Abstract
The expression of the vitamin K–dependent γ-glutamyl carboxylase gene in liver is developmentally regulated. Since the gene product catalyzes an essential post-translational modification of the vitamin K–dependent blood coagulation proteins, the regulation of carboxylase expression is critical for hemostasis. We analyzed the activity of the rat carboxylase gene 5′-regulatory DNA sequences in rat hepatoma cell lines at different states of differentiation. These studies demonstrated that the 2.6-kb 5′-flanking sequence has differentiation-dependent transcriptional activity. Transient gene expression assays, examining the effects of nested deletions and site-directed mutagenesis of putative regulatory sequences, together with electrophoretic mobility shift assays (EMSAs) were used to identify sequences critical for the developmentally regulated transcription of the rat carboxylase gene. We identified a DNA sequence (–76 to –65; GTTCCGGCCTTC) not known to bind to transcription factors, yet which functions as an upstream promoter element. In vivo genomic DNA footprinting confirms the presence of nuclear protein–DNA interactions at this site in the endogenous carboxylase gene in differentiated hepatoma cells. Therefore, this DNA sequence has specific nuclear protein–binding activity and functional properties consistent with a regulatory element that plays a critical role in the developmental expression of the carboxylase gene, and hence the regulation of vitamin K–dependent blood coagulation protein synthesis.
Introduction
The vitamin K–dependent γ-glutamyl carboxylase catalyzes, in what is the only known enzymatic reaction dependent on vitamin K, the posttranslational modification of glutamate to γ-carboxyglutamate (Gla).1 Gla synthesis on vitamin K–dependent proteins (VKDPs) in the endoplasmic reticulum2 requires vitamin K hydroquinone to carboxylate the γ-carbon of glutamate in an oxygen-dependent reaction.1 Gla confers calcium-binding properties to all VKDPs3 and is essential for their biologic activities.4,5 In higher vertebrates, the VKDPs have critical functions in blood coagulation (prothrombin, factor VII, factor IX, factor X, protein C, and protein S), bone formation (osteocalcin), and regulation of extracellular matrix calcification (matrix Gla protein).6-8 Gas6, a cell cycle–regulated receptor tyrosine kinase ligand9 and 2 proline-rich Gla-containing proteins10 are VKDPs whose in vivo activities are not well established. Developmental activities for these VKDPs have been hypothesized11 based on the human teratogenic effects of warfarin, a pharmacological inhibitor of Gla synthesis.12,13 Developmental functions for Gla-containing VKDPs also have been suggested by data developed in a chick embryogenesis model,14 in which vitamin K–dependent signaling events appear to be critical for orderly embryogenesis. Furthermore, mice unable to synthesize Gla due to targeted disruption of the carboxylase gene experience fetal demise or developmental abnormalities with features that resemble the human warfarin embryopathy syndrome,15 indicating that Gla synthesis is essential during mammalian development.
To determine the major sites of Gla synthesis in vivo during mammalian embryogenesis, we examined carboxylase gene expression in postimplantation rat embryos. Molecular analysis at the cellular level by in situ mRNA hybridization demonstrated that at early stages of postimplantation development, carboxylase was expressed predominantly in the neuroectodermal tissues that become the central nervous system and the mesodermal condensations that give rise to bone.11 In contrast, the carboxylase was not detected in the endodermally derived hepatocytes until late in development when the liver matures. The early expression of the carboxylase in developing neural and skeletal tissues, followed by a transition to liver in late embryogenesis, indicates that tissue-specific control of carboxylase expression and, therefore, Gla synthesis is developmentally regulated.
Postnatally, the liver hepatocytes are the major cellular site of carboxylase gene expression.11 The expression of the carboxylase gene is observed in many extrahepatic tissues, however, Northern analyses demonstrate that the highest levels of carboxylase mRNA are found in adult liver.11 Hepatocytes also are the major cellular site of vitamin K–dependent blood coagulation protein synthesis. Hence, this developmentally regulated induction of the carboxylase gene in liver is essential for hemostatic control beginning in late gestation. The sequences and factors that contribute to the regulation of carboxylase gene expression in hepatocytes are unknown. We therefore analyzed the promoter of the carboxylase gene to define elements critical for its expression. We identified a DNA sequence (GTTCCGGCCTTC) not known to be a transcription factor binding site that functions as an upstream promoter element. This DNA sequence binds specific nuclear proteins and is essential for carboxylase expression in cell lines that model hepatic development.
Materials and methods
Reporter plasmids
Luciferase reporter plasmids were generated in pGL3-Basic using the Erase-a-Base system (Promega, Madison, WI). RCBX-Luc16 was used to produce nested deletions from the 5′-end of the 5′-flanking sequence to create RCBX2634 through RCBX18. The sequence of the entire insert in RCBX2634 (–2634 to +75; GenBank accession number AF159140) and the extent of each 5′ deletion were determined by automated sequencing using oligonucleotide primers from GIBCO-BRL (Carlsbad, CA).
Polymerase chain reaction (PCR)–based mutagenesis was used to introduce a unique BglII site immediately upstream of the insert in RCBX64, for insertion of double-stranded oligonucleotides (Table 1) as single or multiple copies in the forward or reverse direction. For experiments that used the pGL2-promoter vector (Promega), oligonucleotides were subcloned into the BglII site in the multiple cloning site upstream of the SV40 early viral promoter sequence. Oligonucleotide-based site-directed mutagenesis with sequential PCR was used to delete the 12-bp sequence (–76 to –65; GTTCCGGCCTTC) in RCBX2634 to create RCBX2634D12 as described.17 DNA sequencing was used to confirm the accuracy of all constructs.
Oligonucleotide sequences used in the study
Name . | DNA Sequence . |
|---|---|
| WT | CTGCGGTTCCGGCCTTCACGA |
| A | CTGCGtggCCGGCCTTCACGA |
| B | CTGCGGTTaatGCCTTCACGA |
| C | CTGCGGTTCCGtaaTTCACGA |
| D | CTGCGGTTCCGGCCggaACGA |
| E | CTGCGtgTCCGGCCTTCACGA |
| F | CTGCGGTgaCGGCCTTCACGA |
| G | CTGCGGTTCatGCCTTCACGA |
| H | CTGCGGTTCCGtaCTTCACGA |
| I | CTGCGGTTCCGGCagTCACGA |
| J | CTGCGGTTCCGGCCTgaACGA |
| K | CgtCGGTTCCGGCCTTCACGA |
| L | CTGatGTTCCGGCCTTCACGA |
| M | CTGCGGTTCCGGCCTTCcaGA |
| N | CTGCGGTTCCGGCCTTCACtc |
Name . | DNA Sequence . |
|---|---|
| WT | CTGCGGTTCCGGCCTTCACGA |
| A | CTGCGtggCCGGCCTTCACGA |
| B | CTGCGGTTaatGCCTTCACGA |
| C | CTGCGGTTCCGtaaTTCACGA |
| D | CTGCGGTTCCGGCCggaACGA |
| E | CTGCGtgTCCGGCCTTCACGA |
| F | CTGCGGTgaCGGCCTTCACGA |
| G | CTGCGGTTCatGCCTTCACGA |
| H | CTGCGGTTCCGtaCTTCACGA |
| I | CTGCGGTTCCGGCagTCACGA |
| J | CTGCGGTTCCGGCCTgaACGA |
| K | CgtCGGTTCCGGCCTTCACGA |
| L | CTGatGTTCCGGCCTTCACGA |
| M | CTGCGGTTCCGGCCTTCcaGA |
| N | CTGCGGTTCCGGCCTTCACtc |
The sense strand sequences of the double-stranded wild-type (WT) and mutant (A through N) synthetic oligonucleotides are shown and correspond to -81 to -61 in the rat carboxylase 5′-flanking region. The 12-bp sequence from -76 to -65 is indicated in bold in each of the oligonucleotide sequences. The mutated bases within the 12-bp sequence of oligonucleotides A through J are indicated in lowercase bold italics. The mutated bases that reside outside the 12-bp sequence of oligonucleotides K through N are indicated in lowercase nonbold italics.
Transient transfection and reporter gene assays
Rat hepatoma H4IIEC3 cells were cultured as described16 ; dedifferentiated rat hepatoma variant cell lines H5, HF1, C2, and C2-Rev7, all derived from H4IIEC3, were obtained from the European Collection of Cell Cultures (Salisbury, Wiltshire, United Kingdom) and cultured as recommended in Coon's Modified Ham's F-12 (BioFluids, Rockville, MD), 2 mM l-glutamine, 1× penicillin/streptomycin, 5% (vol/vol) fetal bovine serum (Hyclone, Logan, UT). Transient transfection of all cell lines were performed at a density of 40 000 cells/cm2 using lipofectin (GIBCO-BRL) as described previously.16 To control for transfection efficiency, cells were cotransfected with a control Renilla luciferase reporter plasmid (pRL-TK; Promega). The cell extracts were prepared and assayed as described.16
Electrophoretic mobility shift assays
Nuclear extracts from cultured rat hepatoma cells were prepared as described.18 Protein concentration was determined using the Bio-Rad Protein Assay (Hercules, CA) with bovine serum albumin as a standard. Rat liver, lung, and brain nuclear extracts were obtained from Active Motif (Carlsbad, CA). For each electrophoretic mobility shift assay (EMSA) reaction, nuclear extract protein (5 μg) and poly(dG-dC)-poly(dG-dC) (1.25 μg, Pharmacia, Uppsala, Sweden) in a total volume of 6 μL was mixed with 5 μL TD (50 mM Tris [tris(hydroxymethyl)aminomethane] and 5 mM dithiothreital [DTT]) and incubated at room temperature for 60 minutes. Each reaction was then supplemented with 1 μL radiolabeled double-stranded DNA probe (1 ng/μL) and incubated for an additional 15 minutes in a final KCl concentration of 50 mM. Samples were then supplemented with 50% (vol/vol) glycerol (3 μL) and analyzed by electrophoresis through nondenaturing 4% polyacrylamide gels in 1 × TGE pH 8.5 (25 mM Tris-Cl, 190 mM Glycine, 1.3 mM EDTA [ethylenediaminetetraacetic acid]) at 4°C. Competition assays were performed by adding unlabeled double-stranded oligonucleotides to the reaction during the last 15 minutes of the initial 60-minute incubation, prior to the addition of radiolabeled probe. All double-stranded oligonucleotides were designed to have 5′-GATC overhangs after annealing, to permit subcloning or radiolabeling with α32P-dCTP (NEN) using the large fragment of DNA polymerase I.
In vivo genomic footprinting
Genomic dimethyl sulfate (DMS) footprinting by ligation-mediated PCR (LM-PCR) was used to analyze the carboxylase gene in the context of native chromatin in H4IIEC3 cells. Cells were grown to 75% confluence (300 000 cells per cm2), at which time monolayers of cells were treated with 0.1% (vol/vol) DMS for 2 minutes at 37°C to methylate DNA in vivo, washed in phosphate-buffered saline (PBS), and immediately lysed to isolate genomic DNA. Alternatively, untreated parallel samples of cells were washed and lysed to isolate DNA for in vitro methylation as described.19 In either case, methylated DNA was purified, cleaved with piperidine, and analyzed using the following nested oligonucleotide primer sets: reverse: 5′-TACCTGAGGCGGGAGCAGCT-3′; 5′-AGCAGCTCGTGCAGAGCCACGGT-3′; 5′-GCAGCTCGTGCAGAGCCACGGTGGAC-3′; forward: 5′-ACTCCTTCGCTCCTGCGCAG-3′; 5′-GCGCAGCCCATCCCTAAAGCTCC-3′; 5′-GCGCAGCCCATCCCTAAAGCTCCTCGC-3′; unidirectional staggered linker primers were described previously.19 Radiolabeled products of in vitro and in vivo LM-PCR reactions were analyzed following electrophoresis through 6% polyacrylamide-urea gels.
Results
Carboxylase promoter activity is dependent on hepatoma cell line differentation
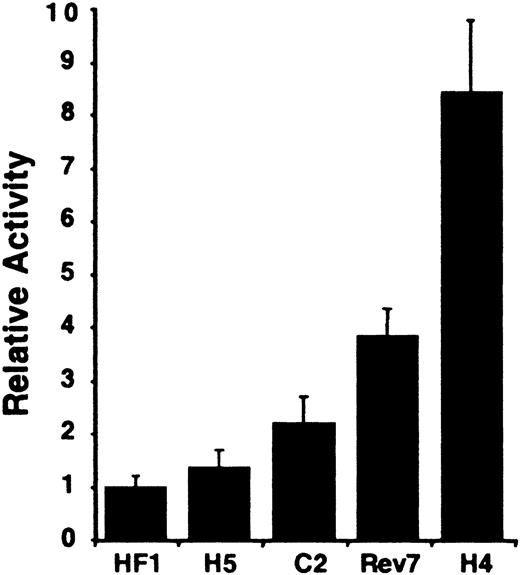
To assess whether transcriptional activity of the carboxylase gene depends on the stage of hepatocyte differentiation, we used transient gene expression assays to analyze carboxylase-mediated reporter gene activity in a series of cell lines that model hepatic development. H4IIEC3 rat hepatoma cells correspond to terminally differentiated hepatocytes.20 They express molecular markers of mature hepatocytes: high levels of albumin, transferrin, inducible tyrosine aminotransferase (TAT) and alanine aminotransferase (AAT), and gluconeogenic enzymes that permit their growth in glucose-free medium.20 In contrast, H5, HF1 and C2 cells are independent dedifferentiated clonal lines derived from H4IIEC3 cells.20-23 These morphologic variants lack or have significantly diminished expression of the aforementioned mature liver markers and of the hepatic transcription factors associated with cell differentiation, C/EBP, HNF-1α, and HNF-4,24-26 distinguishing them from the parental H4IIEC3 cell line. C2-Rev7, a stable revertent cell line derived from C2, displays an intermediate phenotype with normal hepatocyte morphology and partially restored liver-specific and gluconeogenic gene expression23 and expression of the differentiation-associated hepatic transcription factors.24,26 The transcriptional activity of the 2.6-kb 5′-flanking sequence of the carboxylase gene was lowest in the most undifferentiated HF1 and H5 lines (Figure 1). Carboxylase reporter gene activity increased commensurately with markers of differentiation. When compared with HF1 cells, a 2.2-fold increase in reporter gene activity was seen in C2 cells that have low but measurable basal and induced levels of TAT and AAT. A 3.8-fold increase was observed in C2-Rev7 cells, and an 8.4-fold increase was measured in the fully differentiated H4IIEC3 cells. These data demonstrate that the transcriptional activity of the 2.6-kb 5′-flanking sequence parallels the degree of hepatic differentiation.
Rat vitamin K–dependent carboxylase promoter activity in differentiated and dedifferentiated rat hepatoma cell lines. The different cell lines are indicated below the figure. Rev7 indicates C2-Rev7; H4, H4IIEC3. The average activities shown are relative to that measured in the dedifferentiated HF1 cell line variant, n = 3. Error bars indicate SD.
Rat vitamin K–dependent carboxylase promoter activity in differentiated and dedifferentiated rat hepatoma cell lines. The different cell lines are indicated below the figure. Rev7 indicates C2-Rev7; H4, H4IIEC3. The average activities shown are relative to that measured in the dedifferentiated HF1 cell line variant, n = 3. Error bars indicate SD.
An upstream promoter sequence is critical for carboxylase reporter gene expression in H4IIEC3 cells
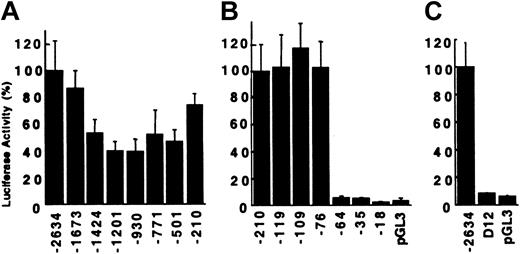
To identify sequences critical for promoter activity of the carboxylase gene, a series of unidirectional 5′-nested deletions of the 2.6-kb regulatory region were generated. The resultant reporter plasmids were transiently transfected into the mature H4IIEC3 cells. Deletion of the DNA sequences from –2634 to –1201 resulted in a 60% loss of activity (Figure 2A). Deletion of an additional 271 bp to –930 had no effect, while deletion to –210 restored activity to 75% of the full-length construct. Additional deletions were analyzed to define the minimal sequences required for activity. Deletion from –210 to –119, including an Sp1 consensus binding site (–127 to –118), had no influence on transactivation (Figure 2B), nor did deletion to –76, including a second Sp1 site (–108 to –99). In contrast, deletion of 12 additional bases (–76 to –65) reduced activity from 103% to 5.7%, corresponding to a 38-fold decrease in reporter gene expression when corrected for 3.1% background activity. RCBX64 was unable to function as a minimal promoter, despite containing carboxylase gene sequences from –64 to +75 that include consensus-binding sites for c-ets-1 (–50 to –41) and Sp1 (–13 to –4). Further deletion to –35 and –18 had no significant effect. The function of this 12-bp sequence from –76 to –65 was analyzed in the context of the full-length reporter gene construct (RCBX2634). Deletion of the 12-bp sequence resulted in a 43-fold decrease in reporter gene activity (Figure 2C), similar to the effect seen in the context of the minimal promoter construct. These data demonstrate a critical function for the 12-bp sequence in the context of both the minimal carboxylase promoter, as well as in the context of the 2.6-kb 5′-flanking sequence.
Transcriptional activity of rat vitamin K–dependent carboxylase mutant reporter plasmids in H4IIEC3 cells. The extent of the 5′-flanking sequence contained in each reporter plasmid is indicated below the figure. (A) Reporter plasmids RCBX2634 through RCBX210. The average normalized luciferase activity relative to RCBX2634 is shown, n = 5. Error bars = SD. (B) Reporter plasmids RCBX210 through RCBX18. The average activity relative to RCBX210 is shown, n = 5. pGL3 = pGL3-Basic without insert. (C) Deletion of the 12-bp sequence (–76 to –65) from the 2634 bp 5′-flanking region (D12) abolishes transcriptional activity of RCBX2634. D12 = RCBX2634D12, n = 3.
Transcriptional activity of rat vitamin K–dependent carboxylase mutant reporter plasmids in H4IIEC3 cells. The extent of the 5′-flanking sequence contained in each reporter plasmid is indicated below the figure. (A) Reporter plasmids RCBX2634 through RCBX210. The average normalized luciferase activity relative to RCBX2634 is shown, n = 5. Error bars = SD. (B) Reporter plasmids RCBX210 through RCBX18. The average activity relative to RCBX210 is shown, n = 5. pGL3 = pGL3-Basic without insert. (C) Deletion of the 12-bp sequence (–76 to –65) from the 2634 bp 5′-flanking region (D12) abolishes transcriptional activity of RCBX2634. D12 = RCBX2634D12, n = 3.
Analysis of the upstream promoter element demonstrates specific nuclear protein–DNA interactions in vitro and in vivo
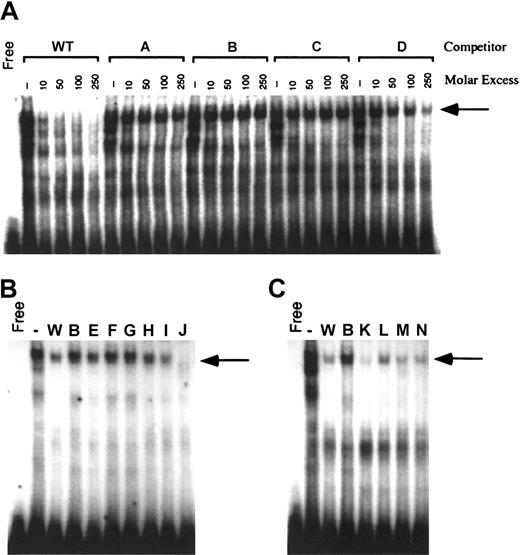
The contribution of the 12-bp promoter element to reporter gene activity suggested that these sequences bound nuclear proteins critical for carboxylase expression. Nuclear proteins from H4IIEC3 cells were therefore used in EMSA analyses with a radiolabeled DNA probe containing the 12-bp sequence. As demonstrated in Figure 3A, incubation of H4IIEC3 nuclear extracts with the wild-type (WT) probe generates several bands with retarded mobility. Self-competition by 10-, 50-, 100-, and 250-fold molar excess of unlabeled WT oligonucleotide efficiently competed for the generation of these complexes, demonstrating their specificity. To define the bases essential for the formation of these complexes, mutant double-stranded oligonucleotides (A, B, C, and D; Table 1) were used in competition assays. Mutations were made in groups of 3 adjacent bases in the 12-bp region shown to be important for transcriptional activity. Purines were substituted with noncomplementary pyrimidines, and pyrimidines were substituted with noncomplementary purines. The mutations in oligonucleotide D moderately attenuated competition for the upper complex, whereas competition for the upper complex was abolished when mutant oligonucleotides A, B, and C were used (Figure 3A, arrow). The lower 2 complexes were equally competed for by all 4 mutant oligonucleotides as shown. To further define the bases in the 12-bp region that are required for the generation of this DNA-protein complex, additional double-stranded mutant oligonucleotides containing sequential 2-bp mutations (E, F, G, H, I, and J; Table 1) were analyzed in competition EMSAs using 100-fold molar excess of oligonucleotide competitors. Mutant J competed efficiently for the upper complex (Figure 3B, arrow). This result is consistent with the partial attenuation of DNA-protein binding that was previously demonstrated with oligonucleotide D. Oligonucleotide D contains mutations in the last 3 bases of the 12-bp sequence (T-10 to G, T-11 to G, C-12 to A). Since the same base substitutions in position 11 and 12 of oligonucleotide J have no effect on competition for the upper complex, we conclude that the partial attenuation of DNA-protein binding by mutations in D is due to the single substitution at T-10. Mutations in oligonucleotides E through H also are critical since they attenuate competition for the upper complex, similar to what was seen with oligonucleotides A through C. Competition EMSAs with oligonucleotides that contain sequential 2-bp mutations outside the region of the 12-bp core sequence (K, L, M, and N; Table 1) reveal they have activities similar to the WT oligonucleotide (Figure 3C), demonstrating that mutations outside the 12-bp region do not interfere with the DNA-protein interactions of the 12-bp motif. Taken together, these data demonstrate that specific residues extending over the first 10 bases in the 12-bp region of the sequence are critical for the DNA-protein interactions that generate the upper complex.
Generation of specific H4IIEC3 nuclear protein–DNA complexes by the sequences from –81 to –61 of the rat carboxylase gene 5′-flanking region. Panel A shows cold competitor wild-type (WT) or mutant oligonucleotides with mutations made in groups of 3 adjacent bases (A, B, C, D) were added in the molar excesses indicated above each lane. Panel B shows cold competitor WT, B, or mutant oligonucleotides with mutations made in groups of 2 adjacent bases (E, F, G, H, I, J) were added in 100-fold molar excess, as indicated above each lane. Panel C shows cold competitor WT, B, or mutant oligonucleotides with mutations made outside the 12-bp core sequence, in groups of 2 adjacent bases (K, L, M, N), were added in 100-fold molar excess, as indicated above each lane. The arrow in all cases indicates the position of the upper complex referred to in “Results” and “Discussion.” Free indicates no added nuclear protein; (–), no added competitor.
Generation of specific H4IIEC3 nuclear protein–DNA complexes by the sequences from –81 to –61 of the rat carboxylase gene 5′-flanking region. Panel A shows cold competitor wild-type (WT) or mutant oligonucleotides with mutations made in groups of 3 adjacent bases (A, B, C, D) were added in the molar excesses indicated above each lane. Panel B shows cold competitor WT, B, or mutant oligonucleotides with mutations made in groups of 2 adjacent bases (E, F, G, H, I, J) were added in 100-fold molar excess, as indicated above each lane. Panel C shows cold competitor WT, B, or mutant oligonucleotides with mutations made outside the 12-bp core sequence, in groups of 2 adjacent bases (K, L, M, N), were added in 100-fold molar excess, as indicated above each lane. The arrow in all cases indicates the position of the upper complex referred to in “Results” and “Discussion.” Free indicates no added nuclear protein; (–), no added competitor.
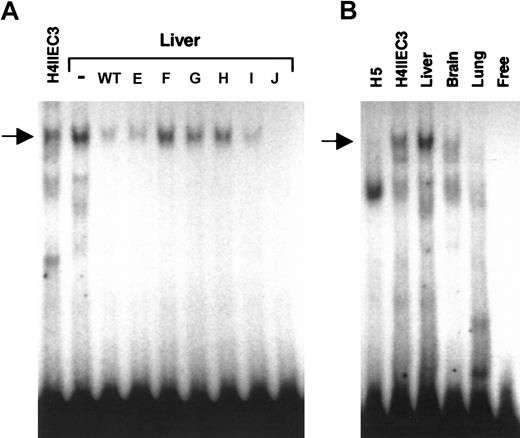
Since sequences in the 12-bp region of the wild-type DNA probe were important for generating a specific DNA-protein complex with nuclear extracts from well-differentiated H4IIEC3 cells, we evaluated the ability of this probe to bind proteins in nuclear extracts from adult rat liver, as well as from dedifferentiated H5 cells, and rat brain and lung to determine if the protein-DNA interactions were restricted to cells with a mature liver phenotype. EMSA analysis with rat liver nuclear extract revealed a prominent DNA-protein complex that comigrated with the H4IIEC3 upper complex and that had an identical competition pattern using wild-type and mutant oligonucleotides (Figure 4A, arrow). Comparative EMSA analysis using the same amount of nuclear extract from each source revealed that the upper complex was absent in the nuclear protein extract from H5 cells and rat lung; however, a weak signal that comigrated with the upper complex was observed with nuclear extract from rat brain (Figure 4B, arrow). While the expression of the factor(s) that interact with the carboxylase upstream promoter element may not be restricted to liver, consistent with low-level extrahepatic expression of the carboxylase gene,11 the specific DNA binding activity is clearly enriched in adult liver and in cells such as H4IIEC3 cells with a differentiated hepatic phenotype.
Comparative EMSA analyses of nuclear extracts from H4IIEC3 cells and adult rat liver, brain, lung and dedifferentiated H5 cells, for binding to DNA sequences from –81 to –61 of the rat carboxylase gene 5′-flanking region. Panel A shows cold competitor WT or mutant oligonucleotides (E, F, G, H, I, J) were added in 100-fold molar excess, as indicated above each lane, in EMSAs with adult rat liver nuclear extract. (–) indicates no added competitor. H4IIEC3 nuclear extract without competitor is shown in the left lane, as indicated. The arrow indicates the position of the upper complex referred to in “Results” and “Discussion.” Panel B shows radiolabeled wild-type double-stranded oligonucleotide containing the 12-bp core sequence was used in EMSA analysis with 5 μg total nuclear protein in each sample. The source of nuclear extract is indicated above each lane. The arrow indicates the position of the specific upper complex referred to in “Results” and “Discussion.” Free indicates no added nuclear protein.
Comparative EMSA analyses of nuclear extracts from H4IIEC3 cells and adult rat liver, brain, lung and dedifferentiated H5 cells, for binding to DNA sequences from –81 to –61 of the rat carboxylase gene 5′-flanking region. Panel A shows cold competitor WT or mutant oligonucleotides (E, F, G, H, I, J) were added in 100-fold molar excess, as indicated above each lane, in EMSAs with adult rat liver nuclear extract. (–) indicates no added competitor. H4IIEC3 nuclear extract without competitor is shown in the left lane, as indicated. The arrow indicates the position of the upper complex referred to in “Results” and “Discussion.” Panel B shows radiolabeled wild-type double-stranded oligonucleotide containing the 12-bp core sequence was used in EMSA analysis with 5 μg total nuclear protein in each sample. The source of nuclear extract is indicated above each lane. The arrow indicates the position of the specific upper complex referred to in “Results” and “Discussion.” Free indicates no added nuclear protein.
To examine whether the sequences critical for the generation of the upper complex were essential also for functional activity, we evaluated the effect of mutations on carboxylase-mediated transcription in transient gene expression assays. One copy of the wild-type or mutant oligonucleotides was subcloned in the forward orientation into RCBX64. Transient transfection analyses in H4IIEC3 cells demonstrated that the wild-type oligonucleotide increased the activity of RCBX64 by 6.7-fold (Figure 5A), whereas mutant oligonucleotides A, B, and C had no influence on transcriptional activity, and mutant oligonucleotide D increased transcriptional activity 2.7-fold (Figure 5A). The transcriptional activity of these DNA sequences therefore correlates with their ability to bind the nuclear proteins present in the upper complex on EMSA. To refine this analysis, the double-stranded mutant oligonucleotides containing sequential 2-bp mutations (E, F, G, H, I, and J; Table 1) also were evaluated. Mutant oligonucleotides E, F, and G did not enhance transcriptional activity (Figure 5B), whereas oligonucleotides H and I led to a 2.6-fold and 1.6-fold increase, respectively. In contrast, transcriptional activity of mutant oligonucleotide J was analogous to that of the wild-type sequences. The functional effects of these 2-bp mutations, like those of the 3-bp mutations, correlated with their ability to compete for the generation of the upper complex in EMSAs. These data, coupled with the EMSA data, demonstrate that the first 10 bases in the 12-bp region GTTCCGGCCT (–76 to –67) are critical for nuclear protein–DNA interactions and transcriptional activity.
Enhancement of RCBX64 transcriptional activity by double-stranded oligonucleotides containing the 12-bp motif. A single copy of each double-stranded oligonucleotide (Table 1) was subcloned into the BglII site of RCBX64 in the forward direction as indicated below the figure. The transcriptional activity of each plasmid was determined in H4IIEC3 cells. The activity is shown as the average fold-stimulation relative to RCBX64 without an oligonucleotide (–), n = 3. Error bars indicate SD. (A) WT and 3-bp mutants A through D. (B) WT and 2-bp mutants E through J.
Enhancement of RCBX64 transcriptional activity by double-stranded oligonucleotides containing the 12-bp motif. A single copy of each double-stranded oligonucleotide (Table 1) was subcloned into the BglII site of RCBX64 in the forward direction as indicated below the figure. The transcriptional activity of each plasmid was determined in H4IIEC3 cells. The activity is shown as the average fold-stimulation relative to RCBX64 without an oligonucleotide (–), n = 3. Error bars indicate SD. (A) WT and 3-bp mutants A through D. (B) WT and 2-bp mutants E through J.
The effect of copy number and orientation also was evaluated in H4IIEC3 cells. Three copies of the wild-type oligonucleotide increased the activity of RCBX64 by 5.8-fold, whereas 6 copies increased the activity by 9.0-fold (Figure 6A). The observed enhancement of activity was greater than that seen with one copy of the sequence and correlated with copy number. Multiple copies of the oligonucleotide with mutations B or C had no significant influence on reporter gene expression. When the same series of multimerized oligonucleotides was tested in the reverse orientation, a similar result was observed. To evaluate whether these sequences could enhance transactivation by a heterologous viral promoter, they were placed upstream to the SV40 early viral promoter. As shown in Figure 6B, these sequences increased reporter gene activity 2.9-fold, whereas the mutant sequence B had no effect. These data demonstrate that the 12-bp sequence has copy number–dependent and orientation-independent enhancement of the carboxylase minimal promoter and is capable of enhancing expression driven by a heterologous viral promoter.
Transactivation mediated by the 12-bp enhancer is copy number dependent and orientation independent. (A) The double-stranded oligonucleotide, its copy number and orientation (forward, reverse) with respect to the minimal promoter in RCBX64 (–) is indicated. The average activity of each reporter plasmid in H4IIEC3 cells relative to that of RCBX64 is shown, n = 3. Error bars indicate SD. (B) The effect of the 12-bp sequence on a heterologous viral promoter in H4IIEC3 cells was analyzed. WT indicates 1 copy of the wild type 12-bp sequence in the forward direction; Mut, 1 copy of mutant oligonucleotide B in the forward orientation. The average activities of these reporter plasmids are shown relative to that of the parental minimal SV40 viral promoter vector (–), n = 3.
Transactivation mediated by the 12-bp enhancer is copy number dependent and orientation independent. (A) The double-stranded oligonucleotide, its copy number and orientation (forward, reverse) with respect to the minimal promoter in RCBX64 (–) is indicated. The average activity of each reporter plasmid in H4IIEC3 cells relative to that of RCBX64 is shown, n = 3. Error bars indicate SD. (B) The effect of the 12-bp sequence on a heterologous viral promoter in H4IIEC3 cells was analyzed. WT indicates 1 copy of the wild type 12-bp sequence in the forward direction; Mut, 1 copy of mutant oligonucleotide B in the forward orientation. The average activities of these reporter plasmids are shown relative to that of the parental minimal SV40 viral promoter vector (–), n = 3.
To analyze nuclear protein–DNA interactions in the carboxylase promoter in the context of native chromatin, we used in vivo footprinting to complement the functional in vitro studies described above. The technique incorporates ligation-mediated PCR (LM-PCR) to detect specific DMS modifications of guanine residues in genomic DNA. Oligonucleotides were designed to analyze the sense and antisense strands of the proximal promoter, extending from the upstream Sp1 site (–127 to –118) to the transcription start site at +1. The guanine methylation pattern of H4IIEC3 genomic DNA exposed to DMS in vitro reveals all the guanines in the sense strand and antisense strand of the sequence (Figure 7, lanes 2 and 4, respectively; Figure 8). The in vivo guanine methylation pattern of the sense strand reveals strong protection (hypomethylation) of selected guanines (solid circles) in the regions of each of the 3 predicted Sp1 sites (–127 to –118, –108 to –99, and –13 to –4) and at residues –21 and –19 (Figure 7, lane 1; Figure 8). While guanines at –71 and –70 within the 12-bp sequence are not protected, enhanced methylation at guanine –76 (Figure 7, lane 1, plus sign; Figure 8) and hypomethylation at guanine –62 are consistent with nuclear protein interactions at or near the 5′ and 3′ ends of the 12-bp sequence, respectively. Partial protection of guanines within the predicted c-ets-1 binding site (–50, –43, and –42) and 3′ of this site (–33, –31, and –27) also is demonstrated in the sense strand. In vivo analysis of the antisense strand confirmed protection of residues in the 2 upstream Sp1 binding sites (Figure 7, lane 3; Figure 8). The in vivo footprint on the antisense strand also demonstrates protection of guanine –63, immediately 3′ of the 12-bp sequence; enhanced methylation of guanine –68, and protection of guanine –72, each within the 12-bp sequence. The protection and enhancement of additional guanine residues in the region of the putative c-ets-1 site and sequences 3′ of this site in the proximal promoter also are indicated. These findings demonstrate in vivo nuclear protein–DNA interactions in the carboxylase promoter as they occur in H4IIEC3 cells that are actively transcribing the carboxylase gene. Notably, they indicate that the genomic DNA sequences corresponding to the 12-bp promoter element interact with nuclear proteins, supporting a functional in vivo role for these sequences.
Ligation-mediated PCR-based in vivo genomic DNA footprinting of the vitamin K–dependent carboxylase gene promoter in H4IIEC3 cells. Guanine methylation patterns in the sense strand of the gene (lanes 1 and 2). Guanine methylation patterns of the antisense strand of the gene (lanes 3 and 4). In vivo methylation patterns of genomic DNA (lanes 1 and 3, left side of each pair of lanes) are compared with in vitro methylation patterns (lanes 2 and 4, right side of each pair of lanes). Relative hypomethylation of guanines in vivo is indicated with a solid dot (•). Relative hypermethylation of guanines in vivo is indicated with a plus sign (+). The nucleotide number is indicated on the left of each footprint. The start site of transcription at + 1 is indicated with an arrow. Vertical lines indicate sequences in the region of the 12-bp upstream promoter element.
Ligation-mediated PCR-based in vivo genomic DNA footprinting of the vitamin K–dependent carboxylase gene promoter in H4IIEC3 cells. Guanine methylation patterns in the sense strand of the gene (lanes 1 and 2). Guanine methylation patterns of the antisense strand of the gene (lanes 3 and 4). In vivo methylation patterns of genomic DNA (lanes 1 and 3, left side of each pair of lanes) are compared with in vitro methylation patterns (lanes 2 and 4, right side of each pair of lanes). Relative hypomethylation of guanines in vivo is indicated with a solid dot (•). Relative hypermethylation of guanines in vivo is indicated with a plus sign (+). The nucleotide number is indicated on the left of each footprint. The start site of transcription at + 1 is indicated with an arrow. Vertical lines indicate sequences in the region of the 12-bp upstream promoter element.
Sequence of the proximal 5′-flanking region of the vitamin K–dependent carboxylase gene. Transcription factor consensus binding sites are boxed and labeled as indicated. UPE indicates upstream promoter element; a solid dot (•), G hypomethylation as indicated in Figure 7; and a plus sign (+), G hypermethylation as indicated in Figure 7. Numbering is relative to the transcription start site at + 1, indicated with an arrow.
Sequence of the proximal 5′-flanking region of the vitamin K–dependent carboxylase gene. Transcription factor consensus binding sites are boxed and labeled as indicated. UPE indicates upstream promoter element; a solid dot (•), G hypomethylation as indicated in Figure 7; and a plus sign (+), G hypermethylation as indicated in Figure 7. Numbering is relative to the transcription start site at + 1, indicated with an arrow.
Discussion
In this study, we analyzed the promoter and adjacent 5′-flanking sequences of the rat carboxylase gene to identify DNA elements that regulate its expression in differentiated hepatocytes. Using well-characterized cell culture models of hepatocyte differentiation, we demonstrated that the 2.6-kb 5′-flanking sequence of the rat carboxylase gene is capable of directing reporter gene expression in a differentiation stage-specific manner, with maximal expression activity in well-differentiated H4IIEC3 cells. This finding parallels our previous data demonstrating that expression in fetal rat liver increased with development.11 Data generated in the H4IIEC3 cell line using reporter gene constructs containing deletions and point mutations identified an upstream promoter sequence from –76 to –65, relative to the carboxylase transcription start site, that was essential for high-level expression in these cells. This DNA sequence binds to nuclear proteins isolated from the H4IIEC3 cells, and in vivo DMS footprinting confirms that nuclear protein–DNA interactions occur in this region of the endogenous carboxylase gene in the H4IIEC3 cells. The sequence is not only critical for the activity of the minimal carboxylase promoter; but it also is required for the activity of the 2.6-kb 5′-flanking sequence in differentiated hepatoma cells. EMSAs using the wild-type DNA probe containing the 12-bp sequence generated a prominent nuclear protein–DNA complex when extracts from well-differentiated H4IIEC3 cells were used, and high levels of the DNA binding protein(s) also were detected in adult rat liver nuclear extract. However, a prominent complex was not observed using nuclear extracts isolated from the dedifferentiated H5 cells, correlating with a 6-fold decrease in transactivation by these sequences in the H5 cells. While these data indicate that transactivation via the 12-bp upstream promoter element contributes to differentiation-dependent expression of the carboxylase in liver cells, it is likely that additional sequences and transcription factors contribute to the observed differences.
Variability in the expression of liver-enriched transcription factors, regulated by and including HNF-4 and HNF-1, is in part responsible for the variable degree of differentiation that distinguishes the hepatoma cell lines used in our studies.26 It has been shown that the stable reintroduction of HNF4 in dedifferentiated H5 cells results in only partial restoration of liver-specific gene expression,27 with pleiotropic effects on the H5 hepatic cell phenotype.28 Furthermore, cooperative interactions between proteins that bind to regulatory regions separated by many kilobases are known to influence developmentally regulated transcription of liver genes. For example, the HNF3γ gene is transactivated by an HNF1 enhancer located 16 kb downstream of the promoter. Nuclear protein–DNA interactions at this downstream enhancer site transactivate reporter gene expression in hepatoma cells and in developing liver in transgenic mice.29 Thus, although nuclear protein–DNA interactions at the upstream promoter sequence from –76 to –65 are critical for optimal transactivation by carboxylase regulatory elements, they are unlikely to be solely responsible for the developmentally regulated expression of the carboxylase gene in liver cells.
In addition to remote regulatory sequences, nuclear protein–DNA interactions in the proximal 5′-flanking sequence of the carboxylase gene may also exert significant regulatory effects on the upstream promoter element. By analogy to other TATA-less genes with a similar organization of the proximal promoter, carboxylase promoter sequences that bind to basal transcription factors are also likely to be important. However, the in vivo interactions between basal transcription factors that regulate TATA-less genes and factors that bind to gene specific regulatory elements to enhance or repress their transcriptional initiation are poorly understood. Sp1 is known to play an essential role in recruiting the basal transcription factors to TATA-less promoters used by RNA polymerase II.30 Since the carboxylase is a TATA-less gene with several GC boxes in the proximal promoter region, Sp1 is likely to be one of these important factors. Indeed, most mammalian TATA-less promoters have one or more GC boxes in addition to other regulatory elements upstream of the transcription start site, implicating a central role for Sp1 in transcriptional activation of TATA-less genes.31,32 Sp1 is believed to function via multiple protein-protein interactions to stabilize an initiation complex and TFIID on TATA-less promoters through a novel tethering activity.32 Alternatively, Sp1 may activate genes by interactions with cell-specific factors, including liver-enriched factors.33 A model that probes the mechanism of transcriptional activation by Sp1 demonstrates the importance of upstream coactivators32 and leads us to propose that nuclear protein–DNA interactions in the region of the 12-bp element are required for induction of carboxylase expression by Sp1. Although in vivo footprinting of the carboxylase promoter detected nuclear protein–DNA interactions at all 3 Sp1 sites in the sequences studied, the 2 upstream sites were not required for transcriptional activation in H4IIEC3 cells, pointing to a critical role for the downstream Sp1 site. In vivo genomic footprinting confirmed nuclear protein–DNA interactions at the downstream c-ets-1 and the Sp1 sites (–50 to –41 and –13 to –4, respectively), and these sites are present in the same relative locations in other TATA-less promoters, where they are essential for their expression.34,35 Without the 12-bp upstream promoter element, none of these downstream sequences were able to support carboxylase promoter activity. However, the difference in the transcriptional activity of the upstream promoter element in its native context (38-fold effect, Figure 2B) compared to that seen when it is inserted, with linkers, upstream to RCBX64 (6.7-fold effect, Figure 5A) demonstrates that the precise location of the 12-bp element relative to adjacent elements is critical for maximal transcriptional activity. This observation suggests that the protein(s) that binds the 12-bp element may synergize with transcription factors that bind to adjacent elements such as the Sp1 and Ets sites, which were shown to be footprinted in the H4IIEC3 cells in vivo (Figures 7, 8).
Further in vitro and in vivo studies with the carboxylase gene as a model TATA-less promoter will provide important insights into the function of the upstream promoter element and its role in the transactivation of this key regulator of hemostasis. Database analysis indicates that the sequence from –76 to –65 does not contain a mammalian transcription factor consensus site.36 Identification of the factor(s) that binds this element and regulates carboxylase gene transcription will permit us to define the consensus sequence and to explore the aforementioned models and their relevance to other TATA-less genes with tissue-specific developmental regulation.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2002-12-3833.
Supported by National Institutes of Health grants HL60307 and HL42443.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Rajat Deo for technical assistance, and Dr Anny Usheva for advice and helpful discussions.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal