Abstract
May-Hegglin anomaly (MHA), Fechtner syndrome (FTNS), Sebastian syndrome (SBS), and Epstein syndrome (EPS) are a group of rare, autosomal dominant disorders characterized by thrombocytopenia, giant platelets, and Döhle-like inclusion bodies, together with variable manifestations of Alport-like symptoms that include high-tone sensorineural deafness, cataracts, and nephritis. These disorders result from mutations in the MYH9 gene, which encodes for the nonmuscle myosin heavy chain A protein (also known as NMMHC-A). To date 20 different mutations have been characterized for this gene, but no clear phenotype-genotype correlation has been established, and very little is known regarding the molecular pathogenesis of this group of diseases. Here, we describe 2 new families with MHA/FTNS phenotypes that have been characterized in terms of their mutations, protein localization in megakaryocytes, protein expression, and mRNA stability. Our findings suggest that, at least for the Asp1424Asn mutation in the MYH9 gene, the phenotypes result from a highly unstable protein. No abnormalities in protein localization or mRNA stability were observed. We hypothesize that haploinsufficiency of the MYH9 results in a failure to properly reorganize the cytoskeleton in megakaryocytes as required for efficient platelet production.
Introduction
Familial macrothrombocytopenias with leukocyte inclusion bodies are a group of rare autosomal dominant disorders characterized by mild bleeding symptoms, giant platelets, and Döhle-like inclusion bodies in peripheral blood granulocytes. These disorders, which include the May-Hegglin anomaly (MHA; OMIM [Online Mendelian Inheritance in Man], #155100), Sebastian syndrome (SBS; OMIM, #605249), Fechtner syndrome (FTNS; OMIM, #153640), and Epstein syndrome (EPS; OMIM, #153650) all have largely overlapping phenotypes but were previously considered as separate clinical entities.1-4
Biochemical analysis of platelets from patients with MHA revealed no abnormalities in the function of these cells,5,6 leading to the hypothesis that the hematologic phenotype in patients may result from a deficit in the demarcation membranes in megakaryocytes prior to platelet formation.7
MHA and SBS are distinguished from each other by small differences in their inclusion bodies revealed by electron microscopy examination.8 FTNS and EPS, however, manifest a number of nonhematologic traits similar to those observed in Alport syndrome cases, such as nephritis, high-tone sensorineural deafness, and bilateral cataracts, all of which are present with variable expressivity.1,9,10
The discovery that the genetic loci for all of these syndromes mapped to chromosome 22q12.3 –q13.2,11-13 and the identification of mutations in the MYH9 gene for each of them, showed that this group of pathologies represent allelic variations of a single genetic disorder.14-18
MYH9, a 5.8-kb (kilobase) mRNA transcript, encodes for the nonmuscle myosin heavy chain A (also known as NMMHC-A), a large cytoplasmic protein that forms part of the myosin II hexameric complex.19,20 MYH9 consists of an adenosine triphosphatase (ATPase) globular head domain at its N-terminus and a C-terminal tail domain that forms a coiled coil structure on dimerization.
The function of nonmuscular myosin II (MYH9 containing) has not been fully characterized.21 It has been shown to form clusters of minifilaments in the cytoplasm, which concentrate in stress fibers near the periphery of cells and in the cleavage furrow of dividing cells.22 Nonmuscular myosin is involved in processes such as phagocytosis23 and cytokinesis, and in the latter it is thought to drive constriction of the cleavage furrow, as shown elegantly in Dictyostelium discoideum, where myosin II– null cells fail to divide.24 These data are consistent with the originally proposed disease mechanism of impaired thrombopoiesis as a result of defects in cytoskeleton rearrangement in megakaryocytes,7 although this is still a poorly understood process.
The MYH9 gene has subsequently been found to be involved in 2 further disorders, DFNA17 (OMIM, #603622), an autosomal dominant nonsyndromic deafness with no hematologic abnormalities,25 and APSM (OMIM, #153650), a variant of Alport syndrome with macrothrombocytopenia.16
To date, 20 different disease-associated mutations have been found in the MYH9 gene, covering the range of phenotypes represented by the 6 clinical manifestations described in the literature. Four of these mutations have been reported to be recurrent, as evidenced by their de novo origin and haplotype background.16,26,27
To date, no clear phenotype-genotype correlation has been established. Some mutations result in variable phenotypes in different individuals (Glu1841Lys, Arg1933X, Asp1424Asn, Asp1424His, Arg1165Leu, Arg1165Cys), some seem to be associated with purely hematologic manifestations (Thr1155Ile, Asn93Lys, Lys371Asn, Ala95Thr, Asp1424Tyr, del Leu1205-Gln1207; and 3 frameshift mutations: 5779delC, 5774delA, and 5828delG) and some are always associated with the more severe hematologic and Alport-like features (Arg702Cys, Arg702His, Ser114Pro, Ser96Leu).14,16,26,27 However, not enough families are available to make strong conclusions. More research is needed to clarify the mechanisms that lead to one variant or another of the disorder and to provide some insight into the pathophysiology of the disease in the different tissues involved. Another as yet unresolved question is whether the phenotypes result from haploinsufficiency, dominant-negative, or gain of function effects of the mutations.
We have studied 2 new families presenting FTNS-like phenotypes and characterized the nucleotide mutations; we subsequently studied MYH9 protein localization in megakaryocytes, MYH9 protein expression in platelets, and MYH9 mRNA stability to better understand the pathophysiology of this group of disorders.
Patients, materials, and methods
Patients
Figure 1 shows the pedigrees of family 1 (Swiss origin) and family 2 (American origin), studied here. For both families the phenotype was unambiguously ascertained on the basis of a complete hematologic evaluation. Both families presented severe thrombocytopenia (mean values of 18.2 and 8.5 × 109 platelets/L, respectively), as well as characteristic giant platelets and Döhle-like inclusion bodies on blood smear examination (not shown).
Pedigrees of 2 families affected with MHA/FTNS. All heterozygous individuals present macrothrombocytopenia. Deafness and cataracts are only present in some individuals carrying the Asp1424Asn mutation. Shading in the upper right quartile indicates macrothrombocytopenia. Shading in the upper left quartile indicates deafness, and shading in the lower right quartile indicates cataract (see “Patients”). Diagonal lines indicate deceased individuals.
Pedigrees of 2 families affected with MHA/FTNS. All heterozygous individuals present macrothrombocytopenia. Deafness and cataracts are only present in some individuals carrying the Asp1424Asn mutation. Shading in the upper right quartile indicates macrothrombocytopenia. Shading in the upper left quartile indicates deafness, and shading in the lower right quartile indicates cataract (see “Patients”). Diagonal lines indicate deceased individuals.
In family 1, 2 affected sisters developed bilateral cataracts at a young age (II-1 and II-3), whereas the third sister (II-2) and her son (III-4) had high-tone sensorineural deafness. Individuals I-2 and III-2 who presented with thrombocytopenia showed no extrahematologic symptoms. None of the individuals examined for Alport-like symptoms (I-1, II-1, II-2, II-3, III-2, and III-4) showed any signs of nephritis or any other kidney pathology.
In family 2 all members were tested for the presence of Alport-like manifestations. Individuals II-2, III-1, III-3, and IV-1 suffered from sensorineural deafness, but no cataracts or nephritis were observed in any member of this family.
The following analyses were performed on the peripheral blood and bone marrow samples of the members of the two families. Prior informed consent was obtained from the family members studied according to the Declaration of Helsinki. The bone marrow aspiration performed in member III-2 of family 1 was conducted as part of the diagnostic procedure to investigate severe thrombocytopenia. The ethical committee of the Department of Internal Medicine at Geneva University Hospital approved this study.
Nucleotide sequence analysis
Polymerase chain reaction (PCR) primers were designed to amplify all coding exons as well as the 3′-UTR (untranslated region), together with their intron/exon boundaries. We PCR-amplified all exons in one affected individual from each pedigree and an unaffected control subject, in a volume of 25 μL using 30 ng genomic DNA per reaction, 1 unit Taq (Amersham Biosciences, Buckinghamshire, England), and standard PCR conditions. PCR products were verified by standard agarose electrophoresis and purified using the Concert system (Invitrogen, San Diego, CA). Sequencing was performed with an ABI 377 system using the Big Dye terminator sequencing kit (Applied Biosystems, Foster City, CA). Sequences were then aligned and analyzed using Sequencher 4.0.5 (Gene Codes, Ann Arbor, MI).
Potential mutations were verified and tested in the population using the pyrosequencing system28 (Pyrosequencing AB, Uppsala, Sweden). For this step, 8 μL Dynabeads was used for PCR immobilization in a final volume of 40 μL. The rest of the protocol was performed according to manufacturer's instructions.
Microsatellite genotyping
Polymorphic markers were analyzed by PCR using radiolabeled primers. One oligonucleotide primer of each marker was labeled with 5 μCi (0.185 MBq) γ32P-ATP (adenosine triphosphate) with T4 polynucleotide kinase. PCR was performed using standard conditions.
Amplification products were separated by electrophoresis in a 6% denaturing urea/polyacrylamide gel, and genotypes were independently scored by 2 different investigators after autoradiography.
Immunohistochemistry
For immunofluorescence staining, bone marrow smears from 5 different donors and from the patient with MHA (family 1 III-2) were first fixed in 4% paraformaldehyde (10 minutes at room temperature), followed by absolute acetone (3 minutes at –20°C). We used an affinity-purified rabbit polyclonal immunoglobulin (IgG) that recognizes only MYH9 (NMMHC-A) and MYH10 (NMMHC-B)29 (Biomedical Technologies, Stoughton, MA). This step was followed by a TRITC (tetramethylrhodamine-B-isothiocyanate)–conjugated swine antirabbit IgG (Dako, Glostrup, Denmark).The secondary antibody alone was used as a negative control.
The stained cells were analyzed using a confocal laser scanning fluorescence inverted microscope (LSM 410; Carl Zeiss, Jena, Germany) equipped with a helium-neon (He-Ne) laser (excitation wavelength at 543 nm).30 Cells were observed through an oil immersion planeofluar X63/1.4 objective, and the visual field was enhanced by zooming in 2 times.
SDS-PAGE and immunoblotting
For sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), platelet extracts of control subjects (n = 4) and patients (n = 3) were suspended in 0.4 M Tris (tris(hydroxymethyl)aminomethane) HCl, pH 6.8, containing 1% SDS, 1% dithiothreitol, 1 mM phenylmethyl sulfonyl fluoride, 1 mM Nα-p-tosyl-l-arginine methyl ester and boiled for 3 minutes. Protein content was determined according to Bradford.31 Proteins (40 μg) were electrophoresed on a 5% to 20% gradient gel and stained with Coomassie blue. For quantification of total actin, gels were scanned with a computerized scanner (Arcus II; AGFA, Mortsel, Belgium).
Samples were loaded according to their total actin content and were electrophoresed on a 5% to 20% gradient gel. Western blotting was performed using the same MYH9 antibody as earlier, a monoclonal antibody recognizing all actin isoforms (clone IC4; Sigma, St Louis, MO) and a monoclonal antibody specific for α-tubulin (Sigma). Separated proteins were transferred to nitrocellulose filters32 that were incubated with anti-MYH9 (1:1000), antitotal actin (1:10 000), and anti–α-tubulin (1:500) for 2 hours. After 3 washes, a second incubation for 1 hour was performed with goat antirabbit (for MYH9) or antimouse (for actin and α-tubulin) IgG labeled with peroxidase. Enhanced chemiluminescence was used for detection (Amersham Biosciences). Total MYH9 and α-tubulin expression were evaluated by densitometric scanning of the Western blots and expressed as mean percentage of control conditions.
Statistical comparison between different sample groups was performed using an unpaired 2-tailed Student t test.
mRNA stability
Total RNA was extracted from blood of 3 subjects with MHA/FTNS, using the Trizol Reagent (Invitrogen), and cDNA was produced using the Superscript II enzyme (Invitrogen) and an oligo dT primer. PCR using cDNA-specific primers (spanning an intron in genomic DNA; 5′-TGCTGAGGAGGTGAAGAGGA, 3′-GCGACAGAGCCTTGGTCTC), in which the forward primer was labeled with 5′ biotin, was performed under standard conditions, and the product was analyzed by the pyrosequencing method. Briefly, this method (http://www.pyrosequencing.com) works by a series of 4 enzymatic reactions in which the number of nucleotide incorporations is quantitatively measured. Each time a nucleotide is incorporated by DNA polymerase, a pyrophosphate is released (hence the term “pyrosequencing”), which is detected by the sulphorylase and luciferase enzymes, that generate a signal proportional to the number of nucleotides incorporated.
For pyrosequencing, an internal primer (CCAGGTCCACCAGCAGG) was designed 2 nucleotides before the mutation site, so that the 2 mRNA populations could be assayed by quantifying the relative amounts of each allele present in the PCR product. DNA samples from healthy and affected subjects were used as controls.
PCR products were immobilized through the 5′ biotin in the forward primer with Dynabeads (Dynal, Oslo, Norway) by a 15-minute, 65°C incubation in a buffer containing 10 mM Tris-HCl, 2 M NaCl, 1 mM EDTA (ethylenediaminetetraacetic acid), and 0.1% Tween 20. PCR products were then removed from solution using magnetic separation, denatured with NaOH 0.5 M, and washed with 200 mM Tris-acetate, 50 mM magnesium acetate. The remaining single-stranded DNA was then hybridized with the internal “sequencing” primer, by heating the mix to 80°C, and slowly cooling it down to room temperature. Enzyme and substrate mixes were then automatically added to each well, and the reactions proceeded at 28°C, with the sequential addition of single nucleotides in a predetermined order. Luciferase peak heights are proportional to the number of nucleotide incorporations, which has been shown to be very quantitative (5% error rate) in a number of experimental settings.33,34
As an alternative method for measuring the stability of the mutant mRNA, we used real-time quantification of the relative proportion of the mutant versus the “wild-type” allele. For this, we used the ABI 7900 sequence detection system (Applied Biosystems).
Common PCR primers were used to amplify an 81–base pair (bp) fragment spanning the Asp1424Asn mutation (5′GACAAGCTGGAGAAGACCAAGAC, 5′GCTCTGGCGCTGGTGGT). Taqman probes specific for each allele were designed (VIC-TGGACGACCTGCTG and 6FAM-TGGACAACCTGCTGG), and analysis of the results was performed using the SDS 2.0 software (Applied Biosystems). All quantifications were performed in 3 replicates.
Results
Mutation analysis
All 40 exons of the MYH9 gene, including intron/exon junctions, and the full 3′-UTR were sequenced in one affected individual from each pedigree, as well as in an unaffected white control subject. Analysis of aligned sequences revealed a missense mutation in exon 30 of the gene c.4270G>A, which causes a conservative amino acid substitution Asp1424Asn in the rodlike tail domain (data not shown). This mutation, which surprisingly was present in heterozygosity in both pedigrees, was shown to cosegregate with the disease phenotype and to be absent from the general population as revealed by pyrosequencing analysis of 100 population-matched control subjects.
To determine whether the 2 mutations were of independent origin we genotyped 5 microsatellite markers (D22S1147, D22S1142, D22S683, D22S283, D22S445) surrounding an approximate 2-Mb region around the mutation (data not shown).
We concluded that the 2 mutations are of independent origin because (1) the mutation in pedigree 2 is a de novo event in individual II-2, as some of his siblings that share the same haplotype identical by descent do not carry the mutation, and (2) the mutation in each pedigree is present on totally different haplotype backgrounds on either side of the pathogenic mutation.
Immunohistochemistry
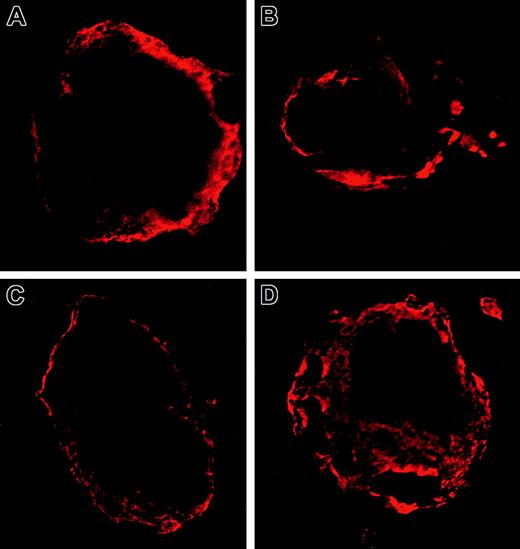
To study whether the Asp1424Asn MYH9 protein variant mislocalizes in megakaryocytes, immunofluorescence using confocal microscopy was performed (Figure 2). No significant differences in localization were observed between megakaryocytes of a patient with MHA/FTNS (Figure 2C-D) and 5 healthy donors (Figure 2A-B), on examination of at least 10 different cells per individual by 2 independent pathologists. In both affected individuals and control subjects we observe a diffuse cytoplasmic staining with a higher intensity toward the periphery of the cell and the cell membrane. There is thus no evidence that the Asp1424Ala mutation causes changes in localization that could be related to the hematologic pathology observed in patients with MHA/FTNS.
MYH9 localization in megakaryocytes. Immunofluorescence micrographs of the bone marrow from a healthy donor (A-B) and the patient with the Asp1424Asn mutation (C-D). Shown are 2 megakaryocytes with a typical staining of their cytoskeleton with MYH9/MYH10 antibodies in one of the 5 donors studied and the patient, respectively. Rhodamine staining. Original magnification × 1000 for all panels.
MYH9 localization in megakaryocytes. Immunofluorescence micrographs of the bone marrow from a healthy donor (A-B) and the patient with the Asp1424Asn mutation (C-D). Shown are 2 megakaryocytes with a typical staining of their cytoskeleton with MYH9/MYH10 antibodies in one of the 5 donors studied and the patient, respectively. Rhodamine staining. Original magnification × 1000 for all panels.
Immunoblotting
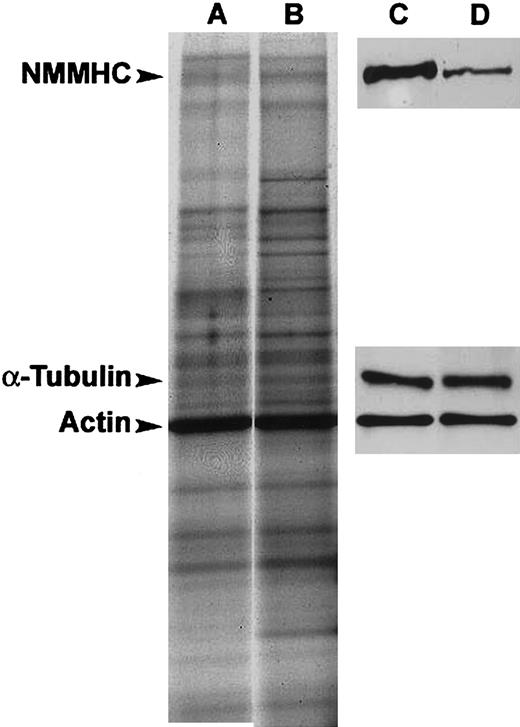
To determine whether there are differences in the steady-state levels of MYH9 protein in patients carrying the Asp1424Asn mutation, we performed a Western blot (Figure 3). The antibody used recognizes both MYH9 and the highly homologous MYH10 protein; however, only the former is expressed in platelets.22 By densitometric analysis of the Western blot, we showed that, as expected, the level of total actin was highly similar in all samples studied (90% ± 6% in 3 patients compared with the 4 control subjects, P = .5). Similarly, the α-tubulin levels did not significantly vary across the samples (129% ± 13% in patients compared with control subjects, P = .4). The level of MYH9, however, markedly decreased, revealing a specific reduction of 49% ± 9% in the amount of steady-state MYH9 protein in platelets of individuals with the Asp1424Asn mutation as compared with control subjects (patients [n = 3], controls [n = 4], P < .01).
Immunoblotting. SDS-PAGE of total protein extracts (A-B) and immunoblots showing MYH9, total actin, and α-tubulin expression (C-D) in a representative control subject (A,C) and a patient with the Asp1424Asn mutation (B,D). Loading was normalized to total actin concentration.
Immunoblotting. SDS-PAGE of total protein extracts (A-B) and immunoblots showing MYH9, total actin, and α-tubulin expression (C-D) in a representative control subject (A,C) and a patient with the Asp1424Asn mutation (B,D). Loading was normalized to total actin concentration.
mRNA stability
To investigate whether the c.4270G>A mutation affects the stability of the MYH9 mRNA transcript, a relative quantification of mutant versus wild-type mRNA molecules from the cDNA of 3 affected individuals was performed by pyrosequencing.
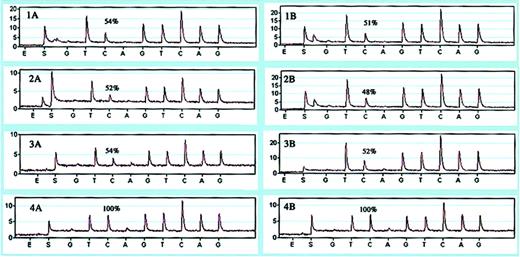
To validate that the pyrosequencing assay at this position is quantitative, we first analyzed the relative abundance of the alleles in DNA from 3 patients heterozygous at this position (Figure 4, panels 1B-3B). We obtained a value of 50.33% ± 2.08% for the wild-type allele represented by the C peak (mutation is G → A, but pyrosequencing was performed in the reverse orientation) that is very close to the expected 50% value, thus showing that the assay is quantitative.
Relative quantification of normal versus mutant alleles. Pyrograms showing the percentage of wild-type allele (first C-peak) in cDNA (1A-4A) and DNA (1B-4B) of 3 patients (1-3) and a control individual (4).
Relative quantification of normal versus mutant alleles. Pyrograms showing the percentage of wild-type allele (first C-peak) in cDNA (1A-4A) and DNA (1B-4B) of 3 patients (1-3) and a control individual (4).
We then performed the same assay on cDNA from 3 affected individuals. Our results show no significant differences in the levels of the 2 mRNA populations, because the mutant mRNA population was present at a relative frequency of 47.66% ± 1.15% (Figure 4, panels 1A-3A). A similar analysis was performed on cDNA and DNA from a control individual, which, as expected, showed 100% of the wild-type allele (Figure 4, panels 4A and 4B, respectively).
To confirm mRNA stability results obtained with the pyrosequencing technique, we designed an allele quantification assay using a real-time PCR detection system. The results of this experiment (Table 1) show that, as with the pyrosequencer, the ratio of the mutant versus the wild-type alleles both in DNA and in cDNA do not significantly deviate from one, demonstrating that their steady-state levels in blood are the same.
Allele ratios quantified by Taqman
Individual . | RNA . | DNA . |
|---|---|---|
| Family 1 II-1 | 1.04 ± 0.004 | 1.15 ± 0.007 |
| Family 1 III-2 | 1.04 ± 0.015 | 1.15 ± 0.031 |
| Family 1 IV-1 | 1.04 ± 0.001 | 1.16 ± 0.012 |
Individual . | RNA . | DNA . |
|---|---|---|
| Family 1 II-1 | 1.04 ± 0.004 | 1.15 ± 0.007 |
| Family 1 III-2 | 1.04 ± 0.015 | 1.15 ± 0.031 |
| Family 1 IV-1 | 1.04 ± 0.001 | 1.16 ± 0.012 |
Discussion
Here, we report 2 additional families with MHA/FTNS. Complete sequencing analysis of the MYH9 gene revealed an Asp1424Asn missense mutation present in heterozygosity in both pedigrees. We determined by haplotype analysis that this mutation arose independently in each family, and the same mutation has been previously described in patients of Japanese and American origin.16,17,27 Recurrent mutations are indicative of amino acid positions that are critical for protein function, because the likelihood that a mutation hits the same position multiple times by chance is very low.35,36 Interestingly, 2 other missense mutations Asp1424His and Asp1424Tyr have also been described at this site, further emphasizing the importance of this residue. The fact that the Asp1424Asn mutation is more common than the other 2 mutations at this residue can be readily explained by the fact that the G→A change occurs in the context of a CG dinucleotide that is known to be particularly mutation prone.37
The tail part of the protein in which residue Asp1424 is located is structurally not very well characterized. However, the numerous 3-dimensional structures of coiled-coil regions available (eg, PDB 1C1G, 1I84, 1D7M), all have a very pronounced distribution of charged and uncharged residues. By trying different alignments with the heptameric repeat of the coiled-coil, residue 1424 is most likely located on the outside surface of the coil, and hence a charge change (Asp→Asn) could disturb the electrostatic interaction between the α-helices as has been previously proposed.15
To gain further insights into the effects of the mutation and how these effects might be related to the MHA/FTNS phenotype, we studied MYH9 localization in megakaryocytes, expression of the protein variant, and relative stability of the mutant mRNA molecules.
Immunofluorescence staining of megakaryocytes in bone marrow smears revealed no significant differences in MYH9 localization between the MHA/FTNS megakaryocytes and those from 5 unrelated control subjects. This finding is interesting because distinct localization patterns and protein inclusions are observed in platelets and neutrophils from affected individuals.17,38,39 This observation suggests that the differences observed in platelet MYH9 localization are most likely a consequence of abnormal thrombopoiesis and not a cause of the disease; this observation is also supported by the fact that platelets from affected individuals are functionally normal.
We then studied the steady-state level of the MYH9 protein in total platelet extracts from patients and control subjects by quantitative Western blot (Figure 3). Our results show a clear and specific decrease in the amount of MYH9 protein in Asp1424Asn platelet extracts as compared with control samples. We observed that, although other proteins such as total actin and α-tubulin remained unchanged, there was a reduction of about 50% in the levels of MYH9 in 3 Asp1424Asn samples as compared with control samples. The reduction observed at the protein level can be either the result of an mRNA effect, such as mRNA instability, or a reduced rate of transcription, or else protein instability as a direct consequence of the amino acid substitution.
Although in principle missense mutations should not affect mRNA stability, several cases have been reported in which this situation is the case.40 To exclude this possibility, we measured the ratio of mutant versus wild-type MYH9 mRNA in blood from affected individuals, using a method based on the pyrosequencing technology.28,33 If the mutant mRNA were unstable or less transcribed, one would expect the ratio between the alleles to deviate from one. As a control for the method, one can use genomic DNA from heterozygous individuals, in which the proportion between the alleles is known to be 1:1. Our data show no decrease in the steady-state levels of the mutant mRNA, because both the normal and the Asp1424Asn mRNAs are present in equal amounts (Figure 4).These results were further confirmed by real-time PCR experiments using allele-specific Taqman probes, in which no differences in the rate of amplification of the 2 alleles could be detected. Taken together, these results strongly suggest that the Asp1424Asn substitution causes the MYH9 protein to become highly unstable, which is consistent with the 3-D protein modeling.
Because we observed no altered localization or aggregation of the mutant protein in megakaryocytes, we hypothesize that the pathology of the disease, at least at the hematologic level, is due to haploinsufficiency rather than to a dominant-negative effect of the mutation and that the decrease in the amount of MYH9 probably interferes with the normal cytoskeletal rearrangements in megakaryocytes necessary for efficient platelet production.
Two important questions that remain concern the molecular mechanisms of mutations in extrahematologic tissues, and the factors that lead to the phenotypic variability of the disease, even within members of the same family,16 as is the case in pedigree 1 of our study. Hence, the presence of genetic-modifying loci needs to be evaluated.
In summary, we have characterized the effects of the MYH9 Asp1424Asn mutation at the cellular localization, protein, and mRNA level, to obtain new insights into the pathophysiology of MHA/FTNS. We hypothesize that haploinsufficiency resulting from the high instability of the mutant MYH9 protein could explain the mutation mechanism, at least at the hematologic level.
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-09-2783.
Supported by grants 32-61845.00 (P.B.) and 31-57149.99 (S.E.A.) from the Swiss National Foundation for Scientific Research.
S.D. and A.R. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked ``advertisement'' in accordance with 18 U.S.C. section 1734.
We thank Dr A. Reymond for advice and critical comments, Dr R. Lyle for assistance with the Taqman-related experiments, Dr C. Rossier for assistance with the sequencing, M. Papasavvas and U. Choudhury for technical support, and J. Ringrose for secretarial help.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal