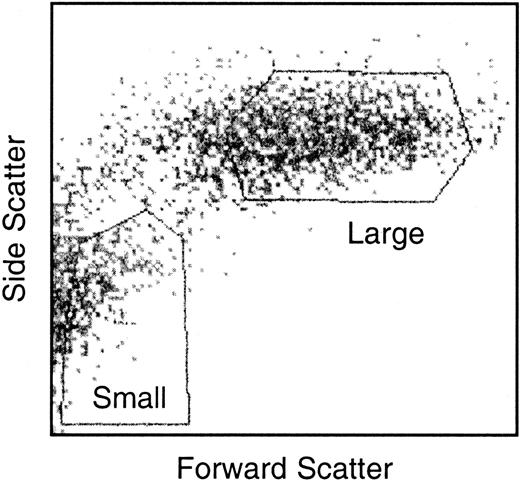
Nonadherent LTC-DCs were sorted into small and large LTC-DC subsets. The collection of pure populations of viable small and large cells was critical. Care was taken when setting gates to collect small LTC-DCs so that nonviable cells and large cells were excluded. Gates used to sort small and large LTC-DCs are shown in Figure 2. Total RNA was extracted from small and large LTC-DCs, and cDNA was synthesized and amplified. The extraction of high-quality RNA was demonstrated by the presence of clear 18S and 28S ribosomal RNA bands and the absence of genomic DNA in an agarose gel (data not shown). In addition, the A260/A280 ratio generated for small and large LTC-DC total RNA was 1.9. Tests conducted during the preparation of cDNA for subtraction also showed that RsaI digestion of cDNA and adaptor ligation of tester cDNA were successful (data not shown).
Gates used to sort nonadherent LTC-DCs into small and large cell subsets. Nonadherent cells were sorted in HBSS/5% FCS/1% glutamine and were collected in sDMEM/25% FCS. Gates shown were chosen as suitable for isolating the best yield of viable small and large LTC-DCs.
Gates used to sort nonadherent LTC-DCs into small and large cell subsets. Nonadherent cells were sorted in HBSS/5% FCS/1% glutamine and were collected in sDMEM/25% FCS. Gates shown were chosen as suitable for isolating the best yield of viable small and large LTC-DCs.
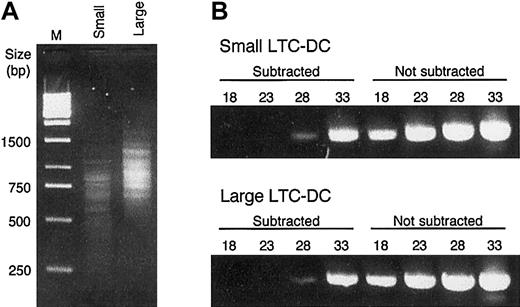
A procedure combining subtractive hybridization and suppressive PCR was used to enrich for differentially expressed genes in small and not large LTC-DCs (small subtracted cDNA) and in large but not small LTC-DCs (large subtracted cDNA). The subtraction protocol allowed only molecules in the test population that did not hybridize with molecules in the control population to be exponentially amplified in subsequent rounds of PCR,24 resulting in a dramatic loss of common background sequences. Aliquots of small and large subtracted cDNA were run on an electrophoresis gel to compare banding patterns. Subtracted cDNA was composed of distinct bands (Figure 3A) rather than the smear produced by nonsubtracted cDNA (data not shown). Small subtracted cDNA was composed of bands ranging in size from 250 bp to 1400 bp, whereas most large subtracted cDNA bands were between 500 bp and 1700 bp (Figure 3A). The variation in banding pattern suggested that different sets of genes were present in the subtracted cDNA populations. Furthermore, PCR analysis of subtracted products using GAPDH-specific primers demonstrated greater than 640-fold elimination of sequences encoding this housekeeping gene (Figure 3B).
Analysis of cDNA subtraction. (A) Electrophoresis of PCR products of subtracted small and large cDNA. After the second round of suppressive PCR, 5 μL product was run on a 2.0% agarose gel. To obtain clear resolution of fine bands, the gel was run at 80 V. DNA size markers (bp) are indicated for the 1-kb DNA ladder in lane M. (B) Comparison of subtracted and nonsubtracted cDNA for small and large LTC-DCs. Subtraction efficiency was judged by comparing the prevalence of the housekeeping gene GAPDH before and after subtraction. After 18, 23, 28, or 33 cycles of PCR using GAPDH 3′ and 5′ primers, 5 μL PCR product was run on a 2.0% agarose gel.
Analysis of cDNA subtraction. (A) Electrophoresis of PCR products of subtracted small and large cDNA. After the second round of suppressive PCR, 5 μL product was run on a 2.0% agarose gel. To obtain clear resolution of fine bands, the gel was run at 80 V. DNA size markers (bp) are indicated for the 1-kb DNA ladder in lane M. (B) Comparison of subtracted and nonsubtracted cDNA for small and large LTC-DCs. Subtraction efficiency was judged by comparing the prevalence of the housekeeping gene GAPDH before and after subtraction. After 18, 23, 28, or 33 cycles of PCR using GAPDH 3′ and 5′ primers, 5 μL PCR product was run on a 2.0% agarose gel.
Small and large subtracted cDNA was cloned into a plasmid vector to generate small and large subtracted cDNA libraries. Then 465 small and 454 large randomly selected clones were screened to detect those differentially expressed. The differential screening procedure was stringent; results are shown in Table 2. Hybridization of 4 different radiolabeled probes was assessed by visual inspection followed by densitometry. 32P-labeled probes were prepared from unsubtracted small- and large-cell cDNA, subtracted small-minus-large cDNA, and subtracted large-minus-small cDNA. A clone was distinguished as positive only if a solid black dot was visualized on the x-ray film because weaker hybridization signals were found to vary between replicates (data not shown). Controls of unrelated cDNA provided in the kit were used to assess the degree of nonspecific binding during hybridization. Neither control hybridized to any of the cDNA probes. Clones were assessed for exclusive specificity, partial specificity, and nonspecific binding according to criteria given in the legend to Table 2. Most (117 of 132) clones were selected because they showed mutually exclusive expression. Clones with partial specificity (15 of 132 selected clones) were selected if they showed at least 10-fold greater hybridization with forward (test) than reverse subtracted probe and 5-fold greater hybridization for unsubtracted probes.
Selection of clones by differential screening
. | No. of clones . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | Screening specificity . | . | . | . | ||||
| Library origin of clones . | Total . | Nil . | Nonspecific . | Partial specificity . | Exclusive specificity . | ||||
| Small subtracted | 465 | 206 | 188 | 7 | 64 | ||||
| Large subtracted | 454 | 352 | 41 | 8 | 53 | ||||
. | No. of clones . | . | . | . | . | ||||
|---|---|---|---|---|---|---|---|---|---|
| . | . | Screening specificity . | . | . | . | ||||
| Library origin of clones . | Total . | Nil . | Nonspecific . | Partial specificity . | Exclusive specificity . | ||||
| Small subtracted | 465 | 206 | 188 | 7 | 64 | ||||
| Large subtracted | 454 | 352 | 41 | 8 | 53 | ||||
Total represents the number of clones picked for differential screening. Screening specificity indicates that plated clones were screened with each of 4 different 32P-labeled probes made from unsubtracted small- and large-cell cDNA, subtracted small minus large cDNA, and subtracted large minus small cDNA. Hybridization was measured by densitometry. Nil indicates not selected because of insufficient binding of subtracted and unsubtracted probes. Nonspecific indicates not selected because of nonspecific binding of probes derived from tester and driver cDNA. Partial specificity indicates selected for sequencing because hybridization with forward (tester) probes was at least 10-fold greater than reverse of subtracted probe and 5-fold for unsubtracted probes using densitometry. Exclusive specificity indicates selected for sequencing because hybridization occurred only with forward (tester) subtracted and unsubtracted probes.
Differential screening produced 71 small subtracted clones and 61 large subtracted clones that were then sequenced and compared to known sequences in public databases. The experimental design chosen was rigorous because no overlap in gene expression was detected in the 2 subtracted populations (Tables 3 and 4). The combined protocol of subtractive hybridization, suppressive PCR, and differential screening was effective in removing commonly expressed genes. Many genes and full-length clones were represented more than once among the subtracted clones. However, because of the nature of the subtraction procedure, the frequency with which clones matched a particular database entry was not an accurate reflection of expression levels for genes within the target population.
Differentially expressed genes in small LTC-DCs
Clone frequency . | Description . | Accession number . | E-value . |
|---|---|---|---|
| Known | |||
| 10 | Thymosin β-4 | X16053 | 0.0 |
| 5 | Human glycoprotein hormone α-subunit | J00152 | e-101 |
| 4 | Human pregnancy-specific β-1-glycoprotein 1 | M93705 | 0.0 |
| 3 | Atp6f, lysosomal ATPase H+ transporter | AB060655 | e-153 |
| 2 | Ferritin light chain | J04716 | 0.0 |
| 2 | Human pregnancy—specific β-1-glycoprotein 4 | M94891 | 0.0 |
| 2 | Epithelial protein lost in neoplasm-a (Eplin) | AF307844 | e-162 |
| 2 | Peroxisomal acyl-CoA oxidase | AF006688 | e-158 |
| 2 | Ribosomal protein S12 | X15962 | e-128 |
| 2 | Cytochrome C oxidase subunit 2 | AF378830 | 0.0 |
| 2 | ATP synthase (F1) ϵ subunit | AK012444 | 0.0 |
| 1 | LAMP-1 | M25244 | 0.0 |
| 1 | Cystatin C (Cst3) | M59470 | e-174 |
| 1 | α-tubulin | BC008117 | 0.0 |
| 1 | CTT ϵ subunit | Z31555 | 0.0 |
| 1 | PCTAIRE-3 | X69026 | e-101 |
| 1 | Thioredoxin interacting factor (Vdup1) | AF282826 | 0.0 |
| 1 | Human Hsp90 | D87666 | 0.0 |
| 1 | ATP synthase (F0) subunit c, P2 | NM026468 | 0.0 |
| 1 | RIKEN clone 2700049122, ribosomal protein P2 | AK012402 | e-178 |
| 1 | RIKEN clone 6530419C17, Nsd1 | AK032683 | 0.0 |
| Unknown | |||
| 1 | Hypothetical protein MGC7474 | NM_024473 | e-144 |
| 1 | RIKEN clone 2410015A15 | AK010484 | e-151 |
| 1 | Similar to KRAB zinc finger protein KR18 | BC040201 | 0.0 |
| 1 | Similar to human putative zinc finger protein C5orf7 | XM_203708 | e-143 |
| 6 | EST matches | — | — |
| 2 | No match | — | — |
| Similar to | |||
| 2 | 60S ribosomal protein L5 | BC005790 | e-59 |
| 1 | Fibroblast growth factor-2 interacting factor (FIF or FT1) | Z67963 | 4e-10 |
| 1 | IMP dehydrogenase | M33934 | 9e-48 |
Clone frequency . | Description . | Accession number . | E-value . |
|---|---|---|---|
| Known | |||
| 10 | Thymosin β-4 | X16053 | 0.0 |
| 5 | Human glycoprotein hormone α-subunit | J00152 | e-101 |
| 4 | Human pregnancy-specific β-1-glycoprotein 1 | M93705 | 0.0 |
| 3 | Atp6f, lysosomal ATPase H+ transporter | AB060655 | e-153 |
| 2 | Ferritin light chain | J04716 | 0.0 |
| 2 | Human pregnancy—specific β-1-glycoprotein 4 | M94891 | 0.0 |
| 2 | Epithelial protein lost in neoplasm-a (Eplin) | AF307844 | e-162 |
| 2 | Peroxisomal acyl-CoA oxidase | AF006688 | e-158 |
| 2 | Ribosomal protein S12 | X15962 | e-128 |
| 2 | Cytochrome C oxidase subunit 2 | AF378830 | 0.0 |
| 2 | ATP synthase (F1) ϵ subunit | AK012444 | 0.0 |
| 1 | LAMP-1 | M25244 | 0.0 |
| 1 | Cystatin C (Cst3) | M59470 | e-174 |
| 1 | α-tubulin | BC008117 | 0.0 |
| 1 | CTT ϵ subunit | Z31555 | 0.0 |
| 1 | PCTAIRE-3 | X69026 | e-101 |
| 1 | Thioredoxin interacting factor (Vdup1) | AF282826 | 0.0 |
| 1 | Human Hsp90 | D87666 | 0.0 |
| 1 | ATP synthase (F0) subunit c, P2 | NM026468 | 0.0 |
| 1 | RIKEN clone 2700049122, ribosomal protein P2 | AK012402 | e-178 |
| 1 | RIKEN clone 6530419C17, Nsd1 | AK032683 | 0.0 |
| Unknown | |||
| 1 | Hypothetical protein MGC7474 | NM_024473 | e-144 |
| 1 | RIKEN clone 2410015A15 | AK010484 | e-151 |
| 1 | Similar to KRAB zinc finger protein KR18 | BC040201 | 0.0 |
| 1 | Similar to human putative zinc finger protein C5orf7 | XM_203708 | e-143 |
| 6 | EST matches | — | — |
| 2 | No match | — | — |
| Similar to | |||
| 2 | 60S ribosomal protein L5 | BC005790 | e-59 |
| 1 | Fibroblast growth factor-2 interacting factor (FIF or FT1) | Z67963 | 4e-10 |
| 1 | IMP dehydrogenase | M33934 | 9e-48 |
The number of clones that match a particular gene is not an accurate reflection of gene expression levels within the tester population because of the nature of the subtraction procedure. Species origin of the genes is Mus musculus, unless indicated otherwise. Database identification codes for gene entry were assigned by GenBank, EMBL, DDBJ, or PDB. Expectation value (E-value) represents the number of different alignments with scores equivalent to or better than the alignment in question and that would be expected to occur in the database by chance alone. E-value is generated by BLAST and is a convenient way to create a significance threshold for reporting results. The lower the E-value, the more significant the match. — indicates not available.
Differentially expressed genes in large LTC-DC
Clone frequency . | Description . | Accession number . | E-value . |
|---|---|---|---|
| Known | |||
| 10 | Cytochrome b-245, β polypeptide | BC003910 | 0.0 |
| 6 | Lysozyme | BC002069 | 0.0 |
| 5 | Vacuolar ATP synthase subunit d | XM143668 | 0.0 |
| 4 | trans-Golgi network protein 2 (TGN38B) | D50032 | 0.0 |
| 2 | Osteopontin | J04806 | 0.0 |
| 2 | Calnexin | BC12408 | 0.0 |
| 1 | CD86 | U39392 | e-137 |
| 1 | CCR1 (MIP-1α receptor) | U28404 | 0.0 |
| 1 | gp49B | U05265 | 0.0 |
| 1 | Cysteine/glutamate transporter xCT | AB022345 | 0.0 |
| 1 | TGF-β—inducible protein (TSC-36) | M91380 | 0.0 |
| 1 | Transcription factor S-2 | M18209 | 0.0 |
| 1 | Complement protein H | M12660 | 0.0 |
| 1 | Regulator of G-protein signaling 18 (Rgs 18) | AF302685 | e-145 |
| 1 | Lymphoid enhancer binding factor | D16503 | 0.0 |
| 1 | Clone MGC: 13853 IMAGE:4212293, sphingosine phosphate lyase 1 | BC026135 | 7e-90 |
| Unknown | |||
| 2 | RIKEN clone E430023L24 | AK088689 | 0.0 |
| 1 | RIKEN clone D130018D01, Ras-related protein RAB-8B homologue | AK051218 | 0.0 |
| 1 | RIKEN clone E430005109, weakly similar to C-MPL binding protein | AK088176 | 0.0 |
| 1 | RIKEN clone 4432405K22, hypothetical protein | AK014477 | e-119 |
| 1 | RIKEN clone 1110005L02 | AK003474 | 0.0 |
| 1 | RIKEN clone 6720435121 | AK020121 | 0.0 |
| 1 | RIKEN clone 4732429D16 | AK028662 | 0.0 |
| 1 | RIKEN clone A730014M21 | AK042676 | e-180 |
| 1 | Chromosome 1q21-23 clone mgs1-423c02 | AC008100 | 0.0 |
| 1 | Clone RP23-277L21 | AC124194 | 0.0 |
| 1 | Similar to chromatin remodeling factor WCRF180 | XM_126922 | 0.0 |
| 3 | EST matches | — | — |
| 1 | No match | — | — |
| Similar to | |||
| 1 | Rat unr protein | X52311 | 6e-63 |
| 1 | RIKEN clone 4930579A11 | NM_029478 | 5e-30 |
Clone frequency . | Description . | Accession number . | E-value . |
|---|---|---|---|
| Known | |||
| 10 | Cytochrome b-245, β polypeptide | BC003910 | 0.0 |
| 6 | Lysozyme | BC002069 | 0.0 |
| 5 | Vacuolar ATP synthase subunit d | XM143668 | 0.0 |
| 4 | trans-Golgi network protein 2 (TGN38B) | D50032 | 0.0 |
| 2 | Osteopontin | J04806 | 0.0 |
| 2 | Calnexin | BC12408 | 0.0 |
| 1 | CD86 | U39392 | e-137 |
| 1 | CCR1 (MIP-1α receptor) | U28404 | 0.0 |
| 1 | gp49B | U05265 | 0.0 |
| 1 | Cysteine/glutamate transporter xCT | AB022345 | 0.0 |
| 1 | TGF-β—inducible protein (TSC-36) | M91380 | 0.0 |
| 1 | Transcription factor S-2 | M18209 | 0.0 |
| 1 | Complement protein H | M12660 | 0.0 |
| 1 | Regulator of G-protein signaling 18 (Rgs 18) | AF302685 | e-145 |
| 1 | Lymphoid enhancer binding factor | D16503 | 0.0 |
| 1 | Clone MGC: 13853 IMAGE:4212293, sphingosine phosphate lyase 1 | BC026135 | 7e-90 |
| Unknown | |||
| 2 | RIKEN clone E430023L24 | AK088689 | 0.0 |
| 1 | RIKEN clone D130018D01, Ras-related protein RAB-8B homologue | AK051218 | 0.0 |
| 1 | RIKEN clone E430005109, weakly similar to C-MPL binding protein | AK088176 | 0.0 |
| 1 | RIKEN clone 4432405K22, hypothetical protein | AK014477 | e-119 |
| 1 | RIKEN clone 1110005L02 | AK003474 | 0.0 |
| 1 | RIKEN clone 6720435121 | AK020121 | 0.0 |
| 1 | RIKEN clone 4732429D16 | AK028662 | 0.0 |
| 1 | RIKEN clone A730014M21 | AK042676 | e-180 |
| 1 | Chromosome 1q21-23 clone mgs1-423c02 | AC008100 | 0.0 |
| 1 | Clone RP23-277L21 | AC124194 | 0.0 |
| 1 | Similar to chromatin remodeling factor WCRF180 | XM_126922 | 0.0 |
| 3 | EST matches | — | — |
| 1 | No match | — | — |
| Similar to | |||
| 1 | Rat unr protein | X52311 | 6e-63 |
| 1 | RIKEN clone 4930579A11 | NM_029478 | 5e-30 |
See Table 3 footnote.
Identity of differentially expressed genes in small LTC-DCs
Small subtracted clones matched 21 known genes and showed similarity to 3 more (Table 3). Thymosin β-4 was identified as the most common transcript found in the small subtracted clone set. Ten clones matched this sequence. Multiple biologic effects have been associated with thymosin β-4. However, it is best documented for its role in the organization of the cytoskeleton.28 Small LTC-DCs were found to express additional genes related to organization of the cytoskeleton, including Cste chaperonin containing TCP (CTT) and heat shock protein 90 (Hsp90). Selected clones from the small subtracted library encoded M-α-2–tubulin and the novel cytoskeletal protein Eplin, suggesting that the expression of cytoskeletal proteins may be functionally significant for small LTC-DCs. Selected small subtracted clones also encoded proteins associated with DC antigen processing, including lysosomeassociated membrane protein (LAMP)–1, Atp6f, and cystatin C. Another transcript represented a homologue of human pregnancy–specific β-1–glycoprotein 1, a protein implicated as an immunomodulator.29
Numerous ribosomal proteins were represented among small subtracted clones. Clones were found to match acidic ribosomal protein P2 and to be similar to 60S ribosomal protein L5. Two clones were also found to match ribosomal protein S12, associated with mitochondria. Mitochondrial transcripts were isolated from the small subtracted cDNA library. These included cytochrome C oxidase subunit 2, adenosine triphosphate (ATP) (F0) synthase subunit c P2 and ATP (F1) synthase ϵ subunit. Five small clones were identified as homologues of the human glycoprotein hormone α-subunit. This protein is the common α-subunit of the 4 glycoprotein hormones.30 However, a free α-subunit is also produced by a variety of tumors and tumor-derived cell lines. This may reflect transcription related to cultured small LTC-DCs because no β-subunit transcripts were isolated. Other differentially expressed transcripts isolated from small LTC-DCs encoded ferritin light chain, peroxisomal acyl-CoA oxidase, PCTAIRE-3, thioredoxin interacting factor (Vdup1), and a homologue to human pregnancy–specific β-1–glycoprotein 4.
Twelve small subtracted clones represented unknown genes, and 4 matched full-length clones in the public database. Three of these matched novel transcripts in the Ensembl mouse genome server and encoded proteins with functional domains. Of the remaining 10 unknown clones, 6 were homologous to known ESTs, and 2 showed no homology with known genes or ESTs. Unknown transcripts will be investigated further because they could represent novel genes important in the differentiation of small into large LTC-DCs.
Identity of differentially expressed genes in large LTC-DCs
Large subtracted clones matched 16 known genes. In addition, 2 large subtracted clones had similarity to sequences in the NCBI nr database. These are listed in Table 4. Large LTC-DCs differentially expressed a number of transcripts encoding proteins of immunologic significance, such as the secreted molecule osteopontin, lysozyme, cytochrome b-245 β-polypeptide, and calnexin. The most commonly matched transcript was cytochrome b-245 β-polypeptide. Ten clones matched this gene. Large LTC-DCs also up-regulated the expression of transcripts encoding a number of immunologically important cell surface proteins. These included the costimulatory molecule CD86, chemokine receptor CCR1 (macrophage inflammatory protein [MIP]–1α receptor), and inhibitory molecule gp49B, which may play a role in the regulation of innate immune responses. Other regulatory molecules isolated from the large subtracted cDNA library included transforming growth factor-β (TGF-β)–inducible protein TSC-36, which has antiproliferative function,31 and Rgs18, which is a novel regulator of G-protein signaling.32 Large LTC-DCs also increased expression of a number of transcripts encoding DNA-binding proteins. These included lymphoid enhancer binding factor, transcription factor S-II, and what appears to be the mouse homologue of rat unr protein. Additional transcripts represented among the large subtracted clones encoded trans-Golgi network protein 2, complement protein H, vacuolar ATP synthase subunit D, and cystine/glutamate transporter xCT.
Sixteen large subtracted clones were isolated representing 15 different unknown transcripts. Twelve clones matched 11 full-length clones in the public database. Of the remaining unknown large clones, all but one showed homology to ESTs in the database. A number of unknown transcripts mapped to predicted novel genes or EST transcripts in the Ensembl mouse genome server. These novel genes contained functionally significant domains including PHD zinc-finger, Ig/MHC, and bHLH domains. One unknown transcript, similar to chromatin remodeling factor WCRF180 and other chromatin remodeling proteins, contained a bromodomain and PHD finger domain.
Verification of differential gene expression using real-time PCR
The expression of several clones of interest was compared in small and large cells by real-time PCR. Five selected large clones were all shown to be expressed in significantly higher levels in large cells over small cells produced in LTCs (Figure 4A). Only 2 of 3 selected small clones showed significantly greater expression in the small-over-large cell population (Figure 4B). The greater expression of thymosin β-4 was consistent with the isolation of 10 clones from the small subtracted cell library.
Real-time PCR analysis of selected clones. Expression of 5 selected large-cell clones and 3 selected small-cell clones was measured by real-time PCR relative to GAPDH in the small- and large-cell populations. The procedure involved analysis of 0.2 to 4 ng small- and large-cell total RNA in triplicate. Data are expressed as the fold expression of each gene in large cells relative to small cells (A) or small cells relative to large cells (B). Histograms represent the fold expression calculated as 2–ΔΔCt, including the range [2–(ΔΔCt +1.96 SE), 2–(ΔΔCt –1.96 SE)]. Gene expression levels in one cell population versus another are significantly different if the range does not overlap the normalized value of 1 (2-sided Z test; P = .05).
Real-time PCR analysis of selected clones. Expression of 5 selected large-cell clones and 3 selected small-cell clones was measured by real-time PCR relative to GAPDH in the small- and large-cell populations. The procedure involved analysis of 0.2 to 4 ng small- and large-cell total RNA in triplicate. Data are expressed as the fold expression of each gene in large cells relative to small cells (A) or small cells relative to large cells (B). Histograms represent the fold expression calculated as 2–ΔΔCt, including the range [2–(ΔΔCt +1.96 SE), 2–(ΔΔCt –1.96 SE)]. Gene expression levels in one cell population versus another are significantly different if the range does not overlap the normalized value of 1 (2-sided Z test; P = .05).
Discussion
Using subtractive hybridization and suppressive PCR, small and large subtracted cDNA libraries were generated to isolate genes differentially expressed in the small and large LTC-DC populations. Small and large subtracted cDNA was found to be enriched at least 640-fold for differentially expressed genes. A stringent screening step was carried out on randomly selected clones from each library so that only differentially expressed transcripts were subsequently selected for sequencing. As a result, there was no overlap in the sample sets of genes isolated from small and large LTC-DC. Real-time PCR was used to verify differential gene expression. This confirmed the suitability of subtracted library screening for isolating genes specific to the homogeneous large-cell population. Confirmation of only 2 of 3 tested small clones by real-time PCR could relate to the greater heterogeneity of the small cell population and perhaps to lower differences in the transcription level specific to housekeeping genes in cells that are small and quiescent. It will be important to verify differential expression of any small clones of interest by real-time PCR before pursuing them further. Small and large LTC-DC subsets differentially express genes that can be related to developmental capacity and function. Consistent with their role as antigen-presenting cells, large LTC-DCs up-regulate the expression of genes representing important immunologic functions, whereas small LTC-DCs, or DC precursors,15 preferentially express genes related to early stages of differentiation.
The small subtracted library contained transcripts for the cytoskeletal protein M-α-2–tubulin. The balancing of monomeric, dimeric, and polymeric forms of tubulin have been found to underlie changes in cell shape and the formation of dendritic processes.33 Small LTC-DCs also specifically express genes related to the organization of the cytoskeleton. Thymosin β-4 prevents the polymerization of actin filaments but supplies a pool of actin monomers when the cell needs filaments.28 It plays an important role in cell morphologic change and migration. In addition, CTT has broad recognition capabilities, but appears to be chiefly involved in the folding of actin and tubulin.34,35 Hsp90 is a molecular chaperone that assists the conformational maturation of many specific targets, including tubulin.36
Actin disassembly is consistent with the absence of dendritic projections on small LTC-DCs and low endocytic capacity for these cells.15 Both of these processes require the integrity of polymerized actin filaments. The expression of proteins related to the cytoskeleton in small LTC-DCs suggests that small cells are entering a phase of morphologic change. This is consistent with previous work showing that small LTC-DCs increase in size and develop dendrites as they differentiate into large LTC-DCs.15 Furthermore, though mitochondrial and ribosomal proteins are ubiquitously expressed in cells, the detection of mitochondrial and ribosomal genes in small LTC-DCs may reflect an increase in energy production and protein synthesis, respectively, before differentiation into large LTC-DCs.
In addition to its role as a chaperone involved in the folding of tubulin, Hsp90 is involved in the transport of antigenic peptides to MHC 1 and to the cell surface.37 Small subtracted clones encode a number of proteins involved in the regulation of MHC molecules and compartments. LAMP-1 colocalizes with lysosomes and MHC 2 compartments,38 and Atp6f is a proteolipid that acidifies early endosomes.39 Cystatin C is an inhibitor of cathepsin S. The up-regulation of cystatin C leads to inefficient Ii chain cleavage, resulting in the transport of MHC 2/Ii chain complexes to lysosomes rather than the plasma membrane.40 High cystatin C expression in small LTC-DCs could contribute to the lack of MHC class 2 observed on the surfaces of these cells. The expression of molecules associated with antigen processing in the small subtracted library could be a reflection of the acquisition of endocytic capacity as some small LTC-DCs begin differentiating into large LTC-DCs.
Consistent with functional capacity as antigen-presenting cells14,15 , large LTC-DCs express transcripts encoding a range of proteins of immunologic importance, including CD86, calnexin, CCR1, gp49B, cytochrome b-245 β polypeptide, lysozyme, and osteopontin. Secretion of osteopontin by large LTC-DCs could enhance TH1 responses, the generation of immunoglobulins, and the proliferation of B cells, or it could induce the cellular chemotaxis of T cells and macrophages.41 Alternate splicing and posttranslational modification (phosphorylation and glycosylation) generate functionally distinct forms of osteopontin. The sequence of large subtracted clones gives no indication of the form of osteopontin represented in large LTC-DCs.
Detection of transcripts for the CD86 costimulatory molecule in large LTC-DCs is consistent with FACS data showing that large LTC-DCs express more CD86 on their cell surfaces than small LTC-DCs. Calnexin is a chaperone protein that contributes to the protection and assembly of MHC 1 and prevents the transportation of incomplete MHC 1 complexes to the cell membrane.42 The ligand for CCR1, MIP-1α, is an inflammatory chemokine, chemotactic for many hematopoietic cell types including immature monocyte-derived DCs.43 Expression of CCR1 may allow LTC-DCs to migrate in response to inflammatory stimuli. Large LTC-DCs also showed specific expression of gp49B, a transmembrane glycoprotein containing an immunoreceptor tyrosine–based inhibitory motif in its cytoplasmic region that functions as an inhibitory molecule.44
Large LTC-DCs show the expression of cytochrome b-245 β polypeptide, a transcript shown to be highly expressed in human DCs but not in monocytes or lymphocytes.45 Cytochrome b-245 is a major component of the NADPH-dependent superoxide-generating system.46 Although most reports find that DCs do not generate superoxide anion,47 production has been demonstrated by human DCs that have phagocytosed Candida albicans.48 Expression of transcripts such as cytochrome b-245 β-polypeptide and lysozyme suggest that large LTC-DCs have microbicidal capabilities. Lysozyme mRNA expression has been observed in a number of myeloid cells, from early Gr-1+ progenitors to activated macrophages and monocyte-derived DCs.20,49,50
Numerous genes isolated during this study are associated with the regulation of cell development and proliferation. Most are not well characterized, though their differential expression by small and large LTC-DCs suggests they are involved in LTC-DC development. Small LTC-DCs specifically express PCTAIRE-3, a serine/threonine-specific protein kinase with unknown cellular function. PCTAIRE-3 belongs to a family of cdc2-related protein kinases that participate in cell cycle regulation and are associated with cell division and differentiation.51 Nsd1 is a nuclear protein that interacts directly with several nuclear receptors and affects the control of cell development, differentiation, and homeostasis.52 Additionally, the overexpression of Eplin has been associated with a decrease in proliferation53 and may contribute to the lower levels of replication seen in small LTC-DCs. Large LTC-DCs up-regulate the expression of several genes that encode regulatory proteins. Rgs18 is a novel regulator of G-protein signaling specific to hematopoietic stem cells and myeloid lineage cells,32 whereas TGF-β–inducible protein is an extracellular glycoprotein with antiproliferative function that may inhibit a growth-factor–like molecule.31 Unr protein is a nucleic-acid–binding protein with unknown function that is developmentally regulated.54 Sphingosine phosphate lyase 1 may also play a role in the regulation of LTC-DC proliferation, differentiation, or programmed cell death through the catalysis of sphingosine phosphate degradation.55
In addition to the isolation of known regulatory genes, novel transcripts were isolated from large LTC-DCs that are candidates for the regulation of DC development. One transcript had similarity to known chromatin-remodeling genes containing a PHD finger domain and a single bromodomain. Bromodomains bind acetylated lysine, and possession of such a domain suggests a role in chromatin remodeling and transcriptional activation.56 RAB-8B is thought to regulate vesicle transport from the Golgi to the plasma membrane. It also induces plasma membrane outgrowth when overexpressed.57 The homologue expressed in large LTC-DC (RIKEN clone D130018D01) may play a similar role, contributing to the morphology and regulation of secretory pathways in these cells. A further unknown transcript (RIKEN clone 4732429D16) maps to a novel sequence in Ensembl that is homologous to human CMRF-35 and natural killer (NK) inhibitory receptor and that contains Ig/MHC and bHLH domains. This may encode a surface receptor involved in the activation of a signaling pathway appropriate to the large LTC-DCs.
Cell surface marker expression identifies LTC-DC as CD11c+CD11b+MHC2loCD8α– myeloid-like DCs. Surface CD8α expression is absent from most cultured DC populations.5 Although it is possible that an in vivo counterpart of LTC-DCs could express CD8α, the expression of other myeloid markers on LTC-DCs foreshadows CD8α– myeloid-like DCs. Furthermore, LTC-DCs do not resemble newly identified B220+ DC subsets because they lack surface B220 and Gr-1.58
The profile of clones detected in small and large LTC-DCs also reflects myeloid-like DCs. Many clones identified in small LTC-DCs, such as α-tubulin, thymosin β-4, cystatin C, Hsp90, ferritin light chain, and ribosomal protein S12, are highly expressed in Gr-1+ myeloid progenitors and immature, but not activated, monocyte-derived DCs.18-20,50 Furthermore, small LTC-DCs express genes associated with early cells that are down-regulated on maturation/activation, consistent with their classification as DC precursors. For example, thymosin β-4 is associated with early cell development from hematopoietic stem cells, myeloid progenitors, and immature DCs.18,50,59
Large LTC-DC also express a range of genes associated with the myeloid lineage. These include osteopontin, CCR1, and CD86, which are highly expressed in monocyte-derived and bone-marrow–derived DCs20,21,45,60 and Rgs18, whose expression is restricted to hematopoietic progenitors and cells of the myeloid lineage.32 CCR1 and osteopontin gene expressions have been associated with myeloid-like and not lymphoid-like human DCs.45 Lysozyme is expressed strongly by macrophages49 and weakly by monocyte-derived DCs.20 It has also been noted that lysozyme transcript levels are up-regulated in monocyte-derived DCs on activation.20 This finding could reflect the comparison between small and large LTC-DCs shown here.
The existence of precursors and progeny within the one LTC provides a unique opportunity to study the development of DC from committed precursors. Gene expression identified here for small and large LTC-DCs is consistent with previous characterization of LTC-DC subset function and development.15 Cell surface marker analysis and differential gene analysis also indicate that LTC-DCs resemble myeloid-like DCs. It is now clear that DC type and maturation state can dictate the type of immune response generated. Novel transcripts isolated from small and large LTC-DCs will be the focus of further study and characterization. All data indicate that these transcripts potentially encode novel proteins important in DC function and development.
Prepublished online as Blood First Edition Paper, May 15, 2003; DOI 10.1182/blood-2002-08-2426.
Supported by grants from the National Health and Medical Research Council of Australia (H.C.O.), the Australian Research Council (H.C.O.), and the Clive and Vera Ramaciotti Foundation of Australia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Terry O'Neill for help with the statistical analysis.

![Figure 4. Real-time PCR analysis of selected clones. Expression of 5 selected large-cell clones and 3 selected small-cell clones was measured by real-time PCR relative to GAPDH in the small- and large-cell populations. The procedure involved analysis of 0.2 to 4 ng small- and large-cell total RNA in triplicate. Data are expressed as the fold expression of each gene in large cells relative to small cells (A) or small cells relative to large cells (B). Histograms represent the fold expression calculated as 2–ΔΔCt, including the range [2–(ΔΔCt + 1.96 SE), 2–(ΔΔCt – 1.96 SE)]. Gene expression levels in one cell population versus another are significantly different if the range does not overlap the normalized value of 1 (2-sided Z test; P = .05).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/5/10.1182_blood-2002-08-2426/6/m_h81734866004.jpeg?Expires=1767766553&Signature=1ADXFqX-OL8jhcM739MVIDZ7bouNkfusHy7Hyr1b9Fa4TPeMuHRuHkt2EUmYiSoHMTCpqfngoTO~qk3lR00a0oHgDjxyGevsM0ELnY02Z1KMubrofhJkGL9INsaCJyAmy0tzgsMiZoKGde3HtXbDFsi2DVBKnwhPdgLgEi5n00JmNQN9gThQqP9BB3WtVfp2kYtCojgeO4sKCKpOdjIse8lsY4UfrwdcIm8XcSA7gatELvoZxeY1UTajK0sHGxpqXQ5LWF4Bw2jbh0WpJPkCgGviZOyhthEPK8izBTM~EyIDPUVYzAUKZgCavwsVIr9ZycnegEDJhXWUakzTm0ugvw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal