Abstract
NKG2D ligands (NKG2DLs) mark malignant cells for recognition by natural killer (NK) cells and cytotoxic T lymphocytes via the activating immunoreceptor NKG2D. This led to the hypothesis that NKG2DLs play a critical role in tumor immune surveillance. The human NKG2DLs MICA and MICB are expressed on tumors of epithelial origin in vivo. For the other recently described set of human NKG2DLs, the UL16-binding proteins (ULBPs), expression in vivo is as yet undefined. In this study we investigated expression and function of NKG2DLs in leukemia using a panel of newly generated NKG2DL-specific monoclonal antibodies. We report that leukemia cells from patients variously express MIC and ULBP molecules on the cell surface with MICA most frequently detected. Patient leukemia cells expressing MICA were lysed by NK cells in an NKG2D-dependent fashion. Sera of patients, but not of healthy donors, contained elevated levels of soluble MICA (sMICA). We also detected increased sMICB levels in patient sera using a newly established MICB-specific enzyme-linked immunosorbent assay. Reduction of leukemia MIC surface expression by shedding may impair NKG2D-mediated immune surveillance of leukemias. In addition, determination of sMICA and sMICB levels may be implemented as a prognostic parameter in patients with hematopoietic malignancies.
Introduction
Natural killer (NK) cells as components of the innate immunity are involved in antiviral and antitumor immune responses.1 Tumor-associated ligands engaging activating NK-cell receptors are largely unknown. An exception is represented by the homodimeric C-type lectin-like receptor NKG2D that has been shown to interact with several highly diversified major histocompatibility complex (MHC) class I–related molecules.2 In mice, NKG2D occurs in 2 isoforms that either exclusively associate with the adaptor protein DAP10 or with both DAP10 and DAP12.3,4 In humans, there is yet only genetic evidence for the latter isoform,3 whereas the DAP10-specific isoform has been intensively characterized5,6 Whereas DAP10 is thought to mediate costimulatory functions, DAP12 directly activates NK cells.7 Among the human NKG2D ligands (NKG2DLs) are the stress-inducible surface glycoproteins, MICA and MICB, which are expressed on many epithelial tumors and upon infection.8-12 More recently, the UL16-binding proteins (ULBPs) have been identified as additional ligands of NKG2D.9,13 ULBPs are members of a multigene family with at least 6 functional members.14 They are expressed on numerous tumor cell lines, but their expression in vivo has yet to be explored.13-15 In the mouse, a family of proteins structurally related to ULBP, the retinoic acid early inducible (RAE-1) molecules function as ligands for NKG2D.16,17 RAE-1 expression has been shown to be induced by carcinogens and to stimulate antitumor activity of γδ T cells.18 RAE-1–transduced cell lines were eliminated in vivo due to NK and CD8 T-cell activity and induced tumor immunity against the parental cell line supporting a role for NKG2D in tumor immune surveillance.19,20 Recently, we demonstrated that MICA is released as a soluble form from the cell surface of tumor cells and can be detected at high levels in sera of patients with gastrointestinal malignancies but not in healthy donors.21 Soluble MICA (sMICA) released by epithelial tumors down-regulates NKG2D surface expression and leads to a functional impairment of tumor antigen–specific cytotoxic lymphocytes.22
To explore the relevance of the NKG2D/NKG2DL system for the immune surveillance of hematopoietic malignancies, we analyzed NKG2DL expression of patient leukemia cells with a panel of newly generated NKG2DL-specific monoclonal antibodies (mAbs). Using these mAbs we also established a highly sensitive enzyme-linked immunosorbent assay (ELISA) to evaluate levels of sMICA and sMICB in sera of patients with hematopoietic malignancies.
Patients, materials, and methods
Patients
Blood samples from patients with hematopoietic malignancies were analyzed at the time of diagnosis prior to therapy. Peripheral blood mononuclear cells (PBMCs) were obtained after Ficoll-Hypaque density gradient centrifugation. Sera were frozen at –80°C until further analysis. PBMCs were analyzed by flow cytometry, used as targets in a standard chromium release assay, or lysed for RNA preparation. All patients gave their written informed consent in accordance with the Helsinki protocol, and the study was performed according to the guidelines of the local ethics committee. All patients presented here were seen between December 2001 and November 2002 at the Department of Internal Medicine II, University Hospital, Eberhard-Karls-University, Tübingen, Germany. Diagnosis was confirmed by morphologic study of bone marrow specimens and flow cytometric immunophenotyping (Table 1). Clinical data were collected the day the blood samples were taken.
Patient characteristics
UPN . | Age, y . | Sex . | Diagnosis . | FAB . | PBB . | Karyotype . | WBC . | Hb . | Plt . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 39 | F | T-NHL | NA | 18 | 46,XX | 99.7 | 9.4 | 260 |
| 2 | 74 | F | AML | M4 | 80 | 46,XX | 117.8 | 9.9 | 277 |
| 3 | 48 | M | AML | M3 | 96 | 46,XY t(15;17) | 38.9 | 9.5 | 13 |
| 4 | 56 | M | AML | M1 | 95 | 46,XY | 17.4 | 7.4 | 18 |
| 5 | 63 | M | AML | M4 | 58 | 46,XY; t(9;22;5) | 87.3 | 9.0 | 28 |
| 6 | 43 | M | AML | M4 | 99 | 46,XY | 72.0 | 13.7 | 32 |
| 7 | 43 | F | AML | M1 | 92 | 47,XX;+8 | 9.0 | 9.0 | 86 |
| 8 | 69 | F | AML | M3 | 43 | 46,XX t(15;17) | 0.9 | 8.1 | 55 |
| 9 | 69 | M | AML | M3 | 38 | 46,XY t(15;17) | 0.7 | 6.6 | 9 |
| 10 | 75 | F | AML | M2 | 66 | 46,XX | 94.0 | 11.1 | 64 |
| 11 | 25 | M | AML | M4 | 5 | 46,XY | 15.8 | 8.6 | 58 |
| 12 | 75 | F | sAML | ND | 42 | 46,XX,5q-7q- | 19.6 | 10.7 | 20 |
| 13 | 60 | F | sAML | M2 | 42 | 46,XX | 34.0 | 8.1 | 75 |
| 14 | 67 | M | sAML | M2 | 12 | 46,XY | 13.1 | 8.4 | 221 |
| 15 | 77 | F | sAML | ND | 4 | 46,XX | 64.6 | 10.0 | 111 |
| 16 | 76 | M | sAML | M4 | 34 | 46,XY | 12.9 | 12.6 | 81 |
| 17 | 60 | F | CMML | NA | 22 | 46,XX | 126.0 | 8.5 | 74 |
| 18 | 27 | F | ALL | NA | 90 | 46,XX | 12.2 | 7.7 | 41 |
| 19 | 73 | F | ALL | NA | 41 | 46,XX | 10.7 | 10.7 | 124 |
| 20 | 30 | M | CML | NA | 57* | 46,XY,t(9;22)(q34;11) | 48.0 | 15.0 | 189 |
| 21 | 41 | F | CML | NA | 47* | 46,XX,der(22)t(9;22)(q34;q11) | 183.1 | 10.5 | 812 |
| 22 | 32 | M | CML | NA | 13* | 46,XY,t(9;22)(q34;11) | 125.3 | 11.6 | 122 |
| 23 | 43 | F | CML | NA | 22* | 46,XX t(9;22) | 11.0 | 10.9 | 31 |
| 24 | 60 | M | CLL | NA | 52 | 46,XY | 20.0 | 14.7 | 775 |
| 25 | 53 | F | CLL | NA | 87 | ND | 126.1 | 13.7 | 142 |
UPN . | Age, y . | Sex . | Diagnosis . | FAB . | PBB . | Karyotype . | WBC . | Hb . | Plt . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 39 | F | T-NHL | NA | 18 | 46,XX | 99.7 | 9.4 | 260 |
| 2 | 74 | F | AML | M4 | 80 | 46,XX | 117.8 | 9.9 | 277 |
| 3 | 48 | M | AML | M3 | 96 | 46,XY t(15;17) | 38.9 | 9.5 | 13 |
| 4 | 56 | M | AML | M1 | 95 | 46,XY | 17.4 | 7.4 | 18 |
| 5 | 63 | M | AML | M4 | 58 | 46,XY; t(9;22;5) | 87.3 | 9.0 | 28 |
| 6 | 43 | M | AML | M4 | 99 | 46,XY | 72.0 | 13.7 | 32 |
| 7 | 43 | F | AML | M1 | 92 | 47,XX;+8 | 9.0 | 9.0 | 86 |
| 8 | 69 | F | AML | M3 | 43 | 46,XX t(15;17) | 0.9 | 8.1 | 55 |
| 9 | 69 | M | AML | M3 | 38 | 46,XY t(15;17) | 0.7 | 6.6 | 9 |
| 10 | 75 | F | AML | M2 | 66 | 46,XX | 94.0 | 11.1 | 64 |
| 11 | 25 | M | AML | M4 | 5 | 46,XY | 15.8 | 8.6 | 58 |
| 12 | 75 | F | sAML | ND | 42 | 46,XX,5q-7q- | 19.6 | 10.7 | 20 |
| 13 | 60 | F | sAML | M2 | 42 | 46,XX | 34.0 | 8.1 | 75 |
| 14 | 67 | M | sAML | M2 | 12 | 46,XY | 13.1 | 8.4 | 221 |
| 15 | 77 | F | sAML | ND | 4 | 46,XX | 64.6 | 10.0 | 111 |
| 16 | 76 | M | sAML | M4 | 34 | 46,XY | 12.9 | 12.6 | 81 |
| 17 | 60 | F | CMML | NA | 22 | 46,XX | 126.0 | 8.5 | 74 |
| 18 | 27 | F | ALL | NA | 90 | 46,XX | 12.2 | 7.7 | 41 |
| 19 | 73 | F | ALL | NA | 41 | 46,XX | 10.7 | 10.7 | 124 |
| 20 | 30 | M | CML | NA | 57* | 46,XY,t(9;22)(q34;11) | 48.0 | 15.0 | 189 |
| 21 | 41 | F | CML | NA | 47* | 46,XX,der(22)t(9;22)(q34;q11) | 183.1 | 10.5 | 812 |
| 22 | 32 | M | CML | NA | 13* | 46,XY,t(9;22)(q34;11) | 125.3 | 11.6 | 122 |
| 23 | 43 | F | CML | NA | 22* | 46,XX t(9;22) | 11.0 | 10.9 | 31 |
| 24 | 60 | M | CLL | NA | 52 | 46,XY | 20.0 | 14.7 | 775 |
| 25 | 53 | F | CLL | NA | 87 | ND | 126.1 | 13.7 | 142 |
UPN indicates uniform patient number; FAB, French-American-British classification; PBB, peripheral blood blasts given in percent of nucleated cells; WBC, white blood count in Giga/L; Hb, hemoglobin in g/dL; Plt, platelets in Giga/L; F, female; M, male; NA, not applicable; and ND, not determined.
Percent immature myeloid cells.
Cell lines and transfectants
C1R cells (human B-cell lymphoma) were cultured in 10% fetal calf serum (FCS)/RPMI 1640, C1R-transfectants in 10% FCS/RPMI 1640 with 1.8 mg G418/mL (PAA Laboratories, Linz, Austria). C1R and the mouse mastocytoma cell line P815 were stably transfected with full-length cDNA encoding MICA*01, MICA*04, MICB*02, ULBP1, ULBP2, or ULBP3, respectively, in RSV.5neo by electroporation (C1R) or with FuGene 6 (P815) (Roche, Mannheim, Germany) according to standard protocols. The MIC and ULBP cDNAs have been described previously.9 Transfectants obtained after G418 selection were sorted using a Vantage cytometer (Becton Dickinson, Mountain View, CA), if necessary. For the production of soluble MIC-containing culture supernatants, C1R transfectants were grown in RPMI 1640 at 106 cells/mL without any additives for 72 hours. The NK cell line NKL, kindly provided by M. J. Robertson (Indiana University School of Medicine, Indianapolis), was grown in RPMI 1640 with 15% FCS and 200 U/mL interleukin 2 (IL-2; Proleukin; Chiron, Ratingen, Germany). Twelve days prior to cellular cytotoxicity assays, NKL cells were cultured without IL-2. All media were supplemented with penicillin (100 IU/mL)/streptomycin (100 μg/mL; Life Technologies, Karlsruhe, Germany), 2 mM l-glutamine, and 1 mM sodium pyruvate.
Reagents
Antimouse IgG2a–horseradish peroxidase (HRP) was from Southern Biotechnology (Birmingham, AL). The goat antimouse phycoerythrin (PE) conjugate was from Jackson ImmunoResearch (West Grove, PA). The anti-NKG2D mAb 8G7C10 (IgG2b) was a gift from Dr Peter Kufer (MicroMet, Munich, Germany). The fluorescein isothiocyanate (FITC)–labeled antihuman CD19, CD33, and CD34 antibodies were from Becton Dickinson.
Real-time reverse transcription–polymerase chain reaction
Total RNA was prepared from PBMCs using TRIZOL (Life Technologies) and reverse transcribed using SuperScript II (Invitrogen, Karlsruhe, Germany) according to the manufacturer's protocol. The resulting cDNA was amplified with NKG2DL and 18S rRNA-specific primer pairs in duplicate (40 cycles: 95°C for 15 seconds and 60°C for 1 minute) and amplification monitored using SYBRGreen chemistry on the ABI PRISM 7000 Sequence Detection System (Applied Biosystems, Weiterstadt, Germany). Primers were selected to flank an intron, where possible, and specificity was validated using cloned NKG2DLs. Data analysis was done by using the ΔCT method for relative quantification. Briefly, threshold cycles (CT) for 18S rRNA (reference) and NKG2DL (sample) were determined in duplicates. We arbitrarily defined the values obtained for healthy donor 1 (HD1) as standard values and determined the relative increase (rI) in relative copy numbers in relation to these standard values according to the formula rI = 2 –[(CT sample – CT reference) – (CT standard sample – CT standard reference)]. Similar amplification efficiencies for NKG2DL and 18S were demonstrated by analyzing serial cDNA dilutions with values of the slope of log cDNA amount versus ΔCT of less than 0.1. Oligonucleotide sequences (forward, reverse) were for 18S rRNA: 5′-CGGCTACCACATCCAAGGAA-3′, 5′-GCTGGAATTACCGCGGCT-3′; MICA: 5′-CCTTGGCCATGAACGTCAGG-3′,5′-CCTCTGAGGCCTCRCTGCG-3′; MICB: 5′-ACCTTGGCTATGAACGTCACA-3′, 5′-CCCTCTGAGACCTCGCTGCA-3′; ULBP1: 5′-GTACTGGGAACAAATGCTGGAT-3′, 5′-AACTCTCCTCATCTGCCAGCT-3′; ULBP2: 5′-TTACTTCTCAATGGGAGACTGT-3′,5′-TGTGCCTGAGGACATGGCGA-3′; ULBP3: 5′-CCTGATGCACAGGAAGAAGAG-3′, 5′-TATGGCTTTGGGTTGAGCTAAG-3′; amplicons were examined on 3% agarose gels for correct size.
Cellular cytotoxicity assay
Cytotoxicity was analyzed by a standard chromium release assay. Target cells were labeled with 50 μCi (1.85 MBq) Na251CrO4 (Amersham, Freiburg, Germany) for 2 hours at 37°C and washed 3 times. In blocking experiments, mAbs were either added at 10 μg/mL (AMO1 and isotype controls) to the target cells during the labeling procedure or as diluted hybridoma supernatant (8G7C10) to the effector cells 30 minutes prior to the assay. Cells were washed and effector cells were titrated on the target cells and incubated for 5 hours at 37°C. Maximum release was determined from target cells lysed in 1% Triton X-100. Percentage of lysis was calculated as follows: 100 × (experimental release – spontaneous release) ÷ (maximum release – spontaneous release). Experiments were performed in duplicates.
Production of soluble MICA*04 and MICB*02 in Escherichia coli
Soluble MIC molecules were produced and purified as previously described.21 In brief, the cDNAs encoding the MICA*04 and MICB*02 ectodomains (Glu1 through Lys276) in pET20b and pET21a (Novagen, Madison, WI), respectively, were introduced in E coli BL21, and MIC production was induced by addition of (isopropryl-β-D-thiogalactopyranoside) IPTG. Inclusion bodies were solubilized and subsequently dialyzed against decreasing concentrations of urea. Refolded MIC molecules were purified by gel filtration, dialyzed against TBS (20 mM Tris [tris(hydroxymethyl)aminomethane]; 150 mM NaCl, pH 8.0) and eventually examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting.
mAbs
NKG2DL-specific mAbs were raised by repeatedly immunizing BALB/c mice with a mixture of P815-MICA*01, P815-MICA*04, and P815-MICB*02, and P815-ULBP1, P815-ULBP2, and P815-ULBP3 transfectants, respectively, according to standard procedures. Hybridoma supernatants were screened by flow cytometry using MIC or ULBP-transfected C1R cells. Hybridomas producing NKG2DL-specific antibodies were subcloned twice. MIC-specific mAbs for the MICA and MICB ELISA were purified by affinity chromatography on a protein A-Sepharose column. The mAb clones AMO1 (anti-MICA), BMO1 (anti-MICB), BAMO1 (anti-MICA/B), AUMO1 (anti-ULBP1), and BUMO2 (anti-ULBP2) were of isotype IgG1; the mAb BMO2 (anti-MICB) and BAMO3 (anti-MICA/B) were IgG2a; and CUMO2 (anti-ULBP3) was IgM.
Flow cytometry
PBMCs were incubated with the NKG2DL-specific mAb or the respective isotype control and then, after washing, with goat antimouse-PE conjugate as secondary reagent. Leukemic cells were selected by staining with anti-CD19, anti-CD33, or anti-CD34 FITC conjugates, respectively, and then analyzed on a FACScan (Becton Dickinson). In case of chronic myeloid leukemia (CML) where no specific marker is applicable, selection was performed according to morphology using forward-scatter and side-scatter settings. Specific fluorescence indices (SFIs) of NKG2DL staining were calculated by dividing median fluorescences obtained with the respective specific mAb by median fluorescences obtained with the corresponding isotype control.
ELISA
For the detection of sMICB, 2 mAbs binding to different MICB epitopes were implemented. Plates were coated with the capture mAb BAMO1 at 2 μg/mL in phosphate-buffered saline (PBS), then blocked by addition of 100 μL 15% bovine serum albumin (BSA) for 2 hours at 37°C and washed. Afterward, the standard (recombinant MICB*02 in 7.5% BSA-PBS) and the sera (after dilution 1:3 in 7.5% BSA-PBS prior to addition to the plates) were added and the plates were incubated for 2 hours at 37°C. After incubation, plates were washed and the detection mAb BMO2 was added at 1 μg/mL 7.5% BSA-PBS for 2 hours at 37°C. After washing, antimouse IgG2a-HRP (1:8000 in 3.5% BSA-PBS) was added for 1 hour at 37°C. Plates were then washed and developed using the TMB peroxidase substrate system (KPL, Gaithersburg, MD). The absorbance was measured at 450 nm. Results shown are means of triplicates.
A MICA sandwich-ELISA similar to the recently described ELISA was used to detect sMICA.21 Here, the mAbs AMO1 and BAMO3 were used at 5 μg/mL and 1 μg/mL, respectively, with recombinant sMICA*04 as a standard.
Results
Patient characteristics
All 25 patients studied received their ultimate diagnosis based on clinical data as well as flow cytometric immunophenotyping and morphologic and cytogenetic data on initial bone marrow studies. The clinical characteristics of each patient are given in Table 1. There were 11 men and 14 women (male-female ratio, 1:1.3); the median age was 60 years with a range from 25 to 77 years. Ten patients had acute myeloid leukemia (AML), 5 had secondary acute myeloid leukemia (sAML) with a prior diagnosis of myelodysplastic syndrome, 1 patient had chronic myelomonocytic leukemia (CMML), 2 had acute lymphatic leukemia (ALL), 4 had chronic myeloid leukemia (CML), 2 had chronic lymphatic leukemia (CLL), and 1 patient had a leukemic form of a T-cell non-Hodgkin lymphoma (T-NHL). Bone marrow aspirates were available on all 25 patients and reviewed by an experienced hematologist. Adequate cytogenetic data were available for 24 of 25 patients (96%) and 14 had a normal karyotype (56%).
Development of NKG2DL mAbs and ELISA
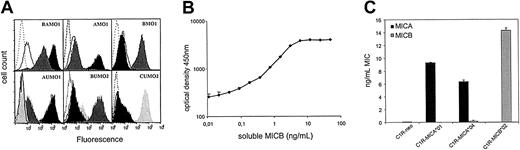
To survey expression of NKG2DL, a panel of mAbs specific for the MIC molecules MICA and MICB and the ULBP molecules ULBP1, ULBP2, and ULBP3 was generated by immunizing BALB/c mice with P815 cells transfected with the respective NKG2DL cDNAs. Specificity of the mAb was validated by staining NKG2DL-transfected C1R cells. The mAb AMO1 detected MICA*01 and MICA*04, but not MICB*02, whereas the mAb BMO1 conversely stained C1R-MICB*02 transfectants, but not MICA-transfected C1R cells (Figure 1A, upper panel and data not shown). Most of the obtained anti-MIC mAb, however, bound both to MICA and MICB in agreement with their high similarity in the extracellular domains. Among them, the mAb BAMO1 and BAMO3 are specific for the α1/α2, and α3 domain,21 respectively, and were included in this study. The mAbs AUMO1, BUMO2, and CUMO2 strongly stained C1R cells transfected with ULBP1, ULBP2, and ULBP3, respectively (Figure 1A, lower panel). Note that C1R cells endogenously express low levels of MICA, MICB, and ULBP2. The mAb AMO1 and BAMO3 had previously been implemented to establish a sandwich ELISA for sMICA,21 which was used after slight modification to increase sensitivity below 10 pg/mL. Similarly, a highly sensitive sandwich MICB ELISA was developed using the mAbs BAMO1 and BMO2 and soluble recombinant MICB*02 as a standard detecting sMICB at concentrations below 10 pg/mL (Figure 1B).
Characterization of NKG2DL mAb and MICB ELISA. (A) C1R-MICA*01 (black histograms), C1R-MICB*02 (gray histograms), and C1R-neo (open histograms, solid line) were stained with BAMO1, AMO1, and BMO1 and an isotype control, respectively (open histograms, dotted line) and analyzed by flow cytometry (upper panel). C1R-ULBP1 (black histograms), C1R-ULBP2 (dark gray histograms), C1R-ULBP3 (light gray histograms), and C1R-neo (open histograms, solid line) were stained with AUMO1, BUMO2, CUMO2, and the respective isotype control (open histograms, dotted line; lower panel). (B) Standardization of the MICB sandwich ELISA. Serial dilutions of sMICB were analyzed in a sandwich of BAMO1 and BMO2 followed by detection with antimouse IgG2a-HRP. The means of 4 replicates including the respective SDs are shown. (C) Supernatants of CIR-neo and the respective MICA and MICB transfectants of C1R were investigated for levels of soluble MIC molecules by the MICA (▪) and MICB sandwich ELISA (▦). The means of duplicates with their SDs are depicted.
Characterization of NKG2DL mAb and MICB ELISA. (A) C1R-MICA*01 (black histograms), C1R-MICB*02 (gray histograms), and C1R-neo (open histograms, solid line) were stained with BAMO1, AMO1, and BMO1 and an isotype control, respectively (open histograms, dotted line) and analyzed by flow cytometry (upper panel). C1R-ULBP1 (black histograms), C1R-ULBP2 (dark gray histograms), C1R-ULBP3 (light gray histograms), and C1R-neo (open histograms, solid line) were stained with AUMO1, BUMO2, CUMO2, and the respective isotype control (open histograms, dotted line; lower panel). (B) Standardization of the MICB sandwich ELISA. Serial dilutions of sMICB were analyzed in a sandwich of BAMO1 and BMO2 followed by detection with antimouse IgG2a-HRP. The means of 4 replicates including the respective SDs are shown. (C) Supernatants of CIR-neo and the respective MICA and MICB transfectants of C1R were investigated for levels of soluble MIC molecules by the MICA (▪) and MICB sandwich ELISA (▦). The means of duplicates with their SDs are depicted.
Whereas BAMO1 and BAMO3 recognize both MICA and MICB, selective specificity for sMICA or sMICB was achieved by combining them with the MICA-specific AMO1 and the MICB-specific BMO2, respectively. Specificity of the ELISA was evaluated with supernatants from C1R-MICA*01, C1R-MICA*04, and C1R-MICB*02 (Figure 1C). When supernatants of C1R-MICA transfectants were analyzed, strong signals in the ELISA for MICA but not for MICB were detected. MICA*01 and MICA*04 are fairly divergent allelic variants, but were similarly detected by the MICA-ELISA suggesting a broad recognition of MICA proteins. Conversely, C1R-MICB supernatants gave rise to strong signals in the MICB-ELISA, but not in the MICA-ELISA, demonstrating that both ELISAs specifically detected the respective MIC proteins. Using these ELISAs, release of soluble MIC protein from various leukemia cell lines was observed (unpublished observations, August 2002).
Expression of NKG2DLs by patient leukemia cells
Blood samples of 25 patients with various hematopoietic malignancies were obtained prior to chemotherapeutic treatment and PBMCs isolated by density gradient centrifugation. To evaluate NKG2DL expression in the patient cells we determined NKG2DL mRNA by reverse transcription–polymerase chain reaction (RT-PCR). Analyses of PBMCs from 13 patients with leukemia and 2 healthy donors for MICA and MICB transcripts uniformly resulted in strong amplicons, demonstrating that MICA and MICB mRNA were present at significant amounts in PBMCs of all donors (data not shown). Therefore, we subsequently used real-time PCR for a more accurate determination of NKG2DL mRNA expression levels. Relative expression levels of the individual NKG2DL mRNA in PBMCs of 4 patients and 2 healthy donors were obtained after standardization with 18S rRNA levels. For MICA, PBMCs of 2 patients (UPN 4 and UPN 20) contained increased mRNA levels compared with healthy donors, whereas no significant differences were observed for MICB transcripts among the 6 donors (Figure 2). For ULBP transcripts, increased expression of ULBP1 transcripts was detected in PBMCs of UPN 5, and of ULBP2 and ULBP3 transcripts in PBMC of UPN 1.
NKG2DL mRNA expression by PBMCs of leukemia patients. RNA from PBMCs of 2 healthy donors (HD1 and HD2) and 4 leukemia patients (UPN 1, UPN 4, UPN 5, UPN 20) was extracted and reverse transcribed. Relative copy numbers of NKG2DL were determined by real-time PCR using NKG2DL-specific primer pairs and normalized with 18S rRNA expression. Relative increase in relative copy numbers was calculated as outlined in “Patients, materials, and methods.” Relative copy numbers of HD1 were defined as 1. Error bars indicate the error propagation taking into account the SDs of the means of the 4 duplicates used for calculation of the relative increase in relative copy numbers.
NKG2DL mRNA expression by PBMCs of leukemia patients. RNA from PBMCs of 2 healthy donors (HD1 and HD2) and 4 leukemia patients (UPN 1, UPN 4, UPN 5, UPN 20) was extracted and reverse transcribed. Relative copy numbers of NKG2DL were determined by real-time PCR using NKG2DL-specific primer pairs and normalized with 18S rRNA expression. Relative increase in relative copy numbers was calculated as outlined in “Patients, materials, and methods.” Relative copy numbers of HD1 were defined as 1. Error bars indicate the error propagation taking into account the SDs of the means of the 4 duplicates used for calculation of the relative increase in relative copy numbers.
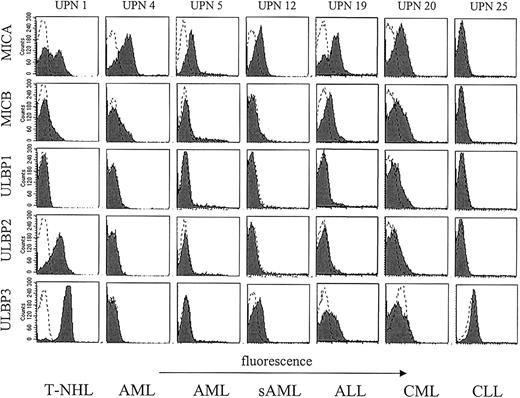
These data suggested a heterogeneous expression of NKG2DL in leukemia. To survey NKG2DL surface expression on patient leukemia cells we performed flow cytometry of freshly isolated patient PBMCs. Neither MIC nor ULBP molecules were detectable on PBMCs of healthy donors with our panel of NKG2DL-specific mAbs. To investigate NKG2DL expression of leukemia cells we selected malignant cells among patient PBMCs by staining for CD19, CD33, or CD34, respectively, or by morphology according to the investigated tumor entity (see Table 1 and “Patients, materials, and methods”). All investigated leukemia cells displayed high levels of MHC class I on the cell surface (data not shown). Analysis of NKG2DL surface expression revealed that leukemia cells of many patients express various NKG2DLs with expression patterns being fairly heterogeneous with regard to individual NKG2DLs. Intensities of NKG2DL cell surface stainings were in general much lower compared with MHC class I surface stainings with the pan-HLA-A, -B, -C mAb W6/32 (data not shown). Therefore, we considered leukemia cells positive for a particular NKG2DL in case of a 2-fold increase of median fluorescence above background. Of the 25 investigated patients, 14 (56%) expressed one or more NKG2DLs at significant levels at the cell surface (Figure 3; Table 2).
NKG2DL surface expression by leukemic cells. PBMCs from 6 patients with different hematopoietic malignancies were stained for NKG2DL expression with AMO1 (MICA), BMO1 (MICB), AUMO1 (ULBP1), BUMO2 (ULBP2), and CUMO2 (ULBP3), respectively, and investigated by FACS. Malignant cells were selected as described in “Patients, materials, and methods.” Shaded histograms represent expression of the indicated NKG2DL, and dotted lines show staining with the respective isotype control.
NKG2DL surface expression by leukemic cells. PBMCs from 6 patients with different hematopoietic malignancies were stained for NKG2DL expression with AMO1 (MICA), BMO1 (MICB), AUMO1 (ULBP1), BUMO2 (ULBP2), and CUMO2 (ULBP3), respectively, and investigated by FACS. Malignant cells were selected as described in “Patients, materials, and methods.” Shaded histograms represent expression of the indicated NKG2DL, and dotted lines show staining with the respective isotype control.
NKG2DL expression and release by patient leukemia cells
UPN . | MICA, pg/mL . | MICB, pg/mL . | ULBP1, SFI . | ULBP2, SFI . | ULBP3, SFI . | sMICA, pg/mL . | nMICA, pg/mL . | sMICB, SFI . | nMICB, SFI . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.0 | 1.8 | 1.0 | 8.5 | 8.2 | 924 | 9.2 | 288 | 2.9 |
| 2 | 3.1 | 3.2 | 2.3 | 1.1 | 1.0 | 885 | 7.5 | 36 | 0.3 |
| 3 | 1.5 | 1.0 | 1.0 | 1.1 | 1.0 | 267 | 6.9 | 336 | 8.6 |
| 4 | 7.3 | 1.2 | 1.0 | 1.0 | 1.0 | 10 296 | 591.7 | 27 | 1.6 |
| 5 | 3.2 | 1.2 | 1.0 | 1.1 | 1.1 | 978 | 11.2 | 183 | 2.1 |
| 6 | 1.4 | 1.5 | 1.3 | 1.7 | 1.0 | 54 | 0.8 | 90 | 1.3 |
| 7 | 1.1 | 1.0 | 1.0 | 1.1 | 1.1 | 75 | 8.3 | 72 | 8.0 |
| 8 | 1.1 | 1.3 | 1.0 | 1.0 | 1.0 | 99 | 110.0 | 126 | 140.0 |
| 9 | 1.2 | 1.6 | 1.7 | 1.7 | 1.0 | 318 | 454.3 | 150 | 214.3 |
| 10 | 1.1 | 10.0 | 1.3 | 1.0 | 1.1 | ND | ND | ND | ND |
| 11 | 2.7 | 1.7 | 1.0 | 3.3 | 1.6 | ND | ND | ND | ND |
| 12 | 2.8 | 1.0 | 1.0 | 1.0 | 2.4 | 642 | 401.3 | 132 | 82.5 |
| 13 | 1.1 | 1.0 | 1.0 | 1.1 | 1.0 | 216 | 6.4 | 180 | 5.3 |
| 14 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 141 | 10.8 | 57 | 4.4 |
| 15 | 1.5 | 1.2 | 1.1 | 1.1 | 1.0 | 114 | 1.8 | 69 | 1.1 |
| 16 | 2.2 | 2.1 | 1.1 | 1.0 | 1.1 | 174 | 13.5 | 69 | 5.3 |
| 17 | 1.4 | 1.2 | 6.4 | 1.9 | 1.0 | 159 | 1.3 | 117 | 0.9 |
| 18 | 4.7 | 4.8 | 3.9 | 1.0 | 6.7 | 321 | 267.5 | 303 | 252.5 |
| 19 | 12.1 | 3.6 | 1.1 | 1.0 | 1.8 | 549 | 51.0 | 66 | 6.2 |
| 20 | 5.6 | 3.1 | 1.9 | 1.8 | 1.0 | 1 173 | 24.4 | 69 | 1.4 |
| 21 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 96 | 0.5 | 153 | 0.8 |
| 22 | 1.7 | 1.3 | 1.8 | 1.0 | 1.3 | 150 | 1.2 | 318 | 2.5 |
| 23 | 1.0 | 1.0 | 1.0 | 1.0 | 2.7 | 216 | 19.6 | 288 | 26.2 |
| 24 | 6.9 | 5.2 | 8.1 | 8.1 | 2.3 | ND | ND | ND | ND |
| 25 | 1.2 | 1.0 | 1.1 | 1.0 | 1.3 | ND | ND | ND | ND |
UPN . | MICA, pg/mL . | MICB, pg/mL . | ULBP1, SFI . | ULBP2, SFI . | ULBP3, SFI . | sMICA, pg/mL . | nMICA, pg/mL . | sMICB, SFI . | nMICB, SFI . |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 5.0 | 1.8 | 1.0 | 8.5 | 8.2 | 924 | 9.2 | 288 | 2.9 |
| 2 | 3.1 | 3.2 | 2.3 | 1.1 | 1.0 | 885 | 7.5 | 36 | 0.3 |
| 3 | 1.5 | 1.0 | 1.0 | 1.1 | 1.0 | 267 | 6.9 | 336 | 8.6 |
| 4 | 7.3 | 1.2 | 1.0 | 1.0 | 1.0 | 10 296 | 591.7 | 27 | 1.6 |
| 5 | 3.2 | 1.2 | 1.0 | 1.1 | 1.1 | 978 | 11.2 | 183 | 2.1 |
| 6 | 1.4 | 1.5 | 1.3 | 1.7 | 1.0 | 54 | 0.8 | 90 | 1.3 |
| 7 | 1.1 | 1.0 | 1.0 | 1.1 | 1.1 | 75 | 8.3 | 72 | 8.0 |
| 8 | 1.1 | 1.3 | 1.0 | 1.0 | 1.0 | 99 | 110.0 | 126 | 140.0 |
| 9 | 1.2 | 1.6 | 1.7 | 1.7 | 1.0 | 318 | 454.3 | 150 | 214.3 |
| 10 | 1.1 | 10.0 | 1.3 | 1.0 | 1.1 | ND | ND | ND | ND |
| 11 | 2.7 | 1.7 | 1.0 | 3.3 | 1.6 | ND | ND | ND | ND |
| 12 | 2.8 | 1.0 | 1.0 | 1.0 | 2.4 | 642 | 401.3 | 132 | 82.5 |
| 13 | 1.1 | 1.0 | 1.0 | 1.1 | 1.0 | 216 | 6.4 | 180 | 5.3 |
| 14 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 141 | 10.8 | 57 | 4.4 |
| 15 | 1.5 | 1.2 | 1.1 | 1.1 | 1.0 | 114 | 1.8 | 69 | 1.1 |
| 16 | 2.2 | 2.1 | 1.1 | 1.0 | 1.1 | 174 | 13.5 | 69 | 5.3 |
| 17 | 1.4 | 1.2 | 6.4 | 1.9 | 1.0 | 159 | 1.3 | 117 | 0.9 |
| 18 | 4.7 | 4.8 | 3.9 | 1.0 | 6.7 | 321 | 267.5 | 303 | 252.5 |
| 19 | 12.1 | 3.6 | 1.1 | 1.0 | 1.8 | 549 | 51.0 | 66 | 6.2 |
| 20 | 5.6 | 3.1 | 1.9 | 1.8 | 1.0 | 1 173 | 24.4 | 69 | 1.4 |
| 21 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 96 | 0.5 | 153 | 0.8 |
| 22 | 1.7 | 1.3 | 1.8 | 1.0 | 1.3 | 150 | 1.2 | 318 | 2.5 |
| 23 | 1.0 | 1.0 | 1.0 | 1.0 | 2.7 | 216 | 19.6 | 288 | 26.2 |
| 24 | 6.9 | 5.2 | 8.1 | 8.1 | 2.3 | ND | ND | ND | ND |
| 25 | 1.2 | 1.0 | 1.1 | 1.0 | 1.3 | ND | ND | ND | ND |
After isolation, malignant cells were selected by staining with anti-CD33, CD34, or CD19 and stained for the indicated NKG2DL. Expression was quantified as specific fluorescence index (SFI) values as described in “Patients, materials, and methods.” Levels of sMICA and sMICB molecules were determined as described in “Patients, materials, and methods.” Serum levels were then divided by the respective white blood count of each patient and are shown as normalized MICA and MICB (nMICA, nMICB). ND indicates not done.
MICA surface expression was detected in many cases (11 of 25 patients), whereas expression of the other NKG2DL was less frequent. MICB was expressed by cells from 7 patients and expression of ULBP molecules was also less frequent with 4 patients expressing ULBP1, 3 expressing ULBP2, and 5 expressing ULBP3 (Table 2). These data demonstrate that patient leukemia cells express NKG2DL at the cell surface to various extents. NKG2DL expression patterns did not correlate with particular leukemic entities, but rather appear to vary among individuals.
NK cell recognition of leukemic cells via NKG2D
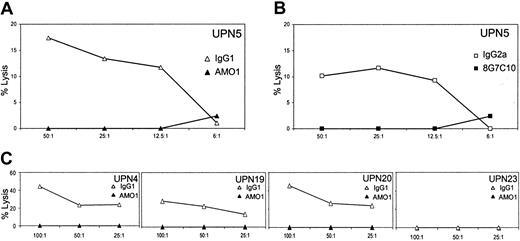
NKG2DL expression has been shown to stimulate antitumor activity of cytotoxic lymphocytes.8,13,19,23 Therefore, we addressed functional significance of NKG2DL expression on freshly isolated leukemia cells with regard to recognition by cytotoxic lymphocytes. To this point, patient cells were subjected to a chromium release assay using the NK cell line NKL. NKL expresses high levels of NKG2D, but only marginal levels of the activating receptors NKp30, NKp44, and NKp46 (our unpublished data, January 2003), and is therefore well suited to study NKG2D-mediated recognition.8 Leukemic cells of UPN 5 were lysed by NKL cells, and this lysis was critically dependent on the MICA/ NKG2D interaction, because it was completely inhibited by addition of anti-MICA (AMO-1; Figure 4A) or anti-NKG2D mAb (Figure 4B). Similar results were obtained with other highly MICA-expressing leukemia cells, namely, of UPN 4, UPN 12, and UPN 20, where lysis was also inhibited by preincubation with anti-MICA (AMO-1), but not by the appropriate isotype control. NKG2DL– leukemia cells of UPN 23 were ignored by NKL cells (Figure 4C). This demonstrates that NKG2DL expression renders patient leukemia cells susceptible for NK cell–mediated cellular cytotoxicity as it has been described for several cell lines of various tissue origins.8,15
NKG2D-mediated lysis of leukemic cells. PBMCs of patients were tested for recognition by NKL cells in a 5-hour chromium release assay. NKG2D-dependent killing was scrutinized by preincubation of UPN 5 PBMCs with anti-MICA mAb AMO1 (▴) or IgG1 isotype control antibodies (▵; A), or NKL cells with anti-NKG2D mAb 8G7C10 (▪) or IgG2a isotype control antibodies (□; B). PBMCs of UPN 4, UPN 12, UPN 20, and UPN 23 were preincubated with anti-MICA mAb AMO1 (▴) or IgG1 isotype control antibodies (▵) and assayed with NKL cells (C).
NKG2D-mediated lysis of leukemic cells. PBMCs of patients were tested for recognition by NKL cells in a 5-hour chromium release assay. NKG2D-dependent killing was scrutinized by preincubation of UPN 5 PBMCs with anti-MICA mAb AMO1 (▴) or IgG1 isotype control antibodies (▵; A), or NKL cells with anti-NKG2D mAb 8G7C10 (▪) or IgG2a isotype control antibodies (□; B). PBMCs of UPN 4, UPN 12, UPN 20, and UPN 23 were preincubated with anti-MICA mAb AMO1 (▴) or IgG1 isotype control antibodies (▵) and assayed with NKL cells (C).
Serum levels of sMICA and sMICB
MICA is released as a soluble form from the cell surface of malignant cells of epithelial origin and is present in sera of patients with gastrointestinal malignancies.21,22 To extend these studies to hematopoietic malignancies we investigated sMICA levels in sera of patients with leukemia. In addition, we used a newly established ELISA to determine serum levels of MICB. Sera of the 9 healthy volunteers contained only low levels of MICA and MICB close to the detection limit of the ELISA, ranging between 9 and 64 pg/mL for MICA and between 6 and 21 pg/mL for MICB with mean values of 34 pg/mL and 10 pg/mL, respectively (Table 2; Figure 5). Sera from 14 patients with AML showed MICA levels between 54 and 978 pg/mL with a mean of 335 pg/mL. One patient with AML (UPN 3) repeatedly presented with extraordinarily high MICA levels of about 10 ng/mL MICA and was not included in the statistical analysis. The levels of MICB were between 27 and 336 pg/mL with the mean being 121 pg/mL. In the 2 patients with ALL the MICA serum levels were 549 pg/mL and 321 pg/mL, MICB levels were 66 pg/mL and 303 pg/mL, respectively. The patient with NHL had serum levels of 924 pg/mL MICA and 288 pg/mL MICB, and the levels in patients with CML ranged between 96 and 1173 pg/mL with a mean of 409 pg/mL for MICA and between 69 and 318 pg/mL with a mean of 207 pg/mL for MICB. No serum had been obtained from the 2 patients with CLL. The differences of the levels of MICA and MICB in sera of healthy donors and patients with AML, ALL, and CML were statistically significant (P < .03) as determined by the Student t test (Table 3). This strong correlation of tumor incidence and elevated sMICA and sMICB levels clearly suggests that MIC molecules are released at significant amounts from leukemia cells in vivo. However, there was no clear correlation between the levels of sMICA or sMICB in serum and the expression of these molecules on the cell surface (Table 2). To evaluate whether a potential correlation was masked due to the highly differing amounts of malignant cells and whether the differences in the levels of sMICA and sMICB were merely due to differing white blood counts in the single patients, we normalized the serum levels of the MIC molecules by dividing with the respective white blood count (nMICA, nMICB). Again, no correlation between nMIC serum levels and the surface expression of the respective NKG2DLs could be determined (Table 2).
Levels of sMICA and sMICB in sera of patients and healthy donors. Serum samples from patients with different malignancies of hematopoietic origin and healthy volunteers were investigated by ELISA for sMICA or sMICB. The data shown are means of triplicates; n indicates number of donors in each group; —, mean of all measurements in the respective group.
Levels of sMICA and sMICB in sera of patients and healthy donors. Serum samples from patients with different malignancies of hematopoietic origin and healthy volunteers were investigated by ELISA for sMICA or sMICB. The data shown are means of triplicates; n indicates number of donors in each group; —, mean of all measurements in the respective group.
Levels of sMICA and sMICB
. | Lowest, pg/mL . | Highest, pg/mL . | Mean, pg/mL . | SD, pg/mL . | No. of samples . |
|---|---|---|---|---|---|
| Healthy | |||||
| MICA | 9 | 64 | 34 | 17 | 9 |
| MICB | 6 | 21 | 10 | 5 | 9 |
| AML | |||||
| MICA | 54 | 978 | 335 | 308 | 14 |
| MICB | 27 | 336 | 121 | 79 | 14 |
| ALL | |||||
| MICA | 321 | 549 | 435 | NA | 2 |
| MICB | 66 | 303 | 185 | NA | 2 |
| CML | |||||
| MICA | 96 | 1173 | 409 | 512 | 4 |
| MICB | 69 | 318 | 207 | 117 | 4 |
| NHL | |||||
| MICA | NA | 924 | NA | NA | 1 |
| MICB | NA | 288 | NA | NA | 1 |
. | Lowest, pg/mL . | Highest, pg/mL . | Mean, pg/mL . | SD, pg/mL . | No. of samples . |
|---|---|---|---|---|---|
| Healthy | |||||
| MICA | 9 | 64 | 34 | 17 | 9 |
| MICB | 6 | 21 | 10 | 5 | 9 |
| AML | |||||
| MICA | 54 | 978 | 335 | 308 | 14 |
| MICB | 27 | 336 | 121 | 79 | 14 |
| ALL | |||||
| MICA | 321 | 549 | 435 | NA | 2 |
| MICB | 66 | 303 | 185 | NA | 2 |
| CML | |||||
| MICA | 96 | 1173 | 409 | 512 | 4 |
| MICB | 69 | 318 | 207 | 117 | 4 |
| NHL | |||||
| MICA | NA | 924 | NA | NA | 1 |
| MICB | NA | 288 | NA | NA | 1 |
For each donor entity, the lowest and the highest value of sMICA and sMICB, the mean and the standard deviation (SD) and the number of donors (samples) is shown. NA indicates not applicable.
Discussion
Expression of NKG2DL is associated with malignant transformation.2,18 Broad expression of MICA/B was described for many epithelial tumors, although MICA and MICB were not discriminated.11 For the other human NKG2DLs, the ULBP1, ULBP2, and ULBP3 proteins, expression in vivo is yet undefined. Analysis of NKG2DL expression in vivo is of particular interest because recent experiments demonstrate that the NKG2D/NKG2DL system stimulates immune surveillance of tumors.19,20,23 Two reports described that tumor cells ectopically expressing NKG2DL were rejected by NK cells and CD8 αβ T cells in a mouse model19,23 and tumor immunity was even gained against the parental cell lines.19 The capacity of the NKG2DL-expressing tumor cell lines to stimulate tumor immunity in vivo was critically dependent on the expression levels of NKG2DL on the tumor cell surface. Because NK cell activity is guided by the balance of activating and inhibitory signals, and an enhanced NKG2DL expression is able to trigger NK cells overcoming inhibitory signals by MHC class I molecules,8,13 even modest NKG2DL down-regulation may critically influence NK cell reactivity.
To date, little is known about the role of the NKG2D/NKG2DL system in leukemia. So far, only expression of MICA on healthy tissues and epithelial tumors has been studied. Recently, Pende and coworkers showed NKG2DL expression for many hematopoietic cell lines and their NKG2D-dependent lysis by NK cells.15,24 However, no data addressing the expression and function of NKG2DL on leukemia cells ex vivo were provided. Another study demonstrated the presence of various NKG2DL mRNAs in leukemia cells ex vivo,25 but, as in our study, NKG2DL mRNA was also detected in PBMCs of healthy donors. However, previous studies13,26 and our own attempts failed to detect NKG2DL surface expression on PBMCs of healthy donors ex vivo. This strongly suggests that NKG2DL mRNA expression does not necessarily translate into NKG2DL surface expression.
In this study, we demonstrate that leukemia cells freshly isolated from patients express NKG2DL at the cell surface. NKG2DL expression patterns vary considerably between individual patients. Pende and coworkers hypothesized that most T-cell leukemia cell lines are characterized by a MICA–ULBP+ phenotype, whereas a MICA–ULBP– phenotype was detected in all AML lines and most B-cell lymphoma lines.15 We found expression of both MIC and ULBP molecules in a T-cell lymphoma and the 2 investigated ALL cases. Among the 15 cases with AML studied we detected significant surface levels of MICA in 6 and of MICB in 3 patients. Significant expression of ULBP molecules was detected in 3 cases with AML. Thus, NKG2DL expression patterns not only differed between individual patients, but there was also no significant correlation between expression of particular NKG2DL and certain leukemic entities. Interestingly, the 2 patients with ALL exhibited high levels of NKG2DL compared with other entities (Figure 3; Table 2). Overall, in 56% of the investigated patients malignant cells expressed one or more ligands for NKG2D on the cell surface. Nonmalignant PBMCs from the same patients did not reveal expression of NKG2DL as determined by selection of the CD19–, CD33–, or CD34– population, respectively, which is in agreement with previous reports that failed to detect NKG2DL expression on PBMCs of healthy individuals.13,26 The patient leukemia cells expressed generally high levels of MHC class I, which did not correlate with NKG2DL expression (data not shown). Detection of NKG2DL mRNA did not always translate into detectable NKG2DL surface expression. For example, UPN 5 showed elevated ULBP1 transcripts, but no ULBP1 molecules were detected with AUMO1 at the cell surface. This may be explained by a regulatory or mutational blockade of surface expression. For example, a Pro→Arg mutation at position 6 of the MICA*10 allele abolishes expression of functional MICA protein.27 In addition, rapid degradation or cleavage of NKG2DL from the cell surface may also account for the failure to detect NKG2DL expression.
In previous studies NKG2DL surface expression led to increased susceptibility of malignant or virus-infected cells to T cell– or NK cell–mediated lysis. Accordingly, NKG2DL expression rendered patient leukemia cells susceptible to lysis by NK cells despite their high MHC class I expression. This indicates that NKG2DL expression on leukemia cells may therefore be relevant for NK cell killing in vivo and extends the current arena for NKG2D function to hematopoietic tumors. In previous studies it has been demonstrated that NK cells can mediate an effect against leukemia cells, which is regulated by activating and inhibitory surface receptors.28 Moreover, data from allogeneic bone marrow transplantation suggest that NK cells may have potent antileukemia activity,29-33 and it has been shown that in patients with AML, NK cell activity correlates with the relapse-free survival in patients.34 These findings may be explained in part by the action of NKG2D.
Recently we demonstrated that MICA is released as a soluble form from the cell surface of epithelial tumor cells due to the activity of metalloproteinases.21 sMICA has also been shown to cause systemic down-regulation of NKG2D and to impair NKG2D functionality in tumor antigen–specific cytotoxic lymphocytes.22 Therefore, release of MICA may not only locally reduce immuno-genicity of tumor cells by diminishing ligand densities on the cell surface, but may also systemically affect NKG2D receptor function of cytotoxic effector cells. In this study we used the ELISA for MICA and a newly established ELISA for MICB to determine the levels of sMICA and sMICB in the sera of the investigated patients with leukemia and healthy volunteers. Whereas only low levels of both sMICA and sMICB close to the detection limit were detected in sera of healthy volunteers, the majority of sera from patients showed significantly elevated levels of sMICA and sMICB as validated by the Student t test. Interestingly, no correlation between surface expression and serum levels of MICA and MICB could be determined (Table 2). To test whether MIC serum levels were dependent on the amount of malignant cells in the blood we normalized the MIC serum values by dividing by the respective white blood count (Table 2). Again this failed to reveal any significant correlation. Also, comparison of MIC mRNA levels and levels of soluble MIC proteins in serum again did not reveal a clear correlation. However, note that for the 2 patients with the highest sMICA levels elevated levels of MICA transcripts were detected (compare UPN 4 and UPN 20 in Figure 2 and Table 2). Furthermore, we found elevated levels of sMICA and sMICB in sera of patients that revealed no surface expression of the respective MIC molecule. Because we demonstrated recently that MICA is shed from the cell surface by metalloproteinases21 and a similar mechanism has been observed for MICB (our unpublished data, October 2002), differing levels or activity of these endopeptidases in the sera of the patients might account for this observation. In addition, there might be an as yet undefined alternative splicing product that encodes soluble MIC molecules. Overall, the impact of NKG2DL surface expression and release in leukemias requires further evaluation. It will be important to determine to what extent the levels of NKG2DL surface expression and release of MIC molecules in patients with leukemia correlate with progress and outcome of hematopoietic malignancies and whether serum levels of MICA can be used as a prognostic marker. However, sMICA may be elevated also in other conditions as in autoimmune processes and during infections, which remains to be investigated. In addition, it is currently under study whether there is also a down-regulation of NKG2D expression on cytotoxic lymphocytes in patients with hematopoietic malignancies.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2003-01-0019.
Supported by grants from the Deutsche Krebshilfe (10-1921-Sa I) and the fortüne Program of the Faculty of Medicine of the Eberhard-Karls-University Tübingen (959-0-0).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
The authors thank Iris Kehrer for excellent technical assistance, Stefan Welte for help with the real-time PCR, and Peter Kufer for the anti-NKG2D antibodies. The authors want to express their gratitude to Graham Pawelec and Peter A. Kiener for critical reading of the manuscript.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal