Abstract
Two functionally distinct subsets of B cells that produce Th1- and Th2-like patterns of cytokines have recently been identified. Interleukin-12 (IL-12) is a critical immunoregulatory cytokine that promotes Th1 differentiation through activation of signal transducer and activator of transcription 4 (STAT4). IL-12 has been reported to induce interferon γ (IFN-γ) production in B cells, but the relevant signaling pathways are poorly documented. Here, in human primary B cells, we found a functional IL-12 receptor (IL-12R) that internalizes following IL-12 binding. IFN-γ and, to a lesser extent, IL-12 positively regulated the IL-12Rβ2 subunit but had no effect on IL-12Rβ1. On examining the effect of IL-12 on STAT4 and T-bet (2 key factors involved in IFN-γ promoter activation), we found that IL-12 induced the phosphorylation and nuclear translocation of STAT4. IL-12-dependent constitutive STAT4 activation was also observed in the Epstein-Barr virus (EBV)-transformed B-cell line RPMI 8866 that spontaneously produces IL-12. T-bet expression has been shown to be dependent on STAT1. IL-12 had no direct effect on STAT1 activation or T-bet expression in primary B cells. In contrast, IL-12-induced IFN-γ led to STAT1 activation, strong expression of T-bet, and IFN-γ expression. IL-12 therefore initiates a cascade of events in B cells, including STAT4 activation, IL-12Rβ2 up-regulation, IFN-γ production, and T-bet up-regulation, potentially leading to Th1-like differentiation. (Blood. 2003;102:4084-4089)
Introduction
In several models of infection, B cells have been shown to influence the development and maintenance of CD4+ and CD8+ T-cell functions; this suggests that B lymphocytes have regulatory functions, some of which may be dependent on cytokine production.1-5 Recently, it was demonstrated that B cells have the potential to differentiate into 2 distinct effector cell populations (Be1 and Be2) that produce interferon-γ (IFN-γ) and interleukin 4 (IL-4), respectively.6 This suggests that the T-helper (Th)1/Th2 paradigm may also apply to B cells. Cross-regulation may exist between Be1/Be2 cells and Th1/Th2 cells.6 However, the signals required for Th1/2-like commitment of B cells are unclear. IL-12, a critical immunoregulatory cytokine that promotes Th1 commitment, may play a significant role in Be1 differentiation. B cells express IL-12 receptor (IL-12R) mRNA.7 IL-12 acts in synergy with IL-18 to induce IFN-γ secretion by these cells.7,8 However, the signaling pathways triggered by IL-12 are poorly known in B cells.7-9
Here, we examined IL-12R expression and investigated the IL-12 signaling pathway in B cells. Our results indicate that human B cells express functional IL-12R and that IL-12 initiates a cascade of molecular events leading to IFN-γ expression and successively involving signal transducer and activator of transcription 4 (STAT4) and T-box expressed in T cells (T-bet), 2 transcription factors critically involved in Th1 development.
Materials and methods
Primary B cells and B-cell lines
Highly purified primary B and CD4+ T cells were obtained from peripheral blood mononuclear cells (PBMCs) of healthy donors following CD19 and CD4 positive selection with a magnetic separation system, according to the manufacturer's instructions (Miltenyi Biotech, Bergish Gladbach, Germany). B-cell purity was always examined by flow cytometry with CD3, CD14, CD16, and CD22 staining (Becton Dickinson, San Jose, CA). The RPMI 8866 Epstein-Barr virus-positive (EBV+) B-cell line was obtained from the European Cell Culture Collection (ECACC, Salisbury, United Kingdom).
mRNA extraction and cDNA synthesis
Purified primary B cells or EBV+ B-cell line (2 × 105/0.2 mL/well) were cultured in 96-well plates in RPMI 1640 medium (Gibco, Carlsbad, CA) supplemented with 10% fetal calf serum (FCS; Biochrom, Berlin, Germany). Primary B cells were activated for up to 72 hours with recombinant human IFN-γ (R&D Systems, Minneapolis, MN), recombinant bioactive IL-12 (R&D Systems), recombinant human IL-18 (R&D Systems), or with the combination of IL-12 plus IL-18. Cytokines were used at a concentration of 100 ng/mL. Cells were also activated for 24 hours with 10 μg/mL anti-CD40 (Diaclone, Besançon, France). RNA was extracted by using the Chomoczinsky method, as modified in the RNAble kit (Eurobio, les Ulis, France). RNA was resuspended in 10 μL diethyl pyrocarbonate (DEPC)-treated water and stored at -80°C until use. cDNAs were obtained by using random hexamers and avian myeloblastosis virus reverse transcriptase (First Strand cDNA Synthesis Kit, Roche Diagnostics, Meylan, France). Reverse transcription-polymerase chain reaction (RT-PCR) controls containing no RNA, and others containing RNA but no reverse transcriptase, were always used and were always negative.
Quantitative PCR
IL-12Rβ1, IL-12Rβ2, T-bet, IFN-γ, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA levels were determined by using Light Cycler-based kinetic quantitative PCR (Roche Diagnostics). IL-12Rβ1, IL-12Rβ2, T-bet, and GAPDH PCR products were detected by using FastStart DNA Master hybridization probes (Roche Diagnostics).
The IFN-γ PCR product was detected by using FastStart DNA Master SYBR Green I (Roche Diagnostics). An external DNA scale was used to quantify cDNA.10-13 To correct for variations in RNA recovery and the reverse transcription yield, the amounts of IL-12R subunit, T-bet, and IFN-γ cDNA were divided by the amount of GAPDH.10-13 Results were expressed as -fold increases of normalized values,10,11 over the level observed with untreated cells (for IL-12R subunit and T-bet) or over the value obtained at the 1-hour time point (for IFN-γ because B cells did not express detectable IFN-γ at rest).
PCR
PCR was run in 50 μL amplification buffer (Gibco BRL) containing 1 μL cDNA, 0.25 mM each primer, and 1 U Taq platinum (Gibco BRL).
Primers and probes
The IL-12Rβ1, IL-12Rβ2, and GAPDH primers have been described elsewhere.10,11 The following primers and probes were used: T-bet 5′ primer: CTAAAGCTCACAAACAACAAGG and 3′ primer: AGAAGCGGCTGGGAACAGGAT; IFN-γ 5′ primer: TCTTGGCTTTTCAGCTCTGCATCG and 3′ primer: GCTGGCGACAGTTCAGCCATCA; IL-12 p40 5′ primer: GAGAAATGGTGGTCCTCACCTGTG and 3′ primer: GAGTGTAGCAGCTCCGCACGTC; IL-12 p35 5′ primer: AGCCTCCTCCTTGTCGCTACC and 3′ primer: GCCTCCACTGTGCTGGTTTTATC; IL-4 5′ primer: TCTCACCTCCCAACTGCTTC and 3′ primer: GCACAGAGTCTTCTGCTCTG; IL-5 5′ primer: TCTGAGGATTCCTGTTCCTG and 3′ primer: TTATCCACTCGGTGTTCATT IL-13 5′ primer: TCGAGAAGACCCAGAGGAT and 3′ primer: AGCACAGGCTGAGGTCTAA; IL-12Rβ1 probes: 5′-CACCTCAGCACCAACCTGGTTATC-Red-640-3′ and 5′-fluorescein-GGGTCTCCCACTCCATACGCAG(phosphate)-3′; IL-12Rβ2 probes: 5′-GCCGATATCTGAGTCGATTAAGCA-Red640-3′ and 5′-fluorescein-GTACCAGTCCCTCATCTCTCCAA(phosphate)-3′; T-bet probes: 5′-Red 640-GCCTACCAGAATGCCGAGATTACT(phosphate)-3′ and 5′-CAAGAAACCCAGTTCATTGCCGTG-fluorescein-3′; GAPDH probes: 5′-ATGGCAACAATATCCACTTTACCAGAG-Red 640-3′ and 5′-fluorescein-AAAAGCCCTGGTGACCAGGCG(phosphate)-3′.
Confocal microscopy
For IL-12/IL-12R studies, cells were incubated with 100 ng/mL recombinant human IFN-γ (R&D Systems) for 24 hours followed by 100 ng/mL recombinant bioactive IL-12 (R&D Systems) for 15 minutes at 4°C or 37°C. Cells were washed, fixed, and permeabilized (Orthopermeafix, Ortho Diagnostic Systems, Raritan, NJ), then incubated with both a mouse monoclonal antihuman IL-12 p70 monoclonal antibody (mAb; 10 μg/mL, R&D Systems) and a polyclonal rabbit antihuman IL-12R antibody (10 μg/mL, Santa Cruz Biotechnology, Santa Cruz, CA). The final incubation step was with both a phycoerythrin (PE)-labeled goat antimouse IgG antibody diluted 1:50 and fluorescein isothiocyanate (FITC)-labeled goat antirabbit IgG (Southern Biotechnology, Birmingham, AL) diluted 1:50. In the corresponding control experiment, anti-IL-12R and anti-IL-12 p70 were replaced by a rabbit IgG isotype control (Santa Cruz Biotechnology) and a mouse IgG1 isotype control (R&D Systems), respectively. For STAT4 confocal microscopy, cells were treated with IL-12 for 15 minutes, washed, fixed and permeabilized, and then incubated with 10 μg/mL of a polyclonal rabbit IgG antihuman STAT4 (Santa Cruz Biotechnology). A second staining step was performed with 10 μg/mL Alexa FluorTM 488-labeled goat antirabbit IgG antibody (Molecular Probes, Montluçon, France). Nuclear staining was performed with propidium iodide diluted 1:50 (Sigma, Saint Quentin Fallavier, France). Cells were analyzed by laser scanning confocal microscopy in an ACAS 570 Interactive Laser Cytometer (Meridian Instruments, Okemos, MI). In some experiments, to neutralize endogenous IL-12 production by B-cell line, 10 μg/mL of a neutralizing monoclonal antihuman IL-12 antibody (mouse IgG1; R&D Systems) was added to the culture medium 1 hour before staining. In the corresponding control experiments, the specific antibodies were replaced by a rabbit IgG isotype control (Santa Cruz Biotechnology).
Western blot
Purified B cells were incubated with RPMI 1640 medium containing 10% FCS and treated with IL-12 (100 ng/mL) or IFN-γ (100 ng/mL) for 15 and 45 minutes at 37°C. They were then washed with phosphate-buffered saline (PBS) containing 50 μM Na3VO4 and resuspended in lysis buffer with a protease inhibitor cocktail (Roche Diagnostics). Cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF) membranes (NEN Research Products, Boston, MA). The membranes were blocked with 5% nonfat milk and probed with rabbit polyclonal IgG anti-pSTAT4 (Zymed Laboratories, San Francisco, CA) or mouse monoclonal IgG anti-pSTAT1 (Santa Cruz Biotechnology). The membranes were washed and then incubated with goat peroxidase-conjugated antirabbit or antimouse IgG (Southern Biotechnology). After washing, bands were visualized by adding Western blotting chemiluminescence luminol reagent (Santa Cruz Biotechnology). The membranes were then washed and reprobed with anti-STAT4 or anti-STAT1 (rabbit polyclonal IgG; Santa Cruz Biotechnology). Densitometric analysis, with background correction, was performed with National Institutes of Health (NIH) Image software. To correct for possible variations in the amount of loaded proteins, values were expressed as pSTAT/STAT ratios. Results were expressed as -fold increases versus untreated cells.
IFN-γ production
Purified primary B cells and CD4+ T cells (1 × 106 per well) were cultured in 48-well plates in a total volume of 500 μL 10% FCS-RPMI 1640 medium. The cells were treated with recombinant bioactive IL-12 (R&D Systems) or IL-12 plus IL-18 (Peprotech, Rocky Hill, NJ) for 48 hours. Cytokines were used at the concentration of 100 ng/mL and were added at the initiation of cultures and 24 hours after. In some experiments, B cells were treated with IL-12 plus Staphylococcus aureus Cowan I (SAC, 1:10 000 vol/vol; Calbiochem, La Jolla, CA) for 48 hours. Control wells included untreated cells and cells treated with IL-18 alone and SAC alone. Supernatants were harvested and assayed for IFN-γ by enzyme-linked immunosorbent assay (ELISA; human IFN-γ Quantikine HS; R&D Systems).
Effect of endogenous IFN-γ on T-bet expression
Purified primary B cells (2 × 105/0.2 mL/well) were treated with IL-12 or IL-12 plus IL-18 as described (see “IFN-γ production”), for up to 72 hours in the presence or absence of 10 μg/mL of a neutralizing monoclonal antihuman IFN-γ antibody (R&D Systems). Antihuman IFN-γ antibody was added at the initiation of culture and 24 hours after. Cells were lysed at the indicated time points and T-bet was quantified by RT-PCR.
Results
A functional IL-12R is expressed and regulated in primary B cells
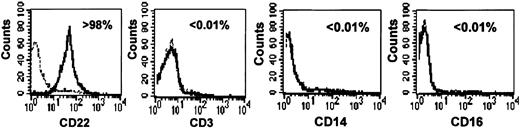
As shown in Figure 1, highly purified primary B cells (> 98% pure) were obtained by using a standardized anti-CD19-coated magnetic microbead procedure. T cell, natural killer (NK) cell, and monocyte contamination was below 0.01%, on the basis of CD3, CD16, and CD14 staining, respectively (Figure 1).
Purity of primary human B-cell preparations. B cells were isolated from PBMCs by CD19 positive selection. Purity was assessed by flow cytometry (see “Materials and methods”) using CD22, CD3, CD16, and CD14 staining to detect B cells, T cells, NK cells, and monocytes, respectively. The percentage of stained cells is indicated in each panel. Dotted lines correspond to isotypic controls.
Purity of primary human B-cell preparations. B cells were isolated from PBMCs by CD19 positive selection. Purity was assessed by flow cytometry (see “Materials and methods”) using CD22, CD3, CD16, and CD14 staining to detect B cells, T cells, NK cells, and monocytes, respectively. The percentage of stained cells is indicated in each panel. Dotted lines correspond to isotypic controls.
As shown in Figure 2A, IL-12Rβ2 mRNA was detected in untreated B cells. Cell activation for 24 hours with IFN-γ and, to a lesser extent, IL-12 significantly up-regulated IL-12Rβ2 mRNA levels, whereas anti-CD40 had no significant effect on constitutive IL-12Rβ2 mRNA expression (Figure 2C). IL-12Rβ1 mRNA was also detected in untreated B cells (Figure 2B). As shown in Figure 2D, activation with IFN-γ, IL-12, or anti-CD40 for 24 hours had no significant effect on IL-12Rβ1 expression. We also investigated IL-12R expression in the EBV-transformed human B-cell line RPMI 8866. This cell line expressed both IL-12Rβ2 and IL-12Rβ1 mRNA (data not shown).
Expression of IL-12R mRNA in primary B cells. IL-12Rβ2 (A) and IL-12Rβ1 (B) PCR curves for untreated purified primary B cells. In panels C and D, quantitative PCR was used to measure IL-12Rβ2 and IL-12Rβ1 mRNA expression (see “Materials and methods”) following activation with IFN-γ, IL-12, or anti-CD40 for 24 hours. Results (means ± SEMs) are expressed as -fold increases in normalized cDNA values of IL-12Rβ2 (C) and IL-12Rβ1 (D) versus the level in untreated cells. 0-PCR indicates PCR controls without cDNA; 0-RNA, RT-PCR controls without RNA; 0-RT, RT-PCR controls with RNA but without reverse transcriptase.
Expression of IL-12R mRNA in primary B cells. IL-12Rβ2 (A) and IL-12Rβ1 (B) PCR curves for untreated purified primary B cells. In panels C and D, quantitative PCR was used to measure IL-12Rβ2 and IL-12Rβ1 mRNA expression (see “Materials and methods”) following activation with IFN-γ, IL-12, or anti-CD40 for 24 hours. Results (means ± SEMs) are expressed as -fold increases in normalized cDNA values of IL-12Rβ2 (C) and IL-12Rβ1 (D) versus the level in untreated cells. 0-PCR indicates PCR controls without cDNA; 0-RNA, RT-PCR controls without RNA; 0-RT, RT-PCR controls with RNA but without reverse transcriptase.
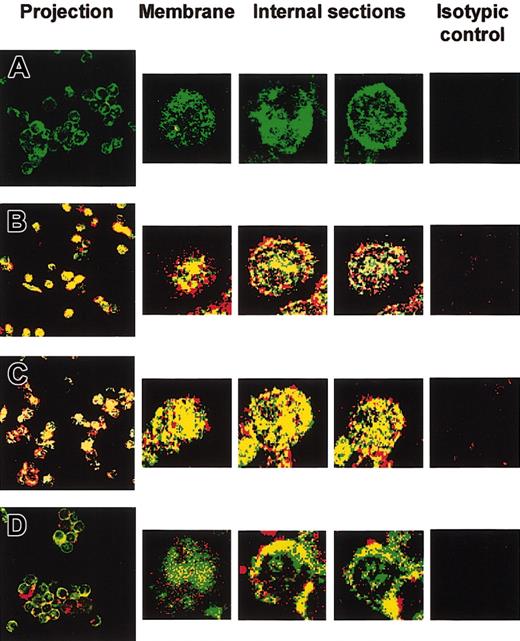
We then examined IL-12/IL-12R complex internalization by using confocal laser scanning microscopy. Primary B cells were treated for 24 hours with IFN-γ to up-regulate IL-12R expression. Cells were then incubated for 15 minutes with IL-12. Internalization of IL-12/IL-12R complexes was examined after dual labeling with anti-IL-12 p70 (red) and anti-IL-12Rβ1 (green). Yellow staining indicated colocalization of IL-12 p70 and IL-12Rβ1. Confocal microscopic analysis of 1-μm optical sections from the surface of untreated cells showed IL-12R expression both at the cell surface and intracellularly (Figure 3A). No endogenous IL-12 was found in untreated (Figure 3A) or IFN-γ-preactivated cells (not shown). Following IL-12 treatment, confocal microscopy showed intracellular colocalization of IL-12 and IL-12Rβ1 in non-preactivated and IFN-γ-preactivated B cells (Figure 3B and C, respectively). IL-12/IL-12R complex internalization was significantly inhibited at 4°C in both non-preactivated (Figure 3D) and IFN-γ-preactivated B cells (not shown), although IL-12 still bound to its cell surface receptor (Figure 3D).
Confocal microscopy of IL-12/IL-12Rβ1 colocalization in B cells. Untreated and 24-hour IFN-γ-activated primary B cells were treated with recombinant IL-12 for 15 minutes at 4 or 37°C, before intracellular staining with anti-IL-12 and anti-IL-12Rβ1. Confocal microscopic analysis of serial 1-μm optical sections was performed from the cell surface to the cell interior. Superimposed IL-12 (red) and IL-12Rβ1 (green) staining patterns are shown. Yellow staining indicates IL-12 and IL-12Rβ1 colocalization (original magnification, × 63). The projection image of the different optical sections is shown. Microscopy confocal analysis of 3 individual serial sections from the surface to the interior of one cell is also shown (5-fold magnification of projection image). (A) Control without IL-12. (B-C) Non-preactivated (B) and 24-hour IFN-γ-preactivated (C) primary B cells were treated with recombinant IL-12 at 37°C. (D) Non-preactivated cells were treated with recombinant IL-12 at 4°C. Isotype controls were used, as described in “Materials and methods,” for each of the 4 experimental conditions.
Confocal microscopy of IL-12/IL-12Rβ1 colocalization in B cells. Untreated and 24-hour IFN-γ-activated primary B cells were treated with recombinant IL-12 for 15 minutes at 4 or 37°C, before intracellular staining with anti-IL-12 and anti-IL-12Rβ1. Confocal microscopic analysis of serial 1-μm optical sections was performed from the cell surface to the cell interior. Superimposed IL-12 (red) and IL-12Rβ1 (green) staining patterns are shown. Yellow staining indicates IL-12 and IL-12Rβ1 colocalization (original magnification, × 63). The projection image of the different optical sections is shown. Microscopy confocal analysis of 3 individual serial sections from the surface to the interior of one cell is also shown (5-fold magnification of projection image). (A) Control without IL-12. (B-C) Non-preactivated (B) and 24-hour IFN-γ-preactivated (C) primary B cells were treated with recombinant IL-12 at 37°C. (D) Non-preactivated cells were treated with recombinant IL-12 at 4°C. Isotype controls were used, as described in “Materials and methods,” for each of the 4 experimental conditions.
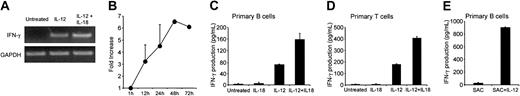
IL-12R was functional because treatment of human B cells with IL-12 led to IFN-γ mRNA and protein expression (Figure 4A-C). IFN-γ mRNA expression in response to IL-12 peaked at 48 hours (Figure 4B). Whereas IL-18 alone had no significant effect on IFN-γ production, it potentiated that triggered by IL-12 (Figure 4C). Similar results were obtained with purified primary T cells (Figure 4D). Activation of B cells with SAC, but not with anti-CD40 (not shown), strongly increased IFN-γ production in response to IL-12 (Figure 4E).
Effect of IL-12 on IFN-γ expression by primary B cells. (A-B) IFN-γ mRNA expression. Purified primary B cells were treated for up to 72 hours with IL-12 or with IL-12 plus IL-18. Corresponding cDNA was amplified with IFN-γ primers. Panel A shows the specific PCR bands for the 48-hour time point. In panel B, IFN-γ mRNA was quantified as indicated in “Materials and methods.” Results (means ± SEMs) are expressed as -fold increases in normalized cDNA values of IFN-γ versus the level at the 1-hour time point, because B cells did not express detectable IFN-γ at rest. (C-E) IFN-γ secretion in supernatants. (C-D) Purified B and T cells were treated for 48 hours with IL-12 or with IL-12 plus IL-18, as described in “Materials and methods.” (E) B cells were treated for 48 hours with SAC plus IL-12, as described in “Materials and methods.” Culture supernatants were harvested and assayed for IFN-γ by ELISA. Similar results were obtained in 3 other experiments.
Effect of IL-12 on IFN-γ expression by primary B cells. (A-B) IFN-γ mRNA expression. Purified primary B cells were treated for up to 72 hours with IL-12 or with IL-12 plus IL-18. Corresponding cDNA was amplified with IFN-γ primers. Panel A shows the specific PCR bands for the 48-hour time point. In panel B, IFN-γ mRNA was quantified as indicated in “Materials and methods.” Results (means ± SEMs) are expressed as -fold increases in normalized cDNA values of IFN-γ versus the level at the 1-hour time point, because B cells did not express detectable IFN-γ at rest. (C-E) IFN-γ secretion in supernatants. (C-D) Purified B and T cells were treated for 48 hours with IL-12 or with IL-12 plus IL-18, as described in “Materials and methods.” (E) B cells were treated for 48 hours with SAC plus IL-12, as described in “Materials and methods.” Culture supernatants were harvested and assayed for IFN-γ by ELISA. Similar results were obtained in 3 other experiments.
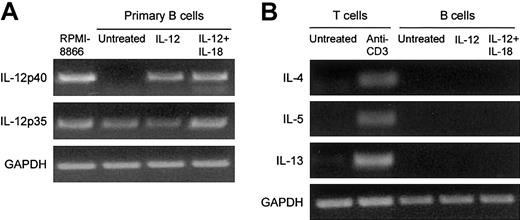
As shown in Figure 5, B cells spontaneously expressed IL-12 p35 but not IL-12 p40 (Figure 5A). IL-12 induced IL-12 p40 mRNA expression by B cells (Figure 5A). In contrast, IL-12 had no effect on the expression of the Th2-like cytokines IL-4, IL-5, and IL-13 (Figure 5B).
Effect of IL-12 on IL-12 and Th2-like cytokine expression by primary B cells. Purified primary B cells were treated for 48 hours with IL-12 or with IL-12 plus IL-18. The corresponding cDNA was amplified with IL-12 p40, IL-12 p35, IL-4, IL-5, and IL-13. The results of PCR product migration are shown in panels A and B. As controls, we used the RPMI 8866 B-cell line for IL-12 p40 expression and purified T cells activated with anti-CD3 for Th2-like cytokines.
Effect of IL-12 on IL-12 and Th2-like cytokine expression by primary B cells. Purified primary B cells were treated for 48 hours with IL-12 or with IL-12 plus IL-18. The corresponding cDNA was amplified with IL-12 p40, IL-12 p35, IL-4, IL-5, and IL-13. The results of PCR product migration are shown in panels A and B. As controls, we used the RPMI 8866 B-cell line for IL-12 p40 expression and purified T cells activated with anti-CD3 for Th2-like cytokines.
Together, these experiments showed the presence of a functional IL-12R in human B cells.
IL-12 induces STAT4 activation in human B cells
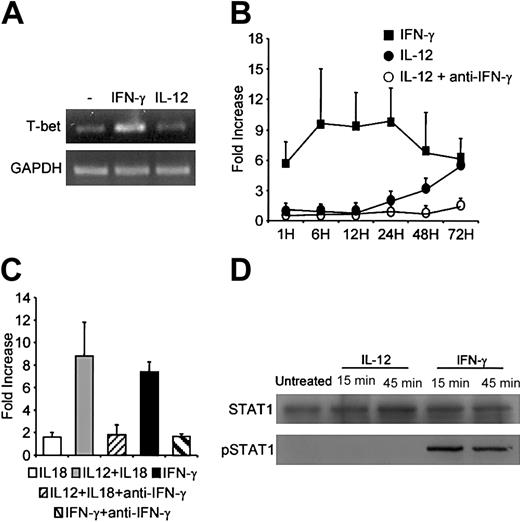
IL-12 induced IFN-γ production by human B cells. The transcription factor STAT4 is involved in IFN-γ promoter activation.14 STAT4 activation includes a tyrosine phosphorylation step. We used Western blotting to determine whether IL-12 affected STAT4 activation in primary B cells. Cell lysates were immunoblotted with antiphosphotyrosine STAT4 (pSTAT4), then reblotted with anti-STAT4. As shown in Figure 6A, B cells constitutively expressed STAT4. A low level of pSTAT4 was found in untreated cells (Figure 6A). After treatment of cells with IL-12 for 15 minutes, densitometric analysis of the bands and measurement of the pSTAT/STAT ratios showed significant tyrosine phosphorylation of STAT4 (Figure 6A). After 45 minutes of IL-12 activation, a slight decrease in the pSTAT4/STAT4 density ratio was observed relative to the 15-minute time point (4.4-fold and 3.3-fold increases over baseline at 15 minutes and 45 minutes of IL-12 activation, respectively). We next used confocal microscopy to determine whether IL-12 induces nuclear translocation of STAT4. Primary B cells were activated with IL-12 and then dually stained with an anti-STAT4 mAb (green spots) and propidium iodide (red) for nuclear staining. As shown in Figure 6Bi, despite the detection of low levels of pSTAT4 by Western blot in untreated cells, no significant constitutive nuclear localization (yellow spots) of STAT4 was found. IL-12 treatment led to nuclear translocation of STAT4 (Figure 6Bii). The B-cell line RPMI 8866 constitutively secretes high levels of IL-1215 and expresses IL-12R. We therefore also examined STAT4 activation in these cells. As shown in Figure 6Ci, STAT4 was present in the nucleus of these cells, indicating constitutive activation. In contrast, following treatment with a neutralizing anti-IL-12 antibody, STAT4 nuclear content fell markedly (Figure 6Cii).
Effect of IL-12 on STAT4 activation. (A) Western blot. B cells were activated with IL-12 for 15 and 45 minutes and then lysed. Western blotting was performed with whole-cell lysates by using antiphospho-STAT4. Membranes were reprobed with anti-STAT4. To correct for variations in the amount of loaded proteins, values are expressed as pSTAT/STAT ratios. Phospho-STAT/STAT levels were determined by densitometric analysis, including correction for background (NIH Image software). Results are expressed as -fold increases versus untreated cells. (B) Confocal microscopy of primary B cells. B cells were activated with IL-12 for 15 minutes. They were then fixed, permeabilized, and stained with anti-STAT4 (i-ii; green) and propidium iodide (nuclear staining, red). Yellow spots indicate nuclear STAT4 localization. (Bi) Untreated cells; (Bii) IL-12-activated cells. The small squares show STAT4 staining (green) without superimposition. In control experiments, anti-STAT4 was replaced by a rabbit IgG isotypic control. (Biii) Isotypic control of IL-12-treated cells. (C) Confocal microscopy of the RPMI 8866 cell line. Cells were extensively washed then treated with neutralizing anti-IL-12 for 1 hour. Cells were fixed, permeabilized, and stained as described. (Ci) Untreated cells; (Cii) Anti-IL-12-treated cells; (Ciii) Isotypic control of untreated cells. For panels A to C, similar results were obtained in 2 other experiments. Original magnification, × 63.
Effect of IL-12 on STAT4 activation. (A) Western blot. B cells were activated with IL-12 for 15 and 45 minutes and then lysed. Western blotting was performed with whole-cell lysates by using antiphospho-STAT4. Membranes were reprobed with anti-STAT4. To correct for variations in the amount of loaded proteins, values are expressed as pSTAT/STAT ratios. Phospho-STAT/STAT levels were determined by densitometric analysis, including correction for background (NIH Image software). Results are expressed as -fold increases versus untreated cells. (B) Confocal microscopy of primary B cells. B cells were activated with IL-12 for 15 minutes. They were then fixed, permeabilized, and stained with anti-STAT4 (i-ii; green) and propidium iodide (nuclear staining, red). Yellow spots indicate nuclear STAT4 localization. (Bi) Untreated cells; (Bii) IL-12-activated cells. The small squares show STAT4 staining (green) without superimposition. In control experiments, anti-STAT4 was replaced by a rabbit IgG isotypic control. (Biii) Isotypic control of IL-12-treated cells. (C) Confocal microscopy of the RPMI 8866 cell line. Cells were extensively washed then treated with neutralizing anti-IL-12 for 1 hour. Cells were fixed, permeabilized, and stained as described. (Ci) Untreated cells; (Cii) Anti-IL-12-treated cells; (Ciii) Isotypic control of untreated cells. For panels A to C, similar results were obtained in 2 other experiments. Original magnification, × 63.
Together, these results indicate that the IL-12 signaling pathway in B cells involves STAT4 activation.
IL-12 has no significant direct effect on STAT1 activation or T-bet expression in B cells
T-bet is a transcription factor whose expression correlates with IFN-γ expression in T cells.16 We investigated T-bet expression in B cells following IL-12 stimulation for up to 72 hours. As shown in Figure 7A, constitutive T-bet mRNA expression was found in primary B cells. IFN-γ induced a rapid increase in T-bet, reaching a plateau between 6 and 24 hours after activation (Figure 7B). In contrast, T-bet expression, in response to IL-12, started to increase significantly only after 48 hours and was inhibited by neutralizing anti-IFN-γ (Figure 7B). This indicated an indirect effect of IL-12, via IFN-γ, on T-bet induction. Similar IFN-γ-dependent T-bet expression was observed following B-cell treatment with the combination of IL-12 plus IL-18 for 48 hours (Figure 7C). T-bet expression was higher than with IL-12 alone because IL-18 potentiates IFN-γ production in response to IL-12 (Figures 4C and 7B-C). Because T-bet expression is STAT1 dependent,17,18 we examined whether IL-12 affected STAT1 activation in primary B cells. As shown in Figure 7D, B cells constitutively expressed STAT1. No detectable pSTAT1 was found in untreated cells (Figure 7D). Significant tyrosine phosphorylation of STAT1 was found after cell treatment with IFN-γ for 15 minutes or 45 minutes (Figure 7D). In contrast, IL-12 had no detectable effect on STAT1 tyrosine phosphorylation.
Effect of IL-12 and IFN-γ on T-bet mRNA expression and STAT1 activation in B cells. (A-C) T-bet mRNA expression. In panels A and B, purified primary B cells were activated for up to 72 hours with IL-12, IFN-γ, or IL-12 plus anti-IFN-γ and then lysed at various time points. mRNA was extracted and reverse transcribed. Panel A shows the PCR bands for the 24-hour time point with T-bet-specific primers. In panel B, T-bet and GAPDH cDNAs were quantified as described in “Materials and methods.” Results (means ± SEMs) are expressed as -fold increases in normalized T-bet cDNA values versus untreated cells. (C) B cells were activated for 48 hours in various conditions as indicated. T-bet mRNA expression was quantified as described. (D) STAT1 activation. Purified primary B cells were treated with IL-12 or IFN-γ for 15 and 45 minutes and then lysed. Western blotting was performed on whole-cell lysates by using antiphospho-STAT1. Membranes were reprobed with anti-STAT1.
Effect of IL-12 and IFN-γ on T-bet mRNA expression and STAT1 activation in B cells. (A-C) T-bet mRNA expression. In panels A and B, purified primary B cells were activated for up to 72 hours with IL-12, IFN-γ, or IL-12 plus anti-IFN-γ and then lysed at various time points. mRNA was extracted and reverse transcribed. Panel A shows the PCR bands for the 24-hour time point with T-bet-specific primers. In panel B, T-bet and GAPDH cDNAs were quantified as described in “Materials and methods.” Results (means ± SEMs) are expressed as -fold increases in normalized T-bet cDNA values versus untreated cells. (C) B cells were activated for 48 hours in various conditions as indicated. T-bet mRNA expression was quantified as described. (D) STAT1 activation. Purified primary B cells were treated with IL-12 or IFN-γ for 15 and 45 minutes and then lysed. Western blotting was performed on whole-cell lysates by using antiphospho-STAT1. Membranes were reprobed with anti-STAT1.
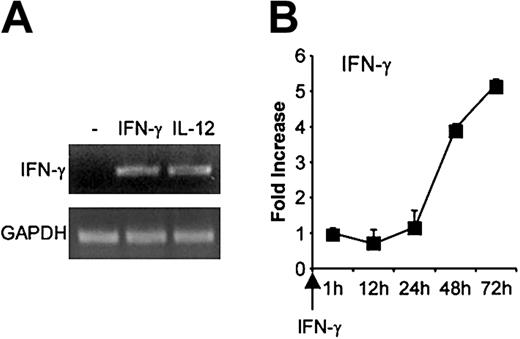
Because IFN-γ up-regulated T-bet expression in primary B cells, we also examined whether IFN-γ influenced its own expression. As shown in Figure 8A, IFN-γ or IL-12 activation of B cells for 48 hours induced detectable IFN-γ mRNA. We then examined the kinetic of IFN-γ mRNA expression in response to IFN-γ for up to 72 hours after activation. IFN-γ started to be detectable 1 hour after activation (not shown), then increased slowly to reach a 5.2-fold increase relative to the 1-hour time point, 72 hours after activation (Figure 8B).
Effect of IFN-γ on its own expression in B cells. Purified primary B cells were activated with IFN-γ for up to 72 hours, then mRNA was extracted and reverse transcribed. cDNA was amplified with IFN-γ primers. (A) PCR bands for the 48-hour time point. (B) IFN-γ mRNA expression was quantified as described. Results (means ± SEMs) are expressed as -fold increases in normalized IFN-γ cDNA values versus the level at the 1-hour time point.
Effect of IFN-γ on its own expression in B cells. Purified primary B cells were activated with IFN-γ for up to 72 hours, then mRNA was extracted and reverse transcribed. cDNA was amplified with IFN-γ primers. (A) PCR bands for the 48-hour time point. (B) IFN-γ mRNA expression was quantified as described. Results (means ± SEMs) are expressed as -fold increases in normalized IFN-γ cDNA values versus the level at the 1-hour time point.
Together, these results suggest that IL-12 acts on B cells, activates STAT4, and induces IFN-γ production. IFN-γ, in turn, activates STAT1, up-regulates T-bet expression, and also induces its own expression in an autocrine fashion.
Discussion
We report that human primary B cells isolated from peripheral blood possess a functional IL-12 receptor that internalizes on IL-12 binding. This receptor is regulated by IFN-γ and, to a lesser extent, by IL-12. IL-12 led to STAT4 activation and IFN-γ production. In turn, IFN-γ triggered a signaling pathway involving STAT1, which led to T-bet up-regulation and IFN-γ expression. These results point to a “successive wave” model of IFN-γ expression following IL-12 activation of B cells.
IL-12R has been identified on several human cell types, including activated T and NK cells, and also macrophages, dendritic cells, and microglial cells.10,19-23 Coexpression of IL-12Rβ1 and IL-12Rβ2 subunits is required to generate IL-12 high-affinity-binding sites, and is also necessary for IL-12 signaling.24 We found that IL-12Rβ2 was positively regulated at the mRNA level by IFN-γ and, to a lesser extent, by IL-12. IL-12 binding to its receptor in primary B cells led to rapid internalization of the corresponding complex. In primary B cells, we observed marked STAT4 tyrosine phosphorylation and nuclear translocation in response to IL-12. We also found IL-12-dependent STAT4 activation in the B-cell line RPMI 8866. STAT4 activation is a key event in the IL-12 signaling cascade in Th1 cells.25-28 STAT4 has been shown to be activated through tyrosine phosphorylation via direct interaction with the phosphotyrosine residue 800-containing peptide located in the cytoplasmic domain of the IL-12Rβ2 subunit.29 This could explain why STAT4 activation in response to IL-12 is shared by various cell types expressing STAT4 and functional IL-12R.27,28,30,31 STAT4 binds the human IFN-γ promoter.14 Activation of this transcription factor is required for IL-12-dependent IFN-γ induction in T cells.14 Our results therefore suggest that IFN-γ production in response to IL-12 in B cells involves STAT4 activation. Combined treatment with IL-12 and IL-18 led to higher IFN-γ production than treatment with IL-12 alone. IL-18 has been shown to act on the IFN-γ promoter by activating the transcription factor AP-1.14 The synergistic action of IL-12 and IL-18 on the IFN-γ promoter may be related to the orchestrated action of STAT4 and AP-1. In addition, IL-12 has been shown to up-regulate IL-18R.7,32 Of interest, activation of B cells with SAC, which acts by interacting with surface immunoglobulin, led to marked up-regulation of IFN-γ production in response to IL-12, suggesting that antigen signal could physiologically amplify IFN-γ secretion by B cells in response to this cytokine.
Another factor that controls IFN-γ expression in T cells is the newly described T-box transcription factor T-bet.16 T-bet transactivates the IFN-γ gene and induces chromatin remodeling of individual IFN-γ alleles.33 Although we detected constitutive expression of this factor in B cells, we observed no IFN-γ transcription, suggesting that the level of T-bet is a limiting factor. Moreover, the T-bet effect on the IFN-γ promoter may require additional inducible factors to be effective. In T cells, T-bet has been shown to cooperate with the inducible homeoprotein Hlx, or with nuclear factor of activated T cells (NFAT1), for optimal induction of IFN-γ.34,35 We found that IL-12 had no significant direct effect on T-bet expression in B cells. This could be related to the absence of STAT1 activation in response to this cytokine. STAT1 (but not STAT4) knock-out mice show defective T-bet induction, pointing to a key role of this transcription factor in the control of T-bet gene expression.17,18 Activation of B cells to IFN-γ production with the combination of IL-12 and IL-18 led to T-bet up-regulation, and this induction was neutralized by anti-IFN-γ. Direct activation of primary B cells by IFN-γ induced STAT1 activation and T-bet up-regulation. T-bet and IFN-γ also regulate each other in T lymphocytes and in antigen-presenting myeloid cells.17 This argues for a model common to several cell types, in which IFN-γ gene regulation involves an autocrine loop. In primary B cells, IFN-γ induced IL-12Rβ2 up-regulation and the transcription of its own gene. This dual effect likely involves T-bet, as ectopic T-bet leads to IL-12Rβ2 expression in T cells from STAT1 knock-out mice.18 Together, these results indicate that STAT4 and T-bet—2 key factors in Th1 differentiation—are also involved in a cascade of molecular events initiated by IL-12 that leads to IFN-γ expression and IL-12Rβ2 up-regulation in B cells. Further studies using gene knock-out mice will be helpful to confirm the roles of STAT4, STAT1, and T-bet in the cascade from IL-12 to IFN-γ in B cells. Interestingly, we found that IL-12 induced IL-12 p40 mRNA expression in B cells. Although this should be confirmed at the protein level, it raises the possibility of an autocrine IL-12 loop in B cells that may amplify or prolong this cytokine action. In contrast, IL-12 had no detectable effect on the expression of the Th2-like cytokines IL-4, IL-13, and IL-5. Our findings are in line with the recent identification of 2 populations of mouse effector B cells (Be1 and Be2) that produce Th1- and Th2-like patterns of cytokines depending on the cytokine environment in which the cells are activated during their primary antigen encounter.6 Mice deficient in T-bet have impaired IgG2a production,36 whereas ectopic T-bet expression gives T-bet-deficient cells the ability to generate IgG2a germline transcripts and to secrete IgG2a.36 This raises the question of whether Be1 cells express a specific immunoglobulin isotype. During initial antigen presentation to B cells within the T-cell area of lymph nodes, IL-12 provided by dendritic cells may trigger a cascade of molecular events in naive B cells possibly leading to Th1-like differentiation, which includes STAT4 activation, IFN-γ production, T-bet up-regulation, IL-12Rβ2 up-regulation, and immunoglobulin isotype switching. Additional signals such as BCR signal or other cytokines derived from dendritic cells or T cells may participate with IL-12 in such a polarization process. Polarized B-cell subsets may, in turn, regulate T-cell differentiation and functions. Our results support the extension of the Th1/Th2 paradigm to B cells and provide new insights into B cell-mediated immune responses.
Prepublished online as Blood First Edition Paper, July 31, 2003; DOI 10.1182/blood-2003-02-0518.
Supported by grants from Association de Recherche contre le Cancer (ARC), Agence Pour la Recherche Contre le SIDA (ANRS), and UQAM.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal