Abstract
Hmgb3 is a member of a family of chromatin-binding proteins that can alter DNA structure to facilitate transcription factor binding. We identified the Hmgb3 cDNA in a subtractive hybridization screen for transcripts that are preferentially expressed in hematopoietic stem cells. We inserted an internal ribosomal entry site–green fluorescence protein cassette into the 3′ untranslated region of the X-linked Hmgb3 locus to identify Hmgb3-expressing cells. In adult mice, Hmgb3 mRNA is detected in bone marrow cells, primitive Lin–, c-kit+, Sca-1+, IL-7Rα– cells, and Ter119+ erythroid cells. We observed that long-term repopulating ability is entirely contained in the subpopulation of Lin–, c-kitHI cells that express Hmgb3. Most common lymphoid and myeloid progenitors express Hmgb3. Introduction of a retrovirus containing the Hmgb3 cDNA into mouse bone marrow stem cells demonstrated that enforced expression of Hmgb3 inhibited B-cell and myeloid differentiation. We conclude that down-regulation of Hmgb3 protein levels is an important step for myeloid and B-cell differentiation.
Introduction
Hematopoietic stem cells (HSCs) are a rare population of cells that predominantly reside within adult bone marrow. They are capable of the exponential proliferation and multilineage differentiation necessary for life-long replenishment of all blood cell types.1 In order to maintain their numbers, HSCs undergo self-renewal, generating progeny that retain the same ability to proliferate and differentiate. The mechanisms by which HSCs make decisions to self-renew or differentiate are still unclear.
We have generated a library of cDNA sequences expressed in adult mouse bone marrow cells depleted of cells expressing lineage-specific surface proteins (Lin–) and expressing high levels of the tyrosine kinase receptor c-kit (c-kitHI), which we previously have reported are enriched for HSCs.2,3 One of the sequences we identified corresponds to mouse Hmgb3, a recently discovered member of a superfamily of high-mobility group DNA-binding proteins.4 Hmgb3 was originally identified in an EST database,4 and the protein is 85%-89% homologous to Hmgb1 and 2. Human HMGB3 is 99% homologous to the mouse gene and has been mapped to X chromosome band q28. Of the 36 Hmgb3 ESTs, 31 were derived from embryonic tissue. Northern analysis demonstrated high levels of Hmgb3 mRNA in 18-dpc mouse embryos, but no detectable mRNA in adult brain, lung, liver, spleen, and kidney.4
The high mobility group superfamily is subdivided into 3 groups. HMGB1, HMGB2, and HMGB3 are included in the HMG-Box family.5 In vertebrates, HMGB1 and 2 are expressed in all cell types. Mammalian HMGB1 and 2 preferentially bind to bent DNA with no sequence specificity.6,7 HMGB1 and 2 can bend linear DNA and appear to play a role in the formation of nucleoprotein complexes by altering chromatin structure to allow factor binding.8,9 HMGB1 and 2 also have been shown to interact with DNA-binding proteins such as Hox and Oct, steroid hormone receptors, RAG1 and 2, Rel, and p53.10 HMGB1 and 2 can either activate or repress transcription. For example, HMGB1 can hinder the interaction between the TATA-binding protein and TFIIB, whereas HMGB2 stabilizes the TFIID-TFIIA complex.11,12 Although nothing is known about the function of Hmgb3, the high homology between Hmgb3 and other HMG-Box family members suggests that Hmgb3 may share some of the same properties with Hmgb1 and 2.
In this report we show that Hmgb3 is expressed in HSCs, primitive progenitor cells, and differentiating erythroid cells in the adult mouse. Analysis of transgenic mice containing a GFP gene inserted into the Hmgb3 locus showed that Lin–, c-kitHI, Hmgb3+ cells were found to be enriched for HSCs, common lymphoid progenitors (CLPs), and common myeloid progenitors (CMPs).13,14 Enforced expression of Hmgb3 in hematopoietic stem cells inhibited B-cell and myeloid differentiation. Our data demonstrate that Hmgb3 expression can act as a marker for long-term repopulating cells and that Hmgb3 functions to inhibit the myeloid differentiation of primitive hematopoietic cells.
Materials and methods
Isolation of lin–, c-kitHI adult bone marrow cells
Mouse bone marrow Lin–, c-kitHI cells were isolated as previously described.3 Briefly, mouse bone marrow harvested from femurs was incubated in a cocktail of rat anti–mouse monoclonal antibodies that recognized the cell surface markers CD4 (clone GK1.5), CD8 (53-6.7), B220 (6B2), Mac-1 (M1/70), Gr-1 (8C5), and Ter119 (Pharmingen, San Diego, CA). Bound cells were removed by magnetic beads coated with goat anti–rat IgG (BioMag beads, Qiagen, Valencia, CA). Lin– cells were then incubated with biotinylated rat anti–mouse anti–c-kit antibody and subsequently stained with streptavidin-phycoerythrin (PE) (Caltag, Burlingame, CA). The use of a biotinylated c-kit antibody allows a greater separation of c-kit+ cells. All cell sorting for this and subsequent procedures was performed on a fluorescence-activated cell sorter (FACS) Vintage SE (Becton Dickinson, San Jose, CA) using 488-nm argon and 633-nm helium-neon lasers.
In vitro nuclear localization assay
A 1.2-kb Hmgb3 cDNA was amplified from MEL cell RNA using the following primers: 5′-AAGCTGCACAG-GGCGAACAATACAGTCAGGA-3′ (GenBank AF022465, bp 7-28) and 5′-CCAAGCTT-ACTTCTCATGCCGGCCGACC-3′ (bp 1242-1261). The primers included PstI (5′) and HindIII (3′) sites to allow cloning into pBS KS+ (Stratagene, La Jolla, CA). The sequence of the amplified cDNA was verified by sequencing. The 1.2-kb Hmgb3 cDNA was excised as a PstI/KpnI fragment and cloned into pEGFP-N2 and pEGFP-C2 (Clontech, Palo Alto, CA) to generate Hmgb3-GFP genes, where the eGFP gene is fused in frame to the 3′ and 5′ ends of the Hmgb3 cDNA, respectively. Both fusion genes are expressed from the cytomegalovirus (CMV) promoter and contain the neomycin resistance gene. The Hmgb3-GFP plasmids and pEGFP-C2 were linearized with NdeI and electroporated into 32D cells using a BioRad GenePulser set at 0.6 kV/25 μFd. Transfected 32D cells were washed and transferred to fresh medium. Twenty-four hours later, G418 (500 mg/mL final active concentration) was added, and selection proceeded for 21 days. Fluorescent microscopy was performed using fluorescein isothiocyanate (FITC) and 4′6-diamidino-2-phenylindole 2HCl (DAPI) filters.
Analysis of Hmgb3 mRNA levels
Northern blot analysis was performed as previously described.15 RNA was isolated using TriZol according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Sources of RNA included bone marrow, spleen, thymus, brain, heart, liver, lung, and kidney of adult C57BL/6 mice and cytotoxic T-lymphocyte (CTLL), mouse erythroleukemia (MEL), and 32D cell lines. The probe used for Northern analysis was a 526-bp BclI-HindIII fragment located within the 3′ untranslated region of the Hmgb3 cDNA.
Cells of erythroid (Ter119+), granulocytic (Gr-1+), monocyte/macrophage (Mac-1+), B-lymphoid (B220+), and T-lymphoid (CD4/8+) lineage were separated by staining with PE-conjugated monoclonal antibodies described in “Isolation of lin–, c-kitHI adult bone marrow cells” followed by FACS sorting. Platelets/megakaryocytes were separated by staining with rat anti–mouse monoclonal antibody for CD41 (Pharmingen, clone MWReg30, FITC conjugated). Mast cells were obtained by culturing whole mouse bone marrow for 3 weeks in murine interleukin-3 (IL-3) (PeproTech, Rocky Hill, NJ). Adherent cells were removed by serial passaging, and cell density was maintained at 2 × 105 cells/mL. Lin–, c-kit–, Sca-1–, and Lin–, c-kit+, Sca-1+, IL-7Rα– bone marrow cells were isolated by staining lineage-depleted bone marrow with allophycocyanin (APC)–conjugated anti–c-kit (2B8: Pharmingen), PE-conjugated anti–Sca-1 (E13-161.7: Pharmingen), and biotinylated anti–IL-7Rα (α734: eBioscience, San Diego, CA). Secondary staining was performed with streptavidin PE-Cy5 (eBioscience). Cells were sorted by FACS based on isotype controls. RNA was isolated from all cell types using TriZol.
Reverse-transcriptase polymerase chain reaction (RT-PCR) was performed as previously described.16 Primers used for PCR amplification of wild-type Hmgb3 cDNA were forward: 5′-CGCTGTGATTGACACATCTC-3′ (GenBank bp 668-687) and reverse 1: 5′-GTACAGTTGACAACTCAAGG-3′ (GenBank bp 1008-1027) (Figure 2B). Primers used for PCR amplification of Hmgb3-KI cDNA were the identical forward primer and reverse 2: 5′-TGATACAGTTTCAGGGCGCTC-3′ (GenBank bp 802-822) (Figure 4B). Amplification of β2-microglobulin cDNA (forward: 5′-TGCTATCCAGAAAACCCCTC-3′, reverse: 5′-GTCATGCTTAACTCTGCAGG-3′) served as a control. Amplifications of Hmgb3 and β2-microglobulin were performed in independent reactions for all lineage-positive samples and in duplex reactions for whole bone marrow, Lin–, c-kit–, Sca-1–, and Lin–, c-kit+, Sca-1+, IL-7Rα– samples. For both conditions, limiting dilutions were used to ensure amplifications of Hmgb3 and β2-microglobulin were performed within the linear range. Reactions were performed for 32 cycles at 94°C for 15 seconds, 58°C for 15 seconds, and 72°C for 30 seconds. Hmgb3 mRNA levels were measured and normalized to β2-microglobulin by performing densitometry on relative band intensities captured on a Phosphor Screen and resolved on a Typhoon 8600 PhosphorImager (Amersham Pharmacia, Piscataway, NJ).
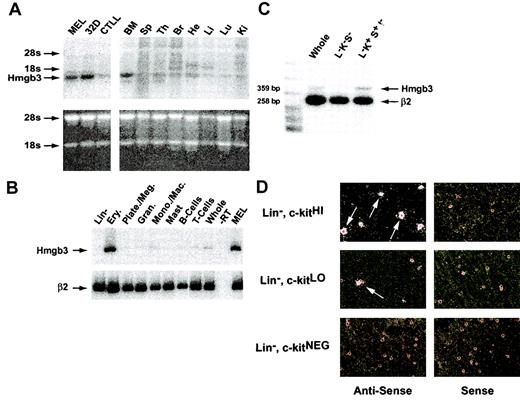
Hmgb3 mRNA levels in adult mouse tissues. (A) Northern blot analysis of Hmgb3 expression in adult mouse tissues. Top: Northern blot analysis of Hmgb3 expression in MEL, 32D cell lines, and CTLLs (left) and in adult mouse bone marrow (BM), spleen (Sp), thymus (Ty), brain (Br), heart (He), Liver (Li), Lung (Lu), and Kidney (Ki) (right). Signals corresponding to Hmgb3 mRNA and location of 18s and 28s rRNAs are indicated by arrows. The 3′ untranslated region was used as a probe (yellow bar, Figure 3, middle panel). Bottom: analysis of RNA loading and integrity. 15 μg RNA was loaded onto an agarose gel and stained with ethidium bromide following electrophoresis. (B) RT-PCR analysis of adult mouse bone marrow populations. RT-PCR analysis of bone marrow populations sorted by lineage: lineage-depleted cells, erythroid cells, platelets/megakaryocytes, granulocytes, monocytes/macrophages, mast cells, B cells, T cells, and whole bone marrow. Signals corresponding to Hmgb3 expression are indicated by black arrows. RT-PCR performed without RT (–RT) was used for a negative control. MEL RNA served as a positive control for Hmgb3 expression. β2-microglobulin (β2) was used for an internal control. (C) Semiquantitative duplex RT-PCR analysis of whole, Lin–, c-kit+, Sca-1–(L–K–S–), and Lin–, c-kit+, Sca-1+, IL-7Rα– (L–K+S+I–) bone marrow cells. Hmgb3 mRNA levels were quantified within the linear range of amplification by densitometry and normalized to β2-microglobulin expression. Amplicon sizes are 359 bp and 258 bp for Hmgb3 and β2-microglobulin, respectively. (D) Hmgb3 mRNA in situ hybridization analysis. All analyses were performed on adult mouse lineage-depleted bone marrow cells sorted based on c-kit protein levels: c-kitHI (upper left), c-kitLO (middle left), and c-kitNEG (lower left). Hybridization was performed with the same probe used for Northern analysis. All antisense hybridizations were performed in tandem with sense control hybridizations (upper, middle, and lower right), and the silver grains visualized by dark field microscopy. Examples of cells positive for Hmgb3 mRNA are indicated by arrows. Original magnification, × 400.
Hmgb3 mRNA levels in adult mouse tissues. (A) Northern blot analysis of Hmgb3 expression in adult mouse tissues. Top: Northern blot analysis of Hmgb3 expression in MEL, 32D cell lines, and CTLLs (left) and in adult mouse bone marrow (BM), spleen (Sp), thymus (Ty), brain (Br), heart (He), Liver (Li), Lung (Lu), and Kidney (Ki) (right). Signals corresponding to Hmgb3 mRNA and location of 18s and 28s rRNAs are indicated by arrows. The 3′ untranslated region was used as a probe (yellow bar, Figure 3, middle panel). Bottom: analysis of RNA loading and integrity. 15 μg RNA was loaded onto an agarose gel and stained with ethidium bromide following electrophoresis. (B) RT-PCR analysis of adult mouse bone marrow populations. RT-PCR analysis of bone marrow populations sorted by lineage: lineage-depleted cells, erythroid cells, platelets/megakaryocytes, granulocytes, monocytes/macrophages, mast cells, B cells, T cells, and whole bone marrow. Signals corresponding to Hmgb3 expression are indicated by black arrows. RT-PCR performed without RT (–RT) was used for a negative control. MEL RNA served as a positive control for Hmgb3 expression. β2-microglobulin (β2) was used for an internal control. (C) Semiquantitative duplex RT-PCR analysis of whole, Lin–, c-kit+, Sca-1–(L–K–S–), and Lin–, c-kit+, Sca-1+, IL-7Rα– (L–K+S+I–) bone marrow cells. Hmgb3 mRNA levels were quantified within the linear range of amplification by densitometry and normalized to β2-microglobulin expression. Amplicon sizes are 359 bp and 258 bp for Hmgb3 and β2-microglobulin, respectively. (D) Hmgb3 mRNA in situ hybridization analysis. All analyses were performed on adult mouse lineage-depleted bone marrow cells sorted based on c-kit protein levels: c-kitHI (upper left), c-kitLO (middle left), and c-kitNEG (lower left). Hybridization was performed with the same probe used for Northern analysis. All antisense hybridizations were performed in tandem with sense control hybridizations (upper, middle, and lower right), and the silver grains visualized by dark field microscopy. Examples of cells positive for Hmgb3 mRNA are indicated by arrows. Original magnification, × 400.
Hmgb3 expression correlates with long-term repopulating ability. (A) Separation of Lin– c-kitHI Hmgb3-KI bone marrow cells into GFP+ and GFP– populations by flow cytometry (top). Sort gates were based on isotype controls (bottom). (B) Duplex RT-PCR performed on RNA isolated from sorted Hmgb3-KI bone marrow (BM) cells sorted into GFP+ and GFP– populations. RT-PCR performed without RT (–RT) was used for a negative control. β2-microglobulin cDNA amplification was used as an internal control. The amplicon sizes are 154 bp and 258 bp for Hmgb3 and β2-microglobulin, respectively. (C) Mean CFU-GM frequencies in Lin–, c-kit+, GFP– (n = 3, 202 colonies counted), and GFP+ (n = 3, 69 colonies counted) populations. Colony-forming assays were performed as described in “Materials and methods.” P values were determined by Student t test. (D) Representative hemoglobin analysis of competitive repopulation of Lin–, c-kitHI, GFP+ versus GFP– cells. Repopulations were performed with mixed doses of either 30 000 Lin–, c-kitHI, GFP+ (n = 5) or 50 000 Lin–, c-kitHI, GFP– (n = 5) and 107 C57BL/6 bone marrow as described in “Materials and methods.” 129/SvJ and C57BL/6 hemoglobin served as assay controls. The βMin, βMaj, (129/SvJ) and βS (C57BL/6) hemoglobins are indicated by black arrows. The average erythroid repopulation by Hmgb3-KI bone marrow (determined as percentage of total Hb that is of 129/SvJ genetic origin) for each population is underneath their respective samples accompanied by the standard deviation. Hb levels were quantified by densitometry.
Hmgb3 expression correlates with long-term repopulating ability. (A) Separation of Lin– c-kitHI Hmgb3-KI bone marrow cells into GFP+ and GFP– populations by flow cytometry (top). Sort gates were based on isotype controls (bottom). (B) Duplex RT-PCR performed on RNA isolated from sorted Hmgb3-KI bone marrow (BM) cells sorted into GFP+ and GFP– populations. RT-PCR performed without RT (–RT) was used for a negative control. β2-microglobulin cDNA amplification was used as an internal control. The amplicon sizes are 154 bp and 258 bp for Hmgb3 and β2-microglobulin, respectively. (C) Mean CFU-GM frequencies in Lin–, c-kit+, GFP– (n = 3, 202 colonies counted), and GFP+ (n = 3, 69 colonies counted) populations. Colony-forming assays were performed as described in “Materials and methods.” P values were determined by Student t test. (D) Representative hemoglobin analysis of competitive repopulation of Lin–, c-kitHI, GFP+ versus GFP– cells. Repopulations were performed with mixed doses of either 30 000 Lin–, c-kitHI, GFP+ (n = 5) or 50 000 Lin–, c-kitHI, GFP– (n = 5) and 107 C57BL/6 bone marrow as described in “Materials and methods.” 129/SvJ and C57BL/6 hemoglobin served as assay controls. The βMin, βMaj, (129/SvJ) and βS (C57BL/6) hemoglobins are indicated by black arrows. The average erythroid repopulation by Hmgb3-KI bone marrow (determined as percentage of total Hb that is of 129/SvJ genetic origin) for each population is underneath their respective samples accompanied by the standard deviation. Hb levels were quantified by densitometry.
To perform in situ hybridization, Lin–, c-kitHI, c-kitLO, and c-kitNEG bone marrow cells were isolated as described in “Isolation of lin–, c-kitHI adult bone marrow cells”. The cells were suspended in collagen and applied to slides. In situ hybridization was performed by Molecular Histology (Rockville, MD) using 35S-labeled sense or antisense RNA transcripts of the 526-bp fragment of the Hmgb3 3′ untranslated region used for Northern analysis as a probe. Silver grains were visualized using dark field microscopy and photography was dark and light field.
Generation of Hmgb3 knock-in (Hmgb3-KI) mice
A 4.1-kb PstI–HpaI fragment containing a region starting 100 bp 5′ of exon 1 and ending 335 bp downstream of the stop codon (EMBL sequence: X chromosome, bp 54786765–54790795) was excised from a 5.5-kb fragment that encompasses the entire Hmgb3 locus. A 1.3-kb NotI–BamHI fragment containing an internal ribosomal entry site (IRES) linked to a green fluorescence protein (GFP) gene (IRES-GFP) was digested from pIRES2-EGFP (Clontech), subcloned into pLITMUS38 (New England Biolabs, Beverly, MA), and subsequently excised with EcoRV and SalI. A triple ligation containing the 4.1-kb PstI–HpaI Hmgb3 fragment, the 1.3-kb EcoRV-SalI IRES-GFP fragment, and PstI-SalI digested pBSII KS+ inserted the IRES-GFP gene into the 3′ untranslated region of Hmgb3. This fragment comprised the 5′ arm of the targeting construct and was excised as a NotI-XhoI fragment and cloned into the matching sites of plasmid AC 641. AC 641 contains a pgk-Neo marker, flanked by loxP sites, the remainder of exon 5, and additional 3′ sequence (EMBL: bp 54791080–54793856), and an HSV-TK gene.
The targeting vector was linearized with NotI and electroporated into TC1 embryonic stem (ES) cells (0.6 kV/25 μFd) as described previously.17 Correctly targeted cells were identified by Southern blot analysis using probes outside the targeting region. Three homologous recombination events were identified among 186 G418 and gancyclovir-resistant ES cell clones. Twelve to eighteen ES cells from clones 81 and 114 were injected into 3.5-day-old C57BL/6 blastocysts. A total of 6 germ line chimeras (4 from 81 and 2 from 114) were bred to 129/SvJ female mice, and F1 generation transgenic mice were identified by Southern blot analysis of tail DNA.
Isolation and differentiation of Hmgb3-KI progenitor cells
Bone marrow cells isolated from male Hmgb3-KI mice were sorted by FACS into GFP+ and GFP– populations for semiquantitative duplex RT-PCR as outlined. Sorted cells also were cultured in complete methylcellulose media (Methocult GF M3434; Stem Cell Technologies, Vancouver, BC, Canada), and colony forming units–culture (CFU-C) were scored after 7 days in culture.
For isolation of common lymphoid progenitors,13 male Hmgb3-KI Lin– bone marrow cells were stained with APC-conjugated anti–c-kit, PE-conjugated anti–Sca-1, and biotinylated anti–IL-7Rα. Secondary staining was performed with streptavidin PE-Cy5. Littermate control Lin– bone marrow cells were stained with the recommended isotype antibodies. Lin–, c-kit+, Sca-1+, IL-7Rα+ cells were sorted into GFP+ and GFP– populations and cultured in MethoCult M3630 (Stem Cell Technologies) containing 10 ng/mL human IL-7 and supplemented with 100 ng/mL murine stem cell factor (SCF) (R&D Systems, Minneapolis, MN) and 20 ng/mL human Flt3L (PeproTech). CFU–pre-B colonies were scored after 7-10 days. As a control, the same cells were cultured in Methocult GF M3434 media, supplemented with 20 ng/mL Flt3L, and CFU-Cs were scored after 14 days.
For isolation of common myeloid progenitors,14 male Hmgb3-KI Lin– bone marrow was stained with APC-conjugated anti–c-kit, biotinylated anti–Sca-1 biotinylated anti–IL-7Rα, and either PE-conjugated anti-FcγRII/III (2.4G2) or PE-conjugated anti-CD34 (RAM34) (Pharmingen). Secondary staining was performed with streptavidin PE-Cy5. Littermate control Lin– bone marrow cells were stained with the recommended isotype antibodies. Lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, or Lin–, c-kit+, Sca-1–, IL-7Rα–, CD34+ were sorted into GFP+ and GFP– populations. Each population was cultured in Methocult GF M3434 supplemented with 20 ng/mL human Flt3L. Granulocyte macrophage colony-forming unit (CFU-GM), erythroid burst-forming unit (BFU-E), and granulocyte-erythrocyte-megakaryocyte-macrophage colony-forming unit (CFU-GEMM) were scored after 7-14 days in culture. As a control, the same cells were cultured in MethoCult M3630 media, supplemented with murine SCF and human Flt3L, and CFU–pre-B colonies were scored after 7-10 days.
Competitive repopulation assay
Lin–, c-kitHI cells were isolated from male Hmgb3-KI bone marrow as described and sorted by FACS into GFP+ and GFP– populations. 30 000 Lin–, c-kitHI, GFP+ or 50 000 Lin–, c-kitHI, GFP– cells were mixed with 1 × 107 male C57BL/6 bone marrow cells injected into lethally irradiated (1000 Gy, Cs137 source) male 129 × C57BL/6 F1 mice (Jackson Laboratory, Bar Harbor, ME). Hemoglobin electrophoresis of peripheral blood proteins and Southern blot analysis of bone marrow, spleen, and thymus DNA were performed after 16 weeks as described.18
Retroviral transduction of murine bone marrow
A 0.6-kb NcoI-BamHI Hmgb3 cDNA fragment, containing the entire Hmgb3 coding region (GenBank: bp 28-628), was cloned into the matching sites in the MFGs retroviral vector (provided by Dr H. Malech, Bethesda, MD) followed by the addition of a BamHI fragment containing an IRES-GFP cassette. A control murine stem cell virus (MSCV) vector containing an IRES-GFP cassette (MGirL22Y) was obtained from Dr Brian Sorrentino (St. Jude Children's Research Hospital).19 Both constructs were introduced into GP+E86 packaging cells by standard techniques, and high titer producer cell clones were isolated by limiting dilution.20
Transduction of mouse bone marrow with either the Hmgb3-IRES-GFP or IRES-GFP retroviral vector was performed as described.21 Briefly, C57BL/6 mice were treated with 5-flurouracil (5-FU: 150 mg/kg, Sigma, St Louis, MO) 48 hours prior to bone marrow harvest. Bone marrow cells were plated at 5 × 105 cells/mL in Dulbecco Modified Eagle Medium (Invitrogen Gibco) supplemented with 15% fetal calf serum (FCS), 4 mM L-glutamine, 20 ng/mL murine IL-3, 100 ng/mL murine IL-6 (PeproTech), and 100 ng/mL murine SCF. Bone marrow cells were collected 48 hours later and cocultured at the same concentration with either Hmgb3-GFP or IRES-GFP producer cells in the same media supplemented with 6 μg/mL polybrene (Sigma). The bone marrow cells were harvested 46 hours later and washed in Hanks Balanced Salt Solution (Invitrogen) supplemented with 2% FCS. To differentiate transduced cells in vitro, harvested cells were sorted based on GFP levels, and both GFP+ and GFP– cells were plated in Methocult GF M3434 supplemented with 20 ng/mL human Flt3L at 1 × 104 cells per plate. For long-term transplantation, 2 × 106 transduced bone marrow cells were injected (unsorted) into the lateral tail vein of female genetically anemic WBB6F1-W/WV mice (Jackson Laboratories).
Thymocytes, splenocytes, bone marrow, and peripheral blood were collected 16 weeks after transplantation and stained with PE-conjugated rat anti–mouse antibodies to CD4, CD8, B220, Mac-1, Gr-1, and Ter119. Positive staining and presence of GFP protein was detected by FACS. Southern blot analysis was performed on DNA isolated from bone marrow, spleen, and thymus tissues using a probe that hybridized to a 750-bp BamHI-EcoRI region from the GFP gene.
Results
Hmgb3 is expressed in primitive adult hematopoietic cells
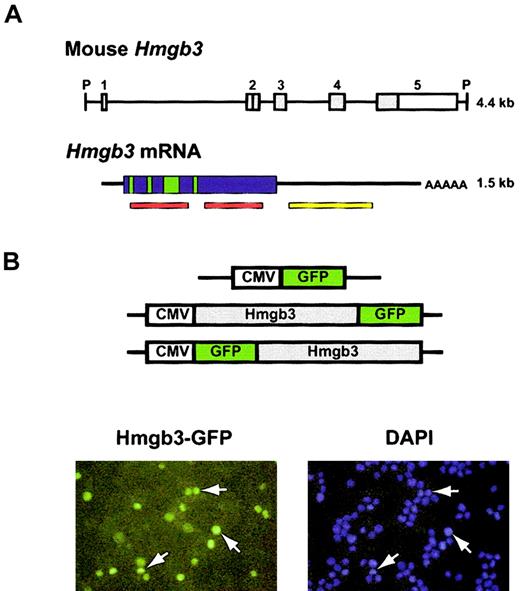
The Hmgb3 locus encodes a 1.5-kb mRNA transcript (Figure 1A). The gene contains 5 exons; only exons 2-5 are translated.4 The protein sequence of Hmgb3 predicted that Hmgb3 should contain nuclear localization signals. We transfected 32D cells with constructs that express Hmgb3 fused with GFP at either the 5′ and 3′ end. (Figure 1B, top). Fluorescence microscopy demonstrated that GFP signals entirely overlap those resulting from DAPI staining, indicating that Hmgb3 is found only in the nucleus (Figure 1B, bottom). No differences were observed with the N-terminal and C-terminal constructs, and GFP was detected in both the nucleus and cytoplasm of cells transfected with the control CMV-GFP construct (data not shown).
Structure of the Hmgb3 gene and protein. (A) Structure of Hmgb3 gene locus, mRNA, and protein. Top: schematic diagram of Hmgb3 gene locus (based on Vaccari et al).4 The entire locus is 4.4 kb flanked by PstI sites (P). There are 5 exons: white boxes represent untranslated regions, gray boxes represent translated regions. Bottom: Hmgb3 mRNA. The transcript is 1.5-kb long. Hmgb3 shares a highly conserved DNA binding domain with Hmgb1 and 2 (red bars). Green boxes represent putative nuclear localization signals based on Hmgb1 and 2 homology. The yellow bar represents the probe used for Northern blotting and in situ hybridization. (B) Nuclear localization of Hmgb3. Upper: constructs used to transfect 32D cells. Hmgb3 cDNA is fused in frame with the GFP gene. Lower: fluorescent microscopy of transfected 32D cells. DAPI staining was used to identify nuclei (blue). Green signals indicate Hmgb3-GFP protein. Some examples of overlapping signals are indicated by white arrows. Original magnification, × 400.
Structure of the Hmgb3 gene and protein. (A) Structure of Hmgb3 gene locus, mRNA, and protein. Top: schematic diagram of Hmgb3 gene locus (based on Vaccari et al).4 The entire locus is 4.4 kb flanked by PstI sites (P). There are 5 exons: white boxes represent untranslated regions, gray boxes represent translated regions. Bottom: Hmgb3 mRNA. The transcript is 1.5-kb long. Hmgb3 shares a highly conserved DNA binding domain with Hmgb1 and 2 (red bars). Green boxes represent putative nuclear localization signals based on Hmgb1 and 2 homology. The yellow bar represents the probe used for Northern blotting and in situ hybridization. (B) Nuclear localization of Hmgb3. Upper: constructs used to transfect 32D cells. Hmgb3 cDNA is fused in frame with the GFP gene. Lower: fluorescent microscopy of transfected 32D cells. DAPI staining was used to identify nuclei (blue). Green signals indicate Hmgb3-GFP protein. Some examples of overlapping signals are indicated by white arrows. Original magnification, × 400.
Northern blot analysis demonstrated that higher levels of Hmgb3 mRNA were present in MEL and 32D cells than in CTLL cells (Figure 2A). In adult mice, there were higher levels of Hmgb3 mRNA in bone marrow than in other tissues, confirming previous findings.4 RT-PCR analysis of RNA from lineage-positive bone marrow cells demonstrated high levels of Hmgb3 mRNA in Ter119+ erythroid cells, and low or absent levels of Hmgb3 mRNA in all other lineage-positive hematopoietic cells (Figure 2B). Hmgb3 mRNA levels within different lineage-negative populations were measured by semiquantitative duplex RT-PCR (Figure 2C). We compared relative levels of Hmgb3 expression in whole bone marrow cells, Lin–, c-kit–, Sca-1–cells, and Lin–, c-kit+, Sca-1+, IL-7Rα– cells, which are enriched for HSCs.13 Hmgb3 expression increased 3-fold in cells enriched for HSCs compared to whole marrow and 12-fold compared to Lin–, c-kit–, Sca-1–cells. The localization of Hmgb3 expression to the most primitive hematopoietic cells within the Lin– fraction was confirmed using in situ hybridization of an antisense riboprobe to Lin–, c-kitHI, Lin–, c-kitLO, and Lin–, c-kitNEG cells. Approximately 50% of Lin–, c-kitHI cells were positive for Hmgb3 expression, while 10% of Lin–, c-kitLO cells and 0% of Lin–, c-kitNEG cells were positive. No hybridization was detected with the sense probe (Figure 2D).
Analysis of Hmgb3 expression in primary hematopoietic cells
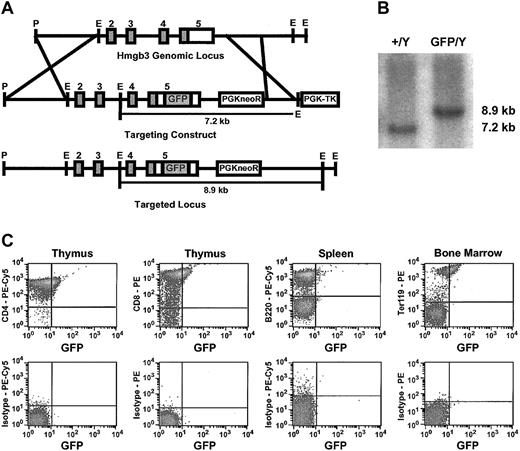
We modified the Hmgb3 locus to insert a GFP gene linked to an IRES in the 3′ untranslated region by homologous recombination in male 129/SvJ ES cells (Hmgb3-KI) (Figure 3A). Because the complete coding sequences for both Hmgb3 and GFP are now part of the same transcript, this strategy allows us to track Hmgb3 expression using GFP without affecting Hmgb3 protein function. Positive ES clones were identified by Southern blot analysis (Figure 3B). Hmgb3-KI mice are phenotypically normal. Since Hmgb3 is an X-linked gene, only male mice hemizygous for the insertion were used in subsequent experiments. FACS analysis performed on male thymocytes, splenocytes, and whole bone marrow demonstrated that 21% to 29% of CD4+ and CD8+ thymocytes expressed GFP but CD4+, CD8+, and B220+ splenocytes and CD4+, CD8+, B220+, Gr-1+, and Mac-1+ bone marrow cells were negative for GFP expression. GFP was expressed in 54% of Ter119+ bone marrow cells (Figure 3C). CD4+, CD8+, B220+, Gr-1+, Mac-1+, and Ter119+ peripheral blood cells did not express GFP (data not shown). FACS analysis of Lin– bone marrow cells demonstrated that approximately 50% of Lin–, c-kitHI cells expressed GFP (Figure 4A).
Generation and characterization of Hmgb3 knock-in mice. (A) Generation of Hmgb3 knock-in (Hmgb3-KI) mice by homologous recombination. Upper: diagram of Hmgb3 gene locus. Middle: targeting construct. An IRES-GFP cassette is inserted into the 3′ untranslated region of the Hmgb3 gene locus. Lower: Hmgb3 gene locus after recombination. (B) Screening of targeted TC1 ES cell clones. Representative Southern blot analysis of genomic DNA isolated from prospective clones digested with EcoRI. The wild-type Hmgb3 locus (+/Y) is contained on a 7.2-kb EcoRI fragment. Homologous recombination (GFP/Y) results in a hemizygous 8.9-kb EcoRI fragment. (C) Expression of Hmgb3-IRES GFP in Hmgb3-KI hematopoietic tissues. Representative results of FACS analysis performed on thymus, spleen, and bone marrow cells isolated from male Hmgb3-KI mice. Cells were stained with antibodies as described in “Materials and methods.” Upper panel: GFP expression in CD4-stained thymocytes, CD8-stained thymocytes, B220-stained splenocytes, and Ter119-stained bone marrow cells isolated from Hmgb3-KI mice. All other samples were negative for GFP expression. Lower panel: respective isotype controls.
Generation and characterization of Hmgb3 knock-in mice. (A) Generation of Hmgb3 knock-in (Hmgb3-KI) mice by homologous recombination. Upper: diagram of Hmgb3 gene locus. Middle: targeting construct. An IRES-GFP cassette is inserted into the 3′ untranslated region of the Hmgb3 gene locus. Lower: Hmgb3 gene locus after recombination. (B) Screening of targeted TC1 ES cell clones. Representative Southern blot analysis of genomic DNA isolated from prospective clones digested with EcoRI. The wild-type Hmgb3 locus (+/Y) is contained on a 7.2-kb EcoRI fragment. Homologous recombination (GFP/Y) results in a hemizygous 8.9-kb EcoRI fragment. (C) Expression of Hmgb3-IRES GFP in Hmgb3-KI hematopoietic tissues. Representative results of FACS analysis performed on thymus, spleen, and bone marrow cells isolated from male Hmgb3-KI mice. Cells were stained with antibodies as described in “Materials and methods.” Upper panel: GFP expression in CD4-stained thymocytes, CD8-stained thymocytes, B220-stained splenocytes, and Ter119-stained bone marrow cells isolated from Hmgb3-KI mice. All other samples were negative for GFP expression. Lower panel: respective isotype controls.
Analysis of Hmgb3 in hematopoietic stem cells
Lin–, c-kit+ cells from Hmgb3-KI mice were sorted based on GFP expression (Figure 4A). Duplex RT-PCR performed on GFP+ and GFP– populations sorted from whole bone marrow demonstrated the presence of the Hmgb3 transcript only in those cells that are GFP+ (Figure 4B). However, the relative stability of Hmgb3 protein compared to GFP is unknown, so it is possible that GFP– cells may contain Hmgb3 protein. Lin–, c-kit+, GFP– cells contained a 2.5-fold higher frequency of CFU-GM than Lin–, c-kit+, GFP+ cells (P < .01, Figure 4C). To analyze HSC activity, Lin–, c-kitHI, GFP+, and Lin–, c-kitHI, GFP– cells were mixed with male C57BL/6 competitor bone marrow before injection into lethally irradiated male 129 × C57BL/6F1 recipients. Polymorphisms in the β-globin locus were used to differentiate between the contributions of Hmgb3-KI and C57BL/6 HSCs; the 129/SvJ strain carries the diffuse allele and the C57BL/6 strain carries the single allele.18 Sixteen weeks after transplantation, hemoglobin electrophoresis was performed on recipients' peripheral blood (Figure 4D). Despite the fact that larger number of Lin–, c-kitHI, GFP– cells were competed against control cells, Lin–, c-kitHI, GFP– cells contributed minimally to long-term repopulation. The smaller number of Lin–, c-kitHI, GFP+ competed against the same number of control cells contributed approximately 28% of the erythroid cells in repopulated mice. Southern blot analysis of bone marrow, spleen, and thymus DNA demonstrated similar results (data not shown).
Analysis of Hmgb3 in common lymphoid progenitors
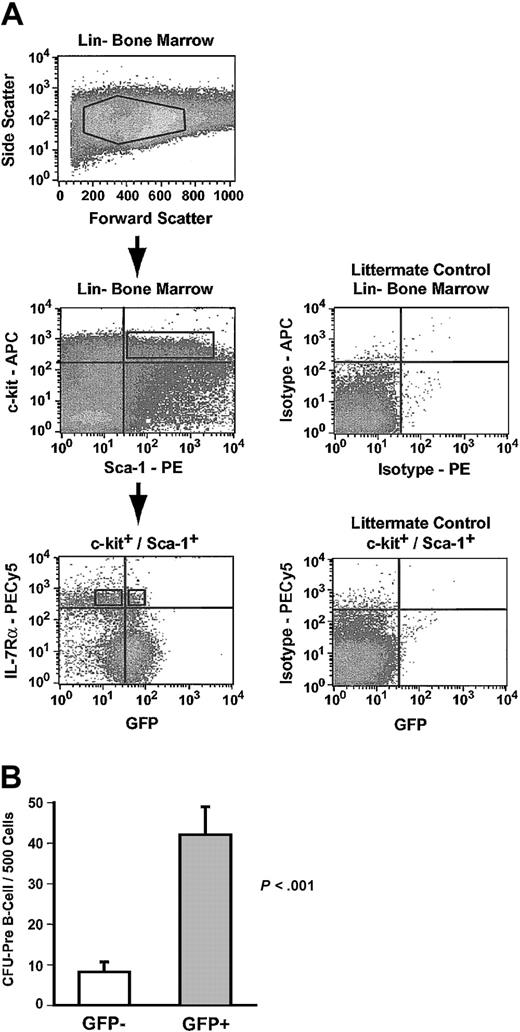
Common lymphoid progenitors (CLPs) express the IL-7 receptor (IL-7Rα), and IL-7Rα expression can be used to discriminate between CLPs and IL-7Rα– HSCs.13,22 CLPs in Hmgb3-KI bone marrow were identified as Lin–, c-kit+, Sca-1+, and IL-7Rα+ cells (0.01% of bone marrow cells) and sorted into GFP+ and GFP– populations (Figure 5A).13 Lin–, c-kit+, Sca-1+, and IL-7Rα+ cells did not form myeloid colonies in culture but were highly enriched for CFU–pre-B activity (Table 1). Approximately 27% of all Lin–, c-kit+, Sca-1+, and IL-7Rα+ cells were GFP+ and GFP+ cells displayed a 5-fold increase (P < .001) in the frequency of CFU–pre-B and a nearly 2-fold increase in absolute CFU–pre-B numbers compared to GFP– cells (Figure 5B; Table 1).
Common lymphoid progenitors express Hmgb3. (A) Isolation of GFP+ and GFP– common lymphoid progenitors by flow cytometry. Upper: Lin– bone marrow isolated from Hmgb3-KI mice. Middle: c-kit and Sca-1 expression in Hmgb3-KI Lin– bone marrow (left). Cells positive for c-kit and Sca-1 were selected based on isotype staining of littermate control Lin– bone marrow cells (right). Lower: IL-7Rα and GFP profiles of c-kit+/Sca-1+ cells (left). Cells were separated into IL-7Rα+ and GFP+/– populations based on isotype staining of littermate control Lin– c-kit+, Sca-1+ bone marrow cells (Right). (B) Mean CFU–pre-B cell frequency in GFP– (n = 8, 66 colonies counted) and GFP+ (n = 7, 294 colonies counted) CLP populations. Values were determined by scoring pre–B cell colonies per 500 cells cultured. P values were determined by Student t test.
Common lymphoid progenitors express Hmgb3. (A) Isolation of GFP+ and GFP– common lymphoid progenitors by flow cytometry. Upper: Lin– bone marrow isolated from Hmgb3-KI mice. Middle: c-kit and Sca-1 expression in Hmgb3-KI Lin– bone marrow (left). Cells positive for c-kit and Sca-1 were selected based on isotype staining of littermate control Lin– bone marrow cells (right). Lower: IL-7Rα and GFP profiles of c-kit+/Sca-1+ cells (left). Cells were separated into IL-7Rα+ and GFP+/– populations based on isotype staining of littermate control Lin– c-kit+, Sca-1+ bone marrow cells (Right). (B) Mean CFU–pre-B cell frequency in GFP– (n = 8, 66 colonies counted) and GFP+ (n = 7, 294 colonies counted) CLP populations. Values were determined by scoring pre–B cell colonies per 500 cells cultured. P values were determined by Student t test.
Frequency and total number of Hmgb3+ and Hmgb3- lymphoid and myeloid CFU-C
. | . | . | CMP . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | CLP . | . | FcγRII/IIILO . | . | CD34+ . | . | |||
. | Hmgb3+ . | Hmgb3- . | Hmgb3+ . | Hmgb3- . | Hmgb3+ . | Hmgb3- . | |||
| Total cells/hind limb | 646.2 | 1747.4 | 4255.1 | 2621.6 | 2685.5 | 908.3 | |||
| CFU-pre-B | |||||||||
| CFU-pre-B/500 CLP (counted colonies) | 42.0 ± 6.8 (294) | 8.3 ± 2.5 (66) | 0 | 0 | 0 | 0 | |||
| Total CLP-derived CFU-pre-B/hind limb | 54.2 | 29.0 | 0 | 0 | 0 | 0 | |||
| CFU-GM | |||||||||
| CFU-GM/500 cells (counted colonies) | 0 | 0 | 11.5 ± 1.9 (69) | 21.0 ± 3.0 (126) | 69.0 ± 4.7 (345) | 72.4 ± 6.2 (362) | |||
| Total CMP-derived CFU-GM/hind limb | 0 | 0 | 97.9 | 110.1 | 370.6 | 131.5 | |||
| BFU-E | |||||||||
| BFU-E/500 cells (counted colonies) | 0 | 0 | 11.5 ± 1.6 (69) | 3.6 ± 1.2 (22) | 21.5 ± 2.1 (107) | 11.4 ± 2.1 (57) | |||
| Total CMP-derived BFU-E/hind limb | 0 | 0 | 97.9 | 18.9 | 115.5 | 20.7 | |||
| CFU-GEMM | |||||||||
| CFU-GEMM/500 cells (counted colonies) | 0 | 0 | 1.3 ± 1.0 (8) | 4.2 ± 1.2 (25) | 7.8 ± 1.7 (40) | 17.2 ± 2.6 (86) | |||
| Total CMP-derived CFU-GEMM/hind limb | 0 | 0 | 11.1 | 22.0 | 41.9 | 31.2 | |||
. | . | . | CMP . | . | . | . | |||
|---|---|---|---|---|---|---|---|---|---|
| . | CLP . | . | FcγRII/IIILO . | . | CD34+ . | . | |||
. | Hmgb3+ . | Hmgb3- . | Hmgb3+ . | Hmgb3- . | Hmgb3+ . | Hmgb3- . | |||
| Total cells/hind limb | 646.2 | 1747.4 | 4255.1 | 2621.6 | 2685.5 | 908.3 | |||
| CFU-pre-B | |||||||||
| CFU-pre-B/500 CLP (counted colonies) | 42.0 ± 6.8 (294) | 8.3 ± 2.5 (66) | 0 | 0 | 0 | 0 | |||
| Total CLP-derived CFU-pre-B/hind limb | 54.2 | 29.0 | 0 | 0 | 0 | 0 | |||
| CFU-GM | |||||||||
| CFU-GM/500 cells (counted colonies) | 0 | 0 | 11.5 ± 1.9 (69) | 21.0 ± 3.0 (126) | 69.0 ± 4.7 (345) | 72.4 ± 6.2 (362) | |||
| Total CMP-derived CFU-GM/hind limb | 0 | 0 | 97.9 | 110.1 | 370.6 | 131.5 | |||
| BFU-E | |||||||||
| BFU-E/500 cells (counted colonies) | 0 | 0 | 11.5 ± 1.6 (69) | 3.6 ± 1.2 (22) | 21.5 ± 2.1 (107) | 11.4 ± 2.1 (57) | |||
| Total CMP-derived BFU-E/hind limb | 0 | 0 | 97.9 | 18.9 | 115.5 | 20.7 | |||
| CFU-GEMM | |||||||||
| CFU-GEMM/500 cells (counted colonies) | 0 | 0 | 1.3 ± 1.0 (8) | 4.2 ± 1.2 (25) | 7.8 ± 1.7 (40) | 17.2 ± 2.6 (86) | |||
| Total CMP-derived CFU-GEMM/hind limb | 0 | 0 | 11.1 | 22.0 | 41.9 | 31.2 | |||
Total cells is the summation of the number of cells with the CLP or CMP phenotypes that were recovered after FACS sorting from all experiments divided by the number of hind limbs. Cells with the CLP phenotype were estimated to be 0.01% of whole bone marrow per hind limb (Figure 5). Cells with either CMP phenotype were estimated to be 0.008% of whole bone marrow per hind limb (Figure 6). Numbers in parentheses indicate total number of colonies counted. Colony frequencies were determined by dividing the total number of colonies counted by the total number of cells plated and normalizing to number of colonies per 500 cells. Total progenitor-derived CFU-C per hind limb was determined by the formula [frequency] × [total cells/hind limb].
Analysis of Hmgb3 in myeloid progenitors
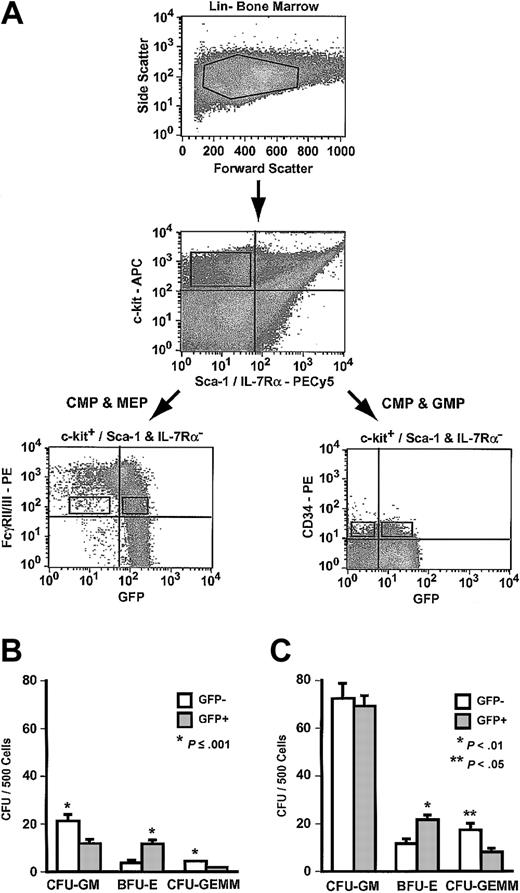
Primitive myeloid progenitors express neither Sca-1 nor IL-7Rα.14 These progenitors in Hmgb3-KI bone marrow were identified as Lin–, c-kit+, Sca-1–, and IL-7Rα– cells. Lin–, c-kit+, Sca-1–, IL-7Rα– cells only formed myeloid colonies in culture; no CFU–pre-B were observed (data not shown). Lin–, c-kit+, Sca-1–, IL-7Rα– progenitors can be further separated into distinct populations by staining with antibodies to FcγRII/III and CD34. The common myeloid progenitor (CMP) exhibits a FcγRII/IIILO and CD34+ phenotype. CMPs can differentiate into one of 2 lineage-restricted progenitors: the megakaryocyte-erythrocyte progenitor (MEP: FcγRII/IIILO and CD34–) and the granulocyte-monocyte progenitor (GMP: FcγRII/IIIHI and CD34+).
To determine if Hmgb3 expression could be used to separate CMPs from MEPs and GMPs, we sorted Lin–, c-kit+, Sca-1–, IL-7Rα– cells from Hmgb3-KI bone marrow based on GFP and either FcγRII/III or CD34 expression. Lin–, c-kit+, Sca-1–, IL-7Rα– FcγRII/IIILO cells (0.008% of bone marrow cells), which contain both CMPs and MEPs but not GMPs, were sorted into GFP+ and GFP– populations (Figure 6A).14 Approximately 89% of Lin–, c-kit+, Sca-1–, IL-7Rα–,FcγRII/IIILO cells expressed Hmgb3. Both FcγRII/IIILO GFP+ and FcγRII/IIILO GFP– cells contained myeloid colony-forming cells of all lineages. The frequency of CFU-GM was 2-fold higher, and the frequency of CFU-GEMM was 3-fold higher in FcγRII/IIILO GFP– cells (P < .01) compared to GFP+ cells (Figure 6B). Conversely, the frequency of BFU-E was 3-fold higher (P = .001) in FcγRII/IIILO GFP+ cells compared to FcγRII/IIILO GFP– cells. The absolute number of colony-forming cells (CFCs) was 7-fold higher in FcγRII/IIILO GFP+ cells compared to FcγRII/IIILO GFP– cells (Table 1).
Analysis of Hmgb3 expression in myeloid progenitors (MP). (A) Isolation of myeloid progenitors by flow cytometry. Upper: Lin– bone marrow isolated from Hmgb3-KI mice. Middle: c-kit and Sca-1/IL-7Rα expression in Hmgb3-KI Lin– bone marrow. Cells positive for c-kit and negative for Sca-1/IL-7Rα were selected based on isotype controls (not shown). Lower: c-kit+/Sca-1 and IL-7Rα– cells were stained with either anti-FcγRII/III or CD34 monoclonal antibodies as described in “Materials and methods.” Left: FcγRII/III and GFP expression in Hmgb3-KI Lin– c-kit+/(Sca-1/IL-7Rα)– cells. Right: CD34 and GFP expression in Hmgb3-KI Lin– c-kit+/(Sca-1/IL-7Rα)– cells. Sort gates were drawn based on isotype controls (not shown). (B) Mean CFU-GM, BFU-E, and CFU-GEMM frequency in FcγRII/IIILO GFP– (n = 6) and GFP+ (n = 6) MP populations. Values were determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured. P values were determined by Student t test. (C) Mean CFU-GM, BFU-E, and CFU-GEMM frequency in CD34+ GFP– (n = 5) and GFP+ (n = 4) MP populations. Values were determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured. P values were determined by Student t test.
Analysis of Hmgb3 expression in myeloid progenitors (MP). (A) Isolation of myeloid progenitors by flow cytometry. Upper: Lin– bone marrow isolated from Hmgb3-KI mice. Middle: c-kit and Sca-1/IL-7Rα expression in Hmgb3-KI Lin– bone marrow. Cells positive for c-kit and negative for Sca-1/IL-7Rα were selected based on isotype controls (not shown). Lower: c-kit+/Sca-1 and IL-7Rα– cells were stained with either anti-FcγRII/III or CD34 monoclonal antibodies as described in “Materials and methods.” Left: FcγRII/III and GFP expression in Hmgb3-KI Lin– c-kit+/(Sca-1/IL-7Rα)– cells. Right: CD34 and GFP expression in Hmgb3-KI Lin– c-kit+/(Sca-1/IL-7Rα)– cells. Sort gates were drawn based on isotype controls (not shown). (B) Mean CFU-GM, BFU-E, and CFU-GEMM frequency in FcγRII/IIILO GFP– (n = 6) and GFP+ (n = 6) MP populations. Values were determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured. P values were determined by Student t test. (C) Mean CFU-GM, BFU-E, and CFU-GEMM frequency in CD34+ GFP– (n = 5) and GFP+ (n = 4) MP populations. Values were determined by scoring CFU-GM, BFU-E, and CFU-GEMM colonies per 500 cells cultured. P values were determined by Student t test.
Lin–, c-kit+, Sca-1–, IL-7Rα– CD34+ cells (0.008% of bone marrow cells), which contain CMPs and GMPs but not MEPs, were sorted into GFP+ and GFP– populations (Figure 6A)14 ; 75% of Lin–, c-kit+, Sca-1–, IL-7Rα– CD34+ cells expressed Hmgb3 (Figure 6A). Both CD34+ GFP+ and CD34+ GFP– cells contained myeloid colony-forming cells of all lineages. The frequency of CFU-GM was identical in both populations, and the frequency of CFU-GEMM was 2-fold higher in CD34+ GFP– cells compared to CD34+ GFP+ cells (P < .05). Conversely, the frequency of BFU-E was 2-fold higher (P < .01) in CD34+ GFP+ cells compared to CD34+ GFP– cells. The absolute number of CFCs was 3-fold higher in CD34+ GFP+ cells compared to CD34+ GFP– cells (Table 1).
Enforced expression of Hmgb3 inhibits myeloid and B-lymphocyte differentiation
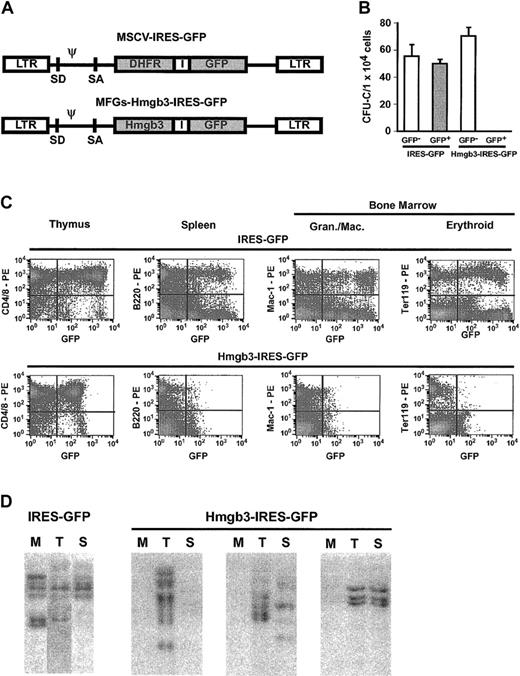
We investigated the effects of enforced expression of Hmgb3 by transducing bone marrow obtained from 5-FU–treated C57BL/6 mice with either a control vector containing an IRES-GFP cassette or an experimental vector containing an Hmgb3-IRES-GFP cassette (Figure 7A). Transduction efficiency was equivalent between the 2 vectors. Transduced marrow was sorted into GFP+ and GFP– populations and plated in methylcellulose. Colonies of all lineages formed from both GFP+ and GFP– cells transduced with the control vector and GFP– cells transduced with Hmgb3-IRES-GFP. However, GFP+ cells transduced with Hmgb3-IRES-GFP were unable to form colonies (Figure 7B). To determine the effects of Hmgb3 overexpression in vivo, transduced (but unsorted) bone marrow was transplanted into genetically anemic WBB6F1-W/WV mice.23 Bone marrow cellularities and peripheral blood counts of animals who received transplants of bone marrow cells transduced with either vector were equivalent. Sixteen weeks after transplantation, FACS analysis was performed on thymocytes, splenocytes, bone marrow, and peripheral blood. Representative results are shown in Figure 7C. In mice who received transplants of bone marrow transduced with the control IRES-GFP vector, GFP was detected in 10% to 62% of CD4+ and CD8+ thymocytes, B220+ splenocytes, Mac-1+, Gr-1+, Ter119+, and Lin– bone marrow cells (Figure 7C). GFP also was expressed in 10% to 71% of all lineages of peripheral blood cells. In mice who received transplants of Hmgb3-IRES-GFP–transduced bone marrow, GFP was detected in approximately 30% of CD4+ and CD8+ thymocytes and 27% of Lin– bone marrow cells, but GFP was detected in fewer than 1% of B220+ splenocytes, Mac-1+, Gr-1+, and Ter119+ bone marrow cells, and peripheral blood cells (Figure 7C). Southern blot analysis demonstrated many common integration sites for the control IRES-GFP vector in genomic DNA isolated from bone marrow, thymus, and spleen of repopulated mice (Figure 7D). In contrast, integrated GFP sequences were detected only in the thymus and occasionally the spleen DNA in mice transplanted with Hmgb3-IRES-GFP transduced marrow, but no GFP sequences were detected in bone marrow DNA.
Overexpression of Hmgb3 impairs myeloid and B-lymphocyte differentiation. (A) Diagram of Hmgb3-IRES-GFP retroviral vector and the control MSCV MGirL22Y IRES-GFP vector. (B) Mean CFU-C numbers per 1 × 105 cells plated for control IRES-GFP vector GFP– (n = 4; 223 colonies counted) and GFP+ (n = 3; 150 colonies counted) populations and for Hmgb3-IRES-GFP GFP– (n = 6; 424 colonies counted) and GFP+ (n = 6; 0 colonies counted) populations. (C) Representative FACS analysis of peripheral blood from mice who received transplants of either control IRES-GFP vector-transduced marrow (top: n = 8) or Hmgb3-IRES-GFP–transduced bone marrow (bottom: n = 10). Thymocytes, splenocytes, and bone marrow cells were analyzed 16 weeks after transplantation for presence of GFP in T cells, B cells, granulocytes/monocytes/macrophages, and erythroid cells, respectively. Quadrants were drawn based on isotype controls similar to those in Figure 3C. (D) Representative Southern blot analysis of genomic DNA isolated from bone marrow (M), thymus (T), and spleen (S) tissues from control GFP and Hmgb3-IRES-GFP mice 16-weeks after transplantation. 10 μg DNA were digested with EcoRI and loaded into each lane. Hybridization was performed with a probe that recognizes the GFP gene.
Overexpression of Hmgb3 impairs myeloid and B-lymphocyte differentiation. (A) Diagram of Hmgb3-IRES-GFP retroviral vector and the control MSCV MGirL22Y IRES-GFP vector. (B) Mean CFU-C numbers per 1 × 105 cells plated for control IRES-GFP vector GFP– (n = 4; 223 colonies counted) and GFP+ (n = 3; 150 colonies counted) populations and for Hmgb3-IRES-GFP GFP– (n = 6; 424 colonies counted) and GFP+ (n = 6; 0 colonies counted) populations. (C) Representative FACS analysis of peripheral blood from mice who received transplants of either control IRES-GFP vector-transduced marrow (top: n = 8) or Hmgb3-IRES-GFP–transduced bone marrow (bottom: n = 10). Thymocytes, splenocytes, and bone marrow cells were analyzed 16 weeks after transplantation for presence of GFP in T cells, B cells, granulocytes/monocytes/macrophages, and erythroid cells, respectively. Quadrants were drawn based on isotype controls similar to those in Figure 3C. (D) Representative Southern blot analysis of genomic DNA isolated from bone marrow (M), thymus (T), and spleen (S) tissues from control GFP and Hmgb3-IRES-GFP mice 16-weeks after transplantation. 10 μg DNA were digested with EcoRI and loaded into each lane. Hybridization was performed with a probe that recognizes the GFP gene.
Discussion
Hmgb3 is a member of a family of chromatin-binding proteins that was originally discovered as an EST preferentially expressed in embryonic tissues.4 We have shown that Hmgb3 mRNA is present at relatively high levels in adult mouse bone marrow erythroid and primitive progenitor cells. HMGB1 and 2 have been shown to be expressed in all types of cells, but Hmgb3 represents the first example of an HMG-Box family member expressed in specific populations of hematopoietic cells.24
Despite the fact that the relative stability of Hmgb3 protein compared to GFP is unknown, the data from our Hmgb3-KI mouse indicate that Hmgb3 expression can serve as a marker for most HSCs and CLPs. The pattern of Hmgb3 expression in myeloid progenitors is more complex. Both Hmgb3+ and Hmgb3– myeloid progenitors gave rise to CFU-GM, BFU-E, and CFU-GEMM indicating that Hmgb3 expression is not confined to one myeloid lineage or one progenitor type and therefore is not a specific marker of CMPs, GMPs, or MEPs. However, in terms of absolute numbers, more myeloid progenitors express Hmgb3 than do not. The greatest differences in both CFC frequencies and absolute numbers were observed for BFU-E. Hmgb3 is expressed in a subset of Ter119+ bone marrow cells, and Hmgb3+ progenitors were enriched for BFU-E. This was observed in both FcγRII/IIILO progenitors (which contain CMPs and MEPs) and CD34+ progenitors (which contain CMPs and GMPs). We speculate that the Lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO, Hmgb3+ population significantly overlaps with MEPs (Lin–, c-kit+, Sca-1–, IL-7Rα–,FcγRII/IIILO, CD34–). Similarly, the Lin–, c-kit+, Sca-1–, IL-7Rα–, CD34+, Hmgb3+ population, which does not contain MEPs, significantly overlaps with CMPs (Lin–, c-kit+, Sca-1–, IL-7Rα–, FcγRII/IIILO CD34+) that have begun erythroid differentiation.
We propose that Hmgb3 levels must drop to allow normal differentiation program of myeloid progenitors and B cells because enforced overexpression of Hmgb3 inhibited myeloid and B-cell differentiation. The exact stage at which myeloid differentiation is affected by the level of Hmgb3 is not clear. It is possible that it varies between different myeloid lineages, since among lineage-positive myeloid cells in the bone marrow, only Ter119+ cells expressed Hmgb3. However, it also is possible that endogenous Hmgb3 expression is suppressed at the same stage in all myeloid cells, at which point Hmgb3 overexpression inhibits differentiation, and that Hmgb3 is subsequently re-expressed in Ter119+ bone marrow cells. Another possibility is that Hmgb3 is subject to posttranscriptional regulation, similar to SCL in erythroid cells, so that mRNA levels stay constant or even increase as protein levels decrease.25
The observations that CD4+ and CD8+ thymocytes from Hmgb3-KI mice express Hmgb3 but B220+ cells in the spleen and bone marrow do not suggest that Hmgb3 may play a role in the specification of the B-cell lineage in CLP. Both HMGB1 and HMGB2 have been shown to interact with the RAG1 homeodomain and enhance recombination in vitro.26 Given the high degree of homology between HMG-Box family members, it is possible that Hmgb3 can perform a similar function, critical in B-lymphocyte development.
HMG-Box family proteins bind to chromatin in a sequence-independent manner and have been shown to interact with several different types of DNA-binding proteins.10 Of these, the Hox and Oct family members both have been implicated in hematopoiesis.27 HMGB1 has been shown to interact with HOXB1, which is expressed in more differentiated hematopoietic cells, and HOXB3, which has been found in CD34+, CD38–cells.28,29 Although it is unknown whether Hmgb3 can interact with Hox proteins, given the homology between Hmgb1 and 3, we speculate that Hmgb3 can interact with Hox proteins in primitive blood cells. Enforced expression of HOXB6, HOXB7, and HOXB8 in hematopoietic cell lines has been shown to block myeloid differentiation.30-33 We propose that Hmgb3 overexpression may enhance recruitment of these Hox proteins to their cognate binding sites. HoxB6 expression has been demonstrated to play a role in erythropoiesis and the regulation of early erythrocyte progenitors.34,35 The coexpression of Hmgb3 and HoxB6 suggests a cooperative role for these proteins in erythropoiesis.
Prepublished online as Blood First Edition Paper, April 24, 2003; DOI 10.1182/blood-2002-11-3541.
M.J.N. and D.J.C. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal