Abstract
The more distal enhancers of the immunoglobulin heavy-chain 3′ regulatory region, hs3b and hs4, were recently demonstrated as master control elements of germline transcription and class switch recombination to most immunoglobulin constant genes. In addition, they were shown to enhance the accumulation of somatic mutations on linked transgenes. Since somatic hypermutation and class switch recombination are tightly linked processes, their common dependency on the endogenous locus 3′ enhancers could be an attractive hypothesis. VDJ structure and somatic hypermutation were analyzed in B cells from mice carrying either a heterozygous or a homozygous deletion of these enhancers. We find that hs3b and hs4 are dispensable both for VDJ assembly and for the occurrence of mutations at a physiologic frequency in the endogenous locus. In addition, we show that cells functionally expressing the immunoglobulin M (IgM) class B-cell receptor encoded by an hs3b/hs4-deficient locus were fully able to enter germinal centers, undergo affinity maturation, and yield specific antibody responses in homozygous mutant mice, where IgG1 antibodies compensated for the defect in other IgG isotypes. By contrast, analysis of Peyer patches from heterozygous animals showed that peanut agglutinin (PNAhigh) B cells functionally expressing the hs3b/hs4-deficient allele were dramatically outclassed by B cells expressing the wild-type locus and normally switching to IgA. This study thus also highlights the role of germinal centers in the competition between B cells for affinity maturation and suggests that membrane IgA may promote recruitment in an activated B-cell compartment, or proliferation of activated B cells, more efficiently than IgM in Peyer patches.
Introduction
During the antigen-independent steps of B-cell ontogeny, multiple cis-regulatory elements control the tissue- and stage-specific regulation of both transcription and V(D)J rearrangements in the immunoglobulin heavy-chain (IgH) locus, allowing the assembly and expression of a functional heavy chain gene. Interactions with cognate antigens promote recruitment of activated B cells into germinal centers and the occurrence of additional processes that allow the generation of high-affinity antibodies and a switch to the synthesis of non-IgM immunoglobulins with various effector functions.1,2 These processes are supported at the gene level by somatic hypermutation (SHM) of V(D)J exons and class switch recombination (CSR) of constant genes. Although these concurrent processes of Ig maturation can sometimes occur independently,1 they clearly share common requirements, such as transcription of the target genes, and signaling through CD40 and the B-cell receptor (BCR).2 Their common need for the product of the activation-induced deaminase (AID) enzyme was also recently underlined by studies in the human hyper-IgM type 2 immunodeficiency and in knock-out mice.3,4
The 3′ IgH regulatory region includes a large palindromic structure and 4 lymphoid-specific transcriptional enhancers: hs3a, hs1,2, hs3b, and hs4.5-11 These elements are controlled by multiple transcription factors including the E2A, oct, Ets, Maf, and nuclear factor–κB (NF-κB) families and B-cell–specific activator protein (BSAP)/Pax5, which mediate their synergistic activation or repression depending on the B-cell differentiation stage.12 Studies in transfected cell lines have suggested that the 3′ regulatory region could act as a locus control region (LCR).9 This finding was partially confirmed in transgenic mice, in which a reporter gene linked to the 4 3′ enhancers displayed position-independent B-cell–specific expression but no copy-number dependence.13 Transgenic experiments showed that hs1,2 was not sufficient to target SHM onto an associated transcribed sequence.14 Similarly, it has long been known that rearranged VH transgenes carrying the μ gene intronic enhancer (Eμ) are unable to mutate at significant levels unless they recombine with the endogenous IgH locus, which provides a complete set of regulatory elements.15-17 In a recent study, addition of both the hs3b and hs4 3′ IgH elements, but not of hs1,2, resulted in an increase in the SHM level of transgenes driven by an Eμ/pVH cassette and led to the conclusion that hs3b/hs4 elements are pivotal in the somatic mutation process.18
Recent gene targeting experiments where the 3′ part of the IgH LCR encompassing hs3b and hs4 was either replaced with a neomycin resistance gene or deleted showed a major reduction in both germline transcription of IgH constant genes and CSR.19 Meanwhile, targeted animals carried normal numbers of B cells in bone marrow and peripheral lymphoid organs, had normal levels of IgM and IgG1, and displayed germinal centers of normal architecture within all peripheral lymphoid organs.19 We thus wished to check whether these animals could mount efficient humoral primary and secondary responses to specific antigens and whether the hs3b and hs4 enhancers were playing a decisive role in the accumulation of somatic mutations in the endogenous IgH locus.
Materials and methods
Animals and immunization
Animals with a targeted deletion of the hs3b and hs4 IgH 3′ enhancers have been described previously.19 Since the disruption was carried out in a 129 embryonic stem (ES) cell line, it resulted in a mutated a allotype IgH locus. Mice were thus bred with C57/BL6 animals in order to derive heterozygous IgH aΔhs3b/hs4/bwt mice.
For each immunization experiment, batches of 6-week-old mice were used (4 Δ/Δ mice and 4 wild-type (wt) mice and the corresponding nonimmunized control mice). The first immunization was performed with 10 μg ovalbumin (Sigma, St Louis, MO) per animal in 50% complete Freund adjuvant (Sigma), and a second immunization was realized 13 days later with the same amount of antigen in 50% incomplete Freund adjuvant (Sigma). Immunized mice were eye-bled every 1 to 3 times during the immunization protocol, and sera were analyzed for the presence of ovalbumin-specific immunoglobulin isotypes by enzyme-linked immunosorbent assay (ELISA). Immunization experiments were repeated twice using 2 batches of mice.
Antibody determinations
ELISA assays were performed in polycarbonate 96 multiwell plates (Maxisorp, Nunc, Denmark) coated overnight at 4°C with 100 μL of 10 μg/mL ovalbumin solution in 0.05 M Na2CO3 buffer. After 3 successive washing steps in 0.1% Tween20/phosphate-buffered saline (PBS) buffer (washing buffer), a blocking step was performed for 30 minutes at 37°C with 150 μL of 2 mg/mL gelatin (Sigma) in PBS buffer. After 3 washing steps, 50 μL assayed sera or control serum (diluted in the appropriate range) was diluted into successive wells in 2 mg/mL gelatin in PBS buffer and incubated 2 hours at 37°C. The positive control consisted in a pool of sera from ovalbumin-immunized wt mice; the same control sera has been used in all ELISA assays. After 3 washing steps, 100 μL/well of appropriate conjugated antibodies was added and adsorbed during 1.5 hours at 37°C. Alkaline phosphatase–conjugated goat antisera specific for mouse immunoglobulin isotypes IgM, IgG1, IgG2a, and IgG2b (Southern Biotechnologies, Birmingham, AL) were used at 1 μg/mL diluted in 0.1% Tween20/PBS buffer. After washing, alkaline phosphatase (AP) activity was assayed using 150 μL/well of 1 mg/mL AP substrate (Sigma) during 15 minutes at room temperature, enzymatic reactions were stopped with 30 μL/well of 3 M NaOH, and optic density was measured at 400 nm in a Spectracount photometer (Packard, Meriden, CT).
Dilutions of the control serum allowed to draw a titration curve. A 1:100 dilution of each serum was compared with the titration curve obtained on the same multiwell plate and allowed the quantification of ovalbumin-specific antibodies in arbitrary units. Each ELISA titration was realized twice.
Cell cytometry and sorting procedures
Spleens were recovered, disrupted, and filtrated on a nylon mesh in PBS supplemented with 10% fetal calf serum (FCS). After erythrocyte lysis using distilled water, cell suspension was filtrated, washed twice in PBS, and adjusted at 2 × 106 cells per mL in 10% FCS/PBS. Cell samples (50 μL) were incubated for 30 minutes at 4°C with various antibodies. Anti-IgM antibodies were either the fluorescein isothiocyanate (FITC)–conjugated monoclonal RS31 anti-IgMa, the MB86 anti–IgMb-FITC (kind gifts from Dr Claudia Berek), or a biotinylated anti–mouse IgM followed with streptavidin-FITC (Southern Biotechnologies); they were used in conjunction with either biotin-conjugated peanut agglutinin (PNA; Sigma) followed with SpectralRed (SPRD)–conjugated streptavidin (Southern Biotechnologies) or with anti–B220-SPRD. The latter was also used in conjunction with FITC-conjugated PNA (Sigma) to analyze germinal center B cells. Following each staining, cells were washed twice in PBS containing 1% BSA. Stained cell suspensions were analyzed on a Coulter XL apparatus (Beckman Coulter, Fullerton, CA).
For cell sorting, Peyer patches (PPs) were recovered by dissection from the small intestine. Cell suspension was prepared by pressing PPs through a nylon mesh; cells were washed 3 times in cold Dulbecco modified Eagle medium containing 10% FCS (Invitrogen, Gaithersburg, MD). Cell suspensions were adjusted at 106 cells per mL after elimination of clumps of dead cells. Cells were incubated with anti–B220-SPRD for 30 minutes at 4°C. After 2 washes with PBS containing 10% FCS, FITC-conjugated PNA was added and incubated on ice for a further 30 minutes, before washing and resuspension in medium. Purification of B220+PNAhigh and B220+PNAlow cells was realized on a FACSVantage (Becton Dickinson, Franklin Lakes, NJ) as previously described.20 The purification rates of these cell populations were constantly higher than 95%.
DNA extraction and amplification
Genomic DNA was extracted from PNAhigh and PNAlow B cells using the QIAamp Tissue Kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. Genomic DNA (2 μL) was amplified by PCR using the following primers and multistep programs: the forward primer VH7183 5′-CGGTACCAAGAASAMCCTGTWCCTGCAAATGASC-3′ complementary to the third framework region (FR3) of VH7183 family VH segments21 and the backward primer 5′-CAGTAAGCAAACCAGGCACA-3′ corresponding to a consensus intronic sequence between the JH2 and JH3 segments using 1 cycle at 94°C for 5 minutes, 35 cycles (94°C for 45 seconds, 58°C for 45 seconds, and 72°C for 45 seconds), and 1 cycle at 72°C for 7 minutes. PCR was carried out in 50 μLof Taq 1 × PCR buffer containing 200 μM of each deoxynucleoside triphosphate, 100 ng of each primer, and 1 unit Taq polymerase (Pharmacia, Piscataway, NJ).
Cloning and sequencing
The PCR product was separated on a 1.2% agarose gel and cloned into the pCRII-TOPO vector (Invitrogen, Leek, the Netherlands) according to the manufacturer's instructions. Plasmids were isolated using a standard Triton/lysozyme method, and inserts were sequenced from both ends using the M13Reverse and M13(–20) forward primers and an automated laser fluorescent DNA ABI-PRISM 310 sequencer (Perkin-Elmer, Branchburg, NJ).
Results
Homozygous hs3b/hs4 knock-out mice display efficient antigen-specific primary and secondary responses
We previously showed that hs3b/hs4 knock-out mice have a class-switching defect but apparently carry germinal centers of normal size and have normal serum total IgM and IgG1 levels.19 In addition, B cells from these mice did not display any general activation defect when stimulated with a polyclonal activator such as lipopolysaccharide (LPS). However, it was questionable whether these cells were also able to produce high-affinity antibodies when challenged with specific antigens.
Mice were thus specifically immunized with ovalbumin and assayed for the level and kinetics of their humoral response. IgM class antibodies were raised with a normal kinetic, but their titers were reduced by comparison with those of wild-type mice (Figure 1). By contrast, IgG antibodies strongly differed between both groups of mice: although IgG3, IgG2a, and IgG2b class antibodies were undetectable in knock-out mice, specific IgG1s were produced with a normal kinetic and raised levels at least equal to those in normal mice. IgG1 antibodies thus provided an internal control showing that B-cell activation by a specific antigen is not compromised in those mice.
Humoral response of hs3b/hs4-deficient mice immunized with ovalbumin. ELISA analysis of ovalbumin-specific immunoglobulin isotype secretion in sera of 8-week-old immunized mice. Antibody levels are expressed in arbitrary units by comparison with a control serum; time after immunization is indicated in days. Each point is the mean of serum determinations from 4 Δhs3b4 homozygous (gray curve) or wt control animals (black curve). The amount of ovalbumin-specific immunoglobulin was quantified by curve comparison with a pool of sera from ovalbumin-immunized wt mice. This experiment is representative of 2 independent experiments.
Humoral response of hs3b/hs4-deficient mice immunized with ovalbumin. ELISA analysis of ovalbumin-specific immunoglobulin isotype secretion in sera of 8-week-old immunized mice. Antibody levels are expressed in arbitrary units by comparison with a control serum; time after immunization is indicated in days. Each point is the mean of serum determinations from 4 Δhs3b4 homozygous (gray curve) or wt control animals (black curve). The amount of ovalbumin-specific immunoglobulin was quantified by curve comparison with a pool of sera from ovalbumin-immunized wt mice. This experiment is representative of 2 independent experiments.
VDJ junctions have normal structures and are not delayed on the hs3b/hs4-deficient allele
VDJ junction sequences from either homozygous knock-out mice or heterozygous aΔhs3b/hs4/bwt mice were amplified, cloned, and sequenced. Out of 50 sequences from the wild-type b allotype IgH locus and 55 sequences from the targeted a allotype locus, we found that there was no significant difference with regard to the complementarity-determining region 3 (CDR3) length, to the extent of deletion at the V-D or D-J junction or to the number of N nucleotides inserted at the V-D-J junctions (Table 1). D usage also appeared similar when the targeted allele was compared with a wild-type a or b allotype allele: the various D segments were used with frequencies that did not significantly differ from normal, including for the most upstream segments (DSP2.2) and the most downstream segments (DQ52). There was thus no indication of any alteration in the V(D)J recombination process or of any delay that would have resulted in a lower extent of N insertions due to the lack of terminal transferase activity beyong the pro-B cell stage.
Patterns of VDJ junctions cloned from heterozygous aΔhs3b/hs4/bwt mice
. | Wild type . | Δhs3b/hs4 . |
|---|---|---|
| Number of sequences analyzed | 50 | 55 |
| CDR3 length | 35.9 ± 8.0 | 35.0 ± 6.7 |
| Total N nucleotides (VD + DJ) | 9.0 ± 4.2 | 9.5 ± 4.3 |
| PNAhigh, % functional sequences (functional sequences/total) | 86.4 (22/26) | 14.3 (5/28) |
| PNAlow, % functional sequences (functional sequences/total) | 41.7 (10/24) | 40.7 (11/27) |
| D usage, % | ||
| Dsp2,1 | 5.1 | 3.9 |
| Dsp2,2 | 5.1 | 15.7 |
| Dsp2.3 | 0 | 3.9 |
| Dsp2.4 | 0 | 2 |
| Dsp2.5 | 7.7 | 13.7 |
| Dsp2.7 | 2.6 | 3.9 |
| Dsp2,9 | 12.8 | 7.8 |
| Dsp2,x | 7.7 | 0 |
| Dfl16,1 | 30.8 | 35.3 |
| Dfl16.2 | 0 | 2 |
| DQ52 | 17.9 | 5.9 |
| Undetermined | 10.2 | 5.9 |
. | Wild type . | Δhs3b/hs4 . |
|---|---|---|
| Number of sequences analyzed | 50 | 55 |
| CDR3 length | 35.9 ± 8.0 | 35.0 ± 6.7 |
| Total N nucleotides (VD + DJ) | 9.0 ± 4.2 | 9.5 ± 4.3 |
| PNAhigh, % functional sequences (functional sequences/total) | 86.4 (22/26) | 14.3 (5/28) |
| PNAlow, % functional sequences (functional sequences/total) | 41.7 (10/24) | 40.7 (11/27) |
| D usage, % | ||
| Dsp2,1 | 5.1 | 3.9 |
| Dsp2,2 | 5.1 | 15.7 |
| Dsp2.3 | 0 | 3.9 |
| Dsp2.4 | 0 | 2 |
| Dsp2.5 | 7.7 | 13.7 |
| Dsp2.7 | 2.6 | 3.9 |
| Dsp2,9 | 12.8 | 7.8 |
| Dsp2,x | 7.7 | 0 |
| Dfl16,1 | 30.8 | 35.3 |
| Dfl16.2 | 0 | 2 |
| DQ52 | 17.9 | 5.9 |
| Undetermined | 10.2 | 5.9 |
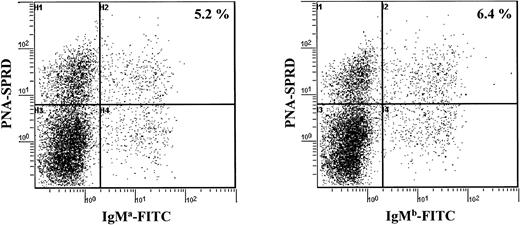
A cell cytometric analysis of splenocytes from aΔhs3b/hs4/bwt mice with allotype-specific antibodies also indicated that similar amounts of B cells expressed the a and the b allotypes, whether the IgM+, PNAlow (naive) or the IgM+, PNAhigh (activated) B-cell pool was considered (Figure 2). This can be taken as another indication that resting B cells expressing the targeted allele, although known to express membrane IgM at a lower density due to the hs3b/hs4 deletion, normally underwent antigen activation in vivo, similarly to what was previously observed in vitro for LPS activation.19 These data show that there is no preferential use of one allele in preswitched B cells. In addition to the similar timing and frequency of full VDJ rearrangements, the balanced expression of both alleles in activated IgM+ peripheral B cells further suggests that the repertoire of cells expressing the targeted allele was as broad as that of cells expressing the wild-type allele.
Analysis of splenocytes for expression of the mutated (a allotype) and the wt (b allotype) IgH alleles. Splenocytes were analyzed simultaneously for their strong or their low expression of PNA and for their expression of either the BCR of IgM a allotype or IgM b allotype.
Analysis of splenocytes for expression of the mutated (a allotype) and the wt (b allotype) IgH alleles. Splenocytes were analyzed simultaneously for their strong or their low expression of PNA and for their expression of either the BCR of IgM a allotype or IgM b allotype.
Homozygous hs3b/hs4 knock-out B cells carry somatically mutated IgH variable genes
An indication that B-cell activation occurs at reasonably good levels in homozygous mice expressing the targeted IgH locus was provided by the observation of specific IgM and IgG1 antibody responses following immunization, confirming that B cells were activated following antigen encounter. However, this observation does not provide any indication about antibody affinity maturation.
In order to appreciate SHM in B cells regardless of their antigen specificity, PP PNAhigh cells from homozygous mutant mice were sorted, and rearranged VDJ junctions were amplified using a VH7183 family FR3 consensus primer together with backward primers located in the intron downstream of JH2 and JH3. Sequence analysis focused on the rearranged JH segment and the following intronic 180–base pair (bp) sequence, a region known to be strongly targeted by the SHM process.22 This sequencing strategy also avoided ambiguities for the assignment of V sequences to their gernline counterpart. Mutations were countered either only in the intronic sequence or in the complete sequence including the JH-encoded fourth framework region (FR4).
VDJ junctions from homozygous mutant animals carried N insertions and displayed normal features. Out of 21 sequences analyzed from PP PNAhigh B cells, somatic mutations were readily observed in homozygous animals and their frequency ranged from 0 to 30 mutations per 1000 bp. Since mutation frequencies obtained in vivo proved to be highly variable in different experiments, data from homozygous mice merely indicated that the process of SHM indeed targeted the IgH locus, but would not have allowed a quantitative assessment of SHM by comparison with normal mice. In a search for a subtle SHM defect that could have been missed by the analysis of homozygous knock-out mice, we undertook the same experiments in heterozygous wt/Δ mice.
The frequency of somatic hypermutation is not lowered on the targeted versus wild-type allele in germinal center B cells from heterozygous (wt/Δhs3b-hs4) mice
A convenient way to discriminate the sequences of both alleles is the use of heterozygous IgHa/b allotype mice in which known allelic differences are located in the JH gernline region. The mutation frequencies of both a knock-out and a wt allele can thus be appreciated in the very same PNAhigh B-cell population that carries an allotype a IgH locus with the hs3b/hs4 deletion and an allotype b control wild-type locus.
We first checked that both allotypes of the IgH locus accumulated mutations at a similar frequency in awt/bwt mice (16.7 and 14.5 mutations per 1000 bp for the a and b allotypes, respectively) (Table 2).
Frequency of somatic mutations observed in the various heterozygous animals assayed (B220+PNAhigh and B220+PNAlow Peyer patch populations)
. | awt/bwt control mice . | . | aΔhs3b/hs4/bwt mice . | . | ||
|---|---|---|---|---|---|---|
| Mutation frequency, per 1000 bp . | a allotype . | b allotype . | a allotype . | b allotype . | ||
| PNAhigh cells | ||||||
| FR4 + intron | 16.7 | 14.5 | 21.75 | 10.84 | ||
| Intron only | 14.7 | 14.2 | 14.0 | 14.0 | ||
| In-frame junctions/total | 17/31 | 11/16 | 5/28 | 22/26 | ||
| Total bp analyzed | 6820 | 3520 | 6160 | 5720 | ||
| PNAlow cells | ||||||
| FR4 + intron | 3 | 0 | 2.9 | 3.0 | ||
| Intron only | 1.9 | 0 | 1.01 | 1.14 | ||
| In-frame junctions/total | 3/3 | 2/4 | 11/27 | 10/24 | ||
| Total bp analyzed | 660 | 880 | 5940 | 5280 | ||
. | awt/bwt control mice . | . | aΔhs3b/hs4/bwt mice . | . | ||
|---|---|---|---|---|---|---|
| Mutation frequency, per 1000 bp . | a allotype . | b allotype . | a allotype . | b allotype . | ||
| PNAhigh cells | ||||||
| FR4 + intron | 16.7 | 14.5 | 21.75 | 10.84 | ||
| Intron only | 14.7 | 14.2 | 14.0 | 14.0 | ||
| In-frame junctions/total | 17/31 | 11/16 | 5/28 | 22/26 | ||
| Total bp analyzed | 6820 | 3520 | 6160 | 5720 | ||
| PNAlow cells | ||||||
| FR4 + intron | 3 | 0 | 2.9 | 3.0 | ||
| Intron only | 1.9 | 0 | 1.01 | 1.14 | ||
| In-frame junctions/total | 3/3 | 2/4 | 11/27 | 10/24 | ||
| Total bp analyzed | 660 | 880 | 5940 | 5280 | ||
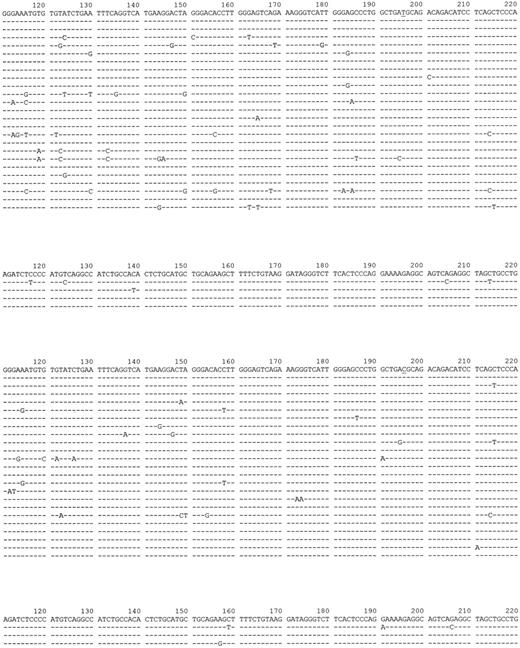
When aΔhs3b/hs4/bwt mice were studied, the first observation gained from analysis of PNAhigh IgH sequences was the presence of a high frequency of hypermutation on the knock-out allele, reaching 21.8 mutations per 1000 bp instead of 10.8 mutations per 1000 bp on the wild-type b allele (Figure 3,Figure 3 and Table 2). This difference was statistically significant but mostly resulted from different frequencies of mutations within the JH/FR4 coding segments. In contrast, a comparison restricted to the 180 bp of intervening sequence following JH indicated the same mutation frequency (14 mutations per 1000 bp) for both the targeted and the wt allele.
FR4/downstream intron sequences obtained from both IgH alleles in aΔhs3b/hs4/bwt mice.
FR4/downstream intron sequences obtained from both IgH alleles in aΔhs3b/hs4/bwt mice.
FR4/downstream intron sequences obtained from both IgH alleles in aΔhs3b/hs4/bwt mice.
FR4/downstream intron sequences obtained from both IgH alleles in aΔhs3b/hs4/bwt mice.
Another striking difference between VDJ sequences from both alleles obtained out of aΔhs3b/hs4/bwt PP B cells, was a strong bias in the proportion of functional sequences. Among PNAhigh B cells, 84.6% of in-frame rearrangements involved the wild-type allele, while reciprocally 85.7% of the out-of-frame rearrangements involved the knock-out allele. This strong disequilibrium was clearly posterior to antigen-driven selection within germinal centers since sequences obtained from PNAlow B cells were balanced (in-frame and out-of-frame junctions included 45% and 47%, respectively, of sequences originating from the wild-type allele). Differences in the activation by antigen were unlikely since we noticed by cytometric analysis that both alleles were expressed with similar frequencies within the PNAhigh IgM-positive B cells. A likely interpretation of this disequilibrium is that, beyond cell activation, cells that efficiently undergo class switching (ie, those expressing a wt IgH locus) have a proliferation advantage within PP germinal centers. It may be worth recalling that PP B cells normally include more than 70% of cells that have switched to the expression of IgA.23
As expected, low mutation frequencies were by contrast observed for control sequences obtained from PNAlow B cells (2.9 and 3.0 per 1000 bp for the mutated and the wild-type alleles, respectively) (Table 2).
Discussion
There has been speculation and conflicting data in the past few years about the potential role of the 3′ IgH region in SHM. Studies in transgenic mice have indicated that the central element of this 3′ LCR, hs1,2, is not able to recruit the SHM machinery even when it is combined with the intronic Eμ enhancer. By contrast, within enhancer combinations, the distal hs3b and hs4 elements appeared to play a pivotal role for the occurrence of SHM onto an associated transgene.18 A previous report clearly showed that the very same 2 downstream elements of the 3′ LCR control CSR through the regulation of gernline transcription from I promoters of the target IgH genes.19 Although CSR and SHM may occur independently, they are often spatially and temporally linked and they rely upon common signaling pathways and enzymatic activities, including the newly described activation-induced deaminase (AID).4 It is now clear that both processes need transcription of target genes, and more precisely it seems that transcription level is directly related to the frequency of those processes. For CSR events, recombination efficiency of a plasmid substrate containing Sμ and Sα regions driven by inducible promoters is quantitatively regulated by switch region transcription in the CH12 cell line.24 The same applies for SHM: in the 18.81 cell line, hypermutation of a green fluorescent protein target driven by an inducible promoter is increased together with transcription.25 Thus, if CSR and SHM seem to be dependent on their respective promoter elements, it is conveivable that those 2 processes could also share a dependency on the same transcriptional enhancers.
In fact, the herein-reported data suggest that joint deletion of the hs3b/hs4 enhancers does not affect SHM of the endogenous IgH locus, showing that CSR and SHM can be uncoupled with regard to their dependence on cis-regulatory elements. The amount of mutations observed on the hs3b/hs4 deleted allele was even higher than on the wild-type allele when mutation of the JH (FR4) region was considered. However, the much higher proportion of nonfunctional sequences obtained from the mutant allele likely accounted for such an increase, since it is known that nonfunctional V regions freely accumulate mutations in the FR segments although FR mutations are counterselected in functional genes.26
These data are reminiscent of those provided by studies carried out in the Ig κ locus. Transfection and transgenic studies have shown that the intronic iEκ and the unique 3′Eκ enhancer both promote transcription of rearranged κ genes and that iEκ is necessary for VκJκ rearrangement.27 Meanwhile, 3′Eκ plays a minor role in κ locus rearrangements but is necessary for optimal expression of rearranged genes.28-30 Transgenic studies first suggested that both the 3′Eκ and the iEκ/MAR were critical for recruiting the SHM machinery on linked V genes.31,32 However, the drop in the mutation frequencies of transgenes lacking 3′Eκ could also have merely resulted from an indirect effect, due to a decreased transcription of these constructs.33,34 In addition, divergent results were obtained from a gene-targeted mutational analysis where a 3′Eκ defect neither altered antigen-specific activation of B cells nor recruitment of the SHM machinery to rearranged κ genes.35 This difference in the need for 3′Eκ with respect to SHM for the endogenous locus versus transgenes likely outlines the role of still unidentified cis-acting elements, which may potentially be redundant in activity to 3′Eκ in the endogenous κ locus and which may have been lacking in transgenes. Similarly, our data about the IgH locus show that although the hs3b/hs4 enhancers may help recruit AID and other potential mutator elements on transgenes randomly inserted in the genome, they are dispensable for SHM within the endogenous locus. If the antigen-driven recruitment of SHM is directly linked to the transcription frequency of Ig genes, then the unaltered occurrence of SHM is consistent with the normal transcription of rearranged VDJ segments observed on a hs3b/hs4 deleted locus in activated B cells.19 While these 2 enhancers play a pivotal role in the control of gernline transcription from I promoters and CSR, their contribution (if any) to SHM in the endogenous locus is thus at the most redundant with that of other regulatory elements of the locus. Regarding the transgenic study that previously suggested h3b/hs4 to recruit SHM, the increase in the mutation frequency could have merely resulted from a nonspecific increase of transcription, which was not monitored in that study.18 Indeed, the hs3b and hs4 elements have been previously shown to undergo strong synergies with other regulatory elements of the IgH locus.36
In addition, this study allowed the characterization of a large number of VDJ junctions from homozygous or heterozygous mutant animals. It clearly showed that the hs3b/hs4 deletion neither affects nor delays VDJ recombination along B-cell lineage differentiation. In heterozygous animals, both the wild-type and the hs3b/hs4-deficient alleles were expressed at similar levels in naive B cells or in IgM+-activated cells. By contrast, PP-activated B cells showed an overwhelming expression of the wild-type allele in which CSR occurred normally. A likely explanation for the preferential outgrowth of cells functionally expressing the wt allele after antigen selection in the PP germinal center is that class switching by itself may provide a selective advantage to switched cells, either at the level of competition for antigen recognition on the surface of follicular dendritic cells and/or at the level of proliferation subsequent to antigen-mediated activation in germinal centers. Indeed, notable differences between immunoglobulins of the various classes reside in their intracellular tails, and it is conceivable that either signaling or antigen internalization may be improved for cells carrying switched isotypes. The hs3b/hs4 deletion may thus constitute a useful model by uncoupling affinity maturation (which is maintained) and class switching (which is severely altered) and may help address such issues.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2002-12-3827.
Supported by grants from Réseau SWITCH, Association pour la Recherche sur le Cancer (grant no. 4403), Ligue Nationale contre le Cancer, and Conseil Régional du Limousin.
C.L.M. and E.P. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Claudia Berek for kindly providing us with the RS31 and MB86 antibodies; Amine Khamlichi for helpful discussion and critical reading of the manuscript; and O. Decourt for his help with the statistical analysis.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal