Abstract
Because of the pivotal role the proteasome plays in apoptosis, inhibitors of this enzyme, such as PS-341, provide a great opportunity for exploring synergy between proteasome inhibition and other apoptosis-inducing agents. Tumor necrosis factor—related apoptosis-inducing ligand (TRAIL) can selectively induce apoptosis in tumor cells. In overnight assays, combinations of PS-341 and TRAIL were much more effective than either agent alone in promoting apoptosis of a murine myeloid leukemia, C1498, and a murine renal cancer, Renca. For C1498 cells, apoptosis sensitization by PS-341 affected neither the activity of nuclear factor κB (NF-κB) nor the levels of most antiapoptotic proteins. However, reductions in the antiapoptotic protein c-FLIP in response to PS-341 were observed in both C1498 and Renca cells. Treatment of normal bone marrow mixed with C1498 tumor cells for 18 hours with a combination of PS-341 and TRAIL resulted in a specific depletion of the tumor cells. Upon transfer to irradiated syngeneic recipient mice, mixtures treated with the PS-341 plus TRAIL combination resulted in enhanced long-term tumor-free survival of mice. These data therefore support the targeting of apoptotic pathways in tumor cells, using combinations of agents such as PS-341 and TRAIL that interact synergistically to preferentially promote tumor cell apoptosis. (Blood. 2003;102:303-310)
Introduction
One of the major problems in cancer therapy is in defining specific agents, or combinations of agents, that are highly toxic to cancer cells yet have limited toxicity on normal nontransformed cells. Recently, a number of novel chemical or biologic agents have been reported to possess these characteristics.1 The proteasome inhibitor PS-341 has been shown to be toxic to multiple solid and hematologic cancer cell lines in vitro and in vivo.1-3 Of significance is the differential effects PS-341 exhibits on these tumor cells over normal cycling cells. The enhanced sensitivity of the cancer cells toward PS-341 provides its therapeutic window, which can be especially large in hematologic malignancies.4 In addition, the therapeutic utility of PS-341 has recently been confirmed in multiple clinical trials focusing on both solid and hematologic cancer.5-7 Treatment of tumor cells with PS-341 results in multiple biologic effects, including inhibition of the cell cycle, increased apoptosis, changes in cell adherence, and inhibition of nuclear factor κB (NF-κB) activation.8 Indeed, the inhibition of NF-κB may even underlay some of the proapoptotic effects of PS-341. Because of the pivotal role the proteasome plays in apoptosis, inhibitors of this enzyme, such as PS-341, provide an opportunity for exploring synergy between proteasome inhibition and other apoptosis-inducing agents.
Tumor necrosis factor (TNF)—related apoptosis-inducing ligand (TRAIL) has also been proposed as a novel anticancer agent because of its ability to induce apoptosis in tumor cells, while having little effect on a range of normal cells.9 Interestingly, on binding to its receptors, TRAIL causes not only Fas-associated death domain-containing protein (FADD) recruitment and caspase activation but can also result in the activation of NF-κB. NF-κB is a key transcriptional regulator of several genes involved in apoptosis.10 NF-κB can activate the transcription of antiapoptotic genes that include cIAP-1; cIAP-2; XIAP; the TNF-receptor—associated factor, TRAF1; the antiapoptotic protein, IEX-IL; as well as the Bcl-2 homologues, A1/Bf1 and Bcl-Xl.11-14 Indeed, transfection of various combinations of cIAP-1, cIAP-2, TRAF-1, and TRAF-2 can protect certain cells against apoptosis mediated by TNF-α or etoposide.11 NF-κB is also reported to increase expression of c-FLIP, which can block extrinsic apoptosis via death receptors.15 Furthermore, the blocking of NF-κB activation by either nondegradable forms of the inhibitor of κ-B (Iκ-B) or proteasome inhibitors can enhance the apoptotic response of some tumor cells to TNF-α or TRAIL in vitro.16-18 However, studies on both human renal carcinoma cells and various transfectants of 293 cells suggest that inhibition of NF-κB activity does not modulate sensitivity to TRAIL in all cells.19,20 Additionally, although the in vitro data are encouraging, no studies to date have described whether combinations of proteasome inhibitors and death ligands such as TRAIL could provide therapeutic benefit in relevant animal models in vivo.
Patients with acute leukemia, lymphomas,21 and testicular cancer22 have been cured by high-dose chemotherapy (HDC). The major dose-limiting toxicity of HDC or radiation therapy is bone marrow suppression; therefore bone marrow transplantation (BMT) has been used following high doses of these regimens.23 This approach has also been applied to breast cancer patients using hematopoietic stem cell transplantation (HSCT),24 although whether this therapy provides significant benefit has proved controversial.25 Nonetheless, relapse remains an important barrier to successful BMT, and one potential problem involves tumor cell contamination of the autologous BM transplant. Since small numbers of tumor cells may persist in any autologous bone marrow inoculum used for hematopoietic rescue, novel methods allowing for the specific purging of these tumor cells from the bone marrow inocula are highly desirable.
Using the murine C1498 acute myeloid leukemia (AML) tumor model and the murine renal carcinoma Renca, we have assessed the effects of PS-341 and TRAIL on the death of these tumor cells in vitro. Furthermore, we have attempted to purge C1498 tumor cells from bone marrow—tumor cell mixtures. These bone marrow—tumor cell mixtures have then been transferred to irradiated syngeneic recipient mice. This model allowed us to estimate the efficacy with which combinations of PS-341 and TRAIL could destroy tumor cells. However, it also allowed for an analysis of the toxicity of the PS-341 plus TRAIL combination, since upon transfer of the bone marrow to irradiated recipients, survival of the donor bone marrow stem cells would be necessary for complete hematopoietic reconstitution.
Materials and methods
Mice
C57BL/6 (B6, H2b) and B6-Ly5.2 congenic mice were obtained from the Animal Production Area of the National Cancer Institute (Frederick, MD). All animals were kept in pathogen-free conditions and used at 8 to 12 weeks of age. Animal care was provided in accordance with the procedures outlined in the Guide for the Care and Use of Laboratory Animals.26
Reagents
Renca, a murine renal cancer of BALB/c origin was provided by Dr R. Wiltrout (NCI-Frederick). C1498, a murine myeloid leukemia cell line of B6 origin, and A20 lymphoma cells (BALB/c) were obtained from the American Type Culture Collection (Rockville, MD). All cell lines were maintained in RPMI-1640 media supplemented with 10% fetal calf serum (FCS), 2 mM l-glutamine, 1 × nonessential amino acids, 1 mM sodium pyruvate, 100 U/mL penicillin, 100 μg/mL streptomycin, 10 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), and 5 × 10-5 M 2-mercaptoethanol (2-ME), pH 7.4 (complete medium). Polyclonal antibodies to cIAP-1 and cIAP-2, TRAIL R2/DR5 (R&D Systems, Minneapolis, MN), XIAP (BD BioSciences, San Diego, CA), Bax and BclXL (Santa Cruz Biotechnology, Santa Cruz, CA), Bcl-2 (Oncogene, San Diego, CA), and a monoclonal antibody to β-actin (Sigma, St Louis, MI) were used as described by the manufacturers. The monoclonal antibody to c-FLIP (clone Dave-2) was purchased from Kamiya Biomedical (Seattle, WA). The proteasome substrate Succinyl-Leu-Leu Val-Tyr-Amido-methyl-coumarin was obtained from Bachem (King of Prussia, PA). The enzyme inhibitors Z-Val-Ala-Asp-(OMe)-CH2F (zVAD-FMK) and Z-Phe-Ala-(OMe)-CH2F (zFA-FMK) were purchased from Enzyme Systems Products (Dublin, CA). Soluble mouse recombinant TRAIL was purchased from Biomol (Plymouth Meeting, PA) and was cross-linked by adding a monoclonal anti-6× histidine antibody (R&D Systems) at a concentration of 2 μg antibody/μg TRAIL. The NF-κB inhibitors SN50 and Helenalin (Biomol) were used as recommended by the manufacturers. PS-341 was obtained from Millennium Pharmaceuticals, Cambridge, MA.
Cytotoxicity assays
Tumor target cells C1498 or Renca were labeled with 111In-labeled Oxine [111In] Ox (Medi-Physics, Silver Spring, MD) as previously described.27 The cells were pretreated with various concentrations of PS-341 for 2 hours prior to the addition of TRAIL. In caspase inhibition studies, the caspase inhibitor zVAD-FMK or control zFA-FMK at a final concentration of 50 μM was also added 2 hours prior to TRAIL. After 18 hours at 37°C, the release of isotope from target cells was calculated as previously described.27 In some assays, as an alternative to isotope labeling, cell viability was assessed by the addition of the novel tetrazolium compound (3-[4,5-dimethylthiazol-2-yl-5]-[3-carboxymethyoxyphenyl]-2-[4-sulfophenyl]-2H tetrazolium) MTS (Promega, Madison, WI) for 4 hours, followed by measuring absorbance at 450 nm on an enzyme-linked immunosorbent assay (ELISA) microplate reader. Apoptosis was also estimated using the Apoptosis Detection Kit (R&D Systems) as described by the manufacturers. C1498 cells were treated for 18 hours with PS-341 and/or TRAIL as described previously. Cells were then isolated and Annexin V staining and propidium iodide uptake were then analyzed on a FACScan as previously described.28 For long-term clonal survival assays, C1498 cells (105 cells/mL) were incubated for 18 hours with PS-341 in the presence or absence of TRAIL. Cells were then washed thoroughly, and various dilutions in 100 μL fresh tissue culture media were incubated at 37°C on flat-bottomed microtiter plates. At various times viable cell numbers from all treatment groups were estimated by MTS staining. A standard curve with fixed numbers of C1498 cells (2500-80 000) was always performed at each time point.
Proteasome inhibition assay
The proteasome assay was performed essentially as previously described.29 Briefly, C1498 cells at 1 × 106 cells/mL or Renca at 1.25 × 105 cells/mL were incubated for various periods of time on Costar 6 tissue-culture plates in the presence or absence of various concentrations of PS-341. Cells were then isolated and washed twice in phosphate-buffered saline (PBS). Cell pellets were extracted by incubation with 200 μL lysis buffer (50 mM HEPES, 5 mM CHAPS [3-[(3-cholamidopropyl)dimethylammonio]-1-propane-sulfonic acid], 0.5 mM EDTA [ethylenediaminetetraacetic acid], 0.035% sodium dodecyl sulfate [SDS], pH 7.5) for one hour on ice. Samples were then centrifuged at 14 000 rpm for 10 minutes in an eppendorf centrifuge and supernatants were isolated. Protein concentrations in each supernatant were then estimated using the bicinchoninic acid (BCA) protein assay. Enzyme assays were performed in 96-well microtiter plates. Protein (50 μg) was added to 90 μL lysis buffer, plates were warmed for 10 minutes at 37°C, and 10 μL of the Succ-Leu-Leu-Val-Tyr-AMC substrate was added to give a final concentration of 150 μM. The resultant fluorescence of the liberated AMC (7-amido-4-methylcoumarin) dye was then measured on a Cytofluor multiwell plate reader series 4000 (Perspective Biosystems, Framington, MA) set on an excitation wavelength of 380 nM and emission of 460 nm.
Analysis of NF-κB activity
C1498 cells or A20 cells were washed twice with Dulbecco phosphate-buffered saline and once with RPMI-1640 without serum. Washed cells were resuspended to 10 × 106/mL in RPMI-1640 without serum. Resuspended cells (0.5 mL) were used for each transfection. The transfection was carried out using ElectroSquarePorator with 250 V at low voltage (LV) mode and 3 pulses with 10-milliseconds pulse length in a 4-mm gap cuvette. Reporter construct (5 μg) was used for each transfection. pTATA-luc was a pGL3-basic—derived construct with TATA minimal promoter. pNF-κB-luc was constructed with 3 copies of NF-κB binding element upstream of the TATA in pTATA-luc. In all transfections, pRL-null was included as an internal control construct in order to correct for differences in transfection and harvesting efficiency. After 36-hour culture in RPMI-1640 with 10% FCS, the transfected cells were equally distributed to either 24-well or 48-well plates in RPMI-1640 without phenol-red containing 5% FCS in the presence of either vehicle or agents at various concentrations and periods of time, as indicated in the figure legends. The treated cells were transferred to 1.5-mL eppendorf microcentrifuge tube and centrifuged at 1300g for 5 minutes. The pelleted cells were washed twice with PBS and then harvested with passive-lysis buffer. Both firefly luciferase and Renilla luciferase activities were measured according to the procedures of the dual-luciferase assay (Promega). Each treatment was performed at least in quadruplicate, and the mean value for each of the treatments on a plate was expressed as a percentage/fold of the change relative to the controls on the same plate.
Western blotting
Western blotting was performed using Novex minigels as previously described.30 Briefly, C1498 or Renca tumor cells were incubated for various periods of time with different concentrations of PS-341. After isolation of the cells and extraction in lysis buffer, gel electrophoresis was performed running 20 μg protein under reducing conditions. After transfer of proteins to polyvinylidene membranes, antibody dilutions were added in overnight incubations at 4°C. The antibody dilutions were anti—bcl-2 (1:20), bax (1:5000), bcl-xL (1:200), TRAIL receptor DR5 (1:1000), cIAP-1 (1:1000), cIAP-2 (1:1000), XIAP (1:250), and anti-FLIP (1:1000). Appropriate dilutions of secondary antibodies conjugated to horseradish peroxidase were then added for one hour and blots were developed using the enhanced chemiluminescence Plus kit (Amersham, Little Chalfont, England).
Transfection of c-FLIP
C1498 cells (2 × 104 cells/well) were plated in Costar 24-well plates 24 hours prior to transfection. The cDNA for a FLAG-tagged murine c-FLIP was kindly provided by Dr J. P. Medema (University of Leiden, the Netherlands). Transfection was performed with 0.8 μg plasmid DNA per well using electroporation as previously described. Transfected cells were cloned by single-cell cloning in 96-well plates under selection with G418 sulfate (Geneticin; 400 μg/mL). Clones were screened for c-FLIP expression by Western blotting as described in the previous section.
Bone marrow—tumor cell transfers
For experiments assessing the antitumor effects of PS-341 and recombinant murine TRAIL (rmTRAIL), replicates a of mixture of 6 × 107 B6-Ly5.2 (CD45.1+) bone marrow cells (BMCs) and 1.2 × 105 C1498 cells were preincubated with or without 20 nM PS-341 for 4 hours in 10 mL Iscove modified Dulbecco medium (IMDM) supplemented with 10% fetal bovine serum, 5 × 10-5 M 2-ME, and 100 U/mL penicillin, 100 μg/mL streptomycin (10% IMDM) at 37°C, 5% CO2. At 4 hours, rmTRAIL was added to appropriate cultures at 1 μg/mL and incubated for an additional 20 hours at 37°C, 5% CO2. Untreated BMCs without C1498 were also cultured as a control. The cocultured cells were then harvested and resuspended in Dulbecco phosphate-buffered saline (DPBS) at 5 × 106 BMCs and 1 × 104 C1498 per 0.2 mL based on the cell numbers at the initiation of the coculture. B6 (CD45.2+) recipients were irradiated at 8 Gy and infused with the cells at 5 × 106 BMCs and 1 × 104 C1498 per mouse (intravenously), and survival of mice was monitored for up to 100 days. To determine the number of tumor cells surviving after these treatments, aliquots from bone marrow—tumor cell cocultures were transferred to RPMI media containing 10% FCS. At various time points viable cell counts were estimated by trypan blue exclusion. Alternatively, tumor cell numbers were estimated at various time points by MTS staining as previously described. Briefly after overnight incubation of bone marrow—C1498 mixtures in the presence or absence of various treatments, cells were pelleted, washed thoroughly, then resuspended in media at various dilutions and incubated for different periods of time at 37°C, 5% CO2. To assess tumor cell number, MTS was added, and tumor cell number at each time point was calculated based on a standard curve generated with C1498 cells. The contribution of bone marrow cells to the reduction of MTS in this assay at the cell dilutions used was negligible.
Hematopoietic reconstitution by donor bone marrow cells
To estimate the effects of PS-341 and TRAIL on BMC reconstitution in vivo, peripheral blood samples from recipient mice were collected in EDTA-treated Microtainer tubes (Becton Dickinson, San Jose, CA) at 14 or 60 days after BMT. Different parameters of hematopoietic recovery were determined by analyzing the blood samples on HEMAVET Multispecies Hematology Analyzer (CDC Technologies, Oxford, CT). In some experiments, the level of donor chimerism in surviving mice was determined at day 95 after BMT by 2-color flow cytometric analysis of splenocytes using fluorescein isothiocyanate—conjugated antimouse CD45.2 and phycoerythrinconjugated antimouse CD45.1 and FACScan (PharMingen, San Diego, CA) to discriminate between donor (CD45.1+) and host (CD45.2+) cells.
Statistics
All experiments were performed at least 3 times with similar findings. The significance of differences between experimental and control groups was determined using the Student t test. The significance of differences between experimental groups for survival studies in vivo was determined by the log-rank test.
Results
Effects of PS-341 and TRAIL tumor apoptosis
Treatment of the leukemia C1498 or the renal carcinoma Renca with PS-341 significantly increased the sensitivity of these cells to cytolysis by TRAIL as assessed by either 18-hour isotope release or MTS staining assays (Figure 1A-B). Concentrations of PS-341 between 5 to 40 nM dramatically sensitized tumor cells to TRAIL-induced apoptosis, and under the conditions used in these experiments, the concentration of TRAIL required for maximal lysis of C1498 cells was 1000 to 2000 ng/mL (data not shown). In addition, similar results were obtained using Annexin V and propiduim iodide staining as a measure of cell death in 18-hour assays (Figure 2). Tumor cells treated with PS-341 plus TRAIL also exhibited many of the morphologic characteristics of apoptosis (data not shown). Furthermore, the caspase inhibitor zVAD-FMK was able to significantly reduce (Renca) or completely abolish (C1498) the effects of PS-341 plus TRAIL (Figure 3). This indicates that the cell death for both tumors was dependent on caspase activation.
Enhancement of tumor cell death by PS-341 plus TRAIL combinations. (A) C1498 cells labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), or 30 nM (□) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” (B) Renca cells were incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), and 40 nM (▴) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.” *P < .01 and **P < .001 by Student t test.
Enhancement of tumor cell death by PS-341 plus TRAIL combinations. (A) C1498 cells labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), or 30 nM (□) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” (B) Renca cells were incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), and 40 nM (▴) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.” *P < .01 and **P < .001 by Student t test.
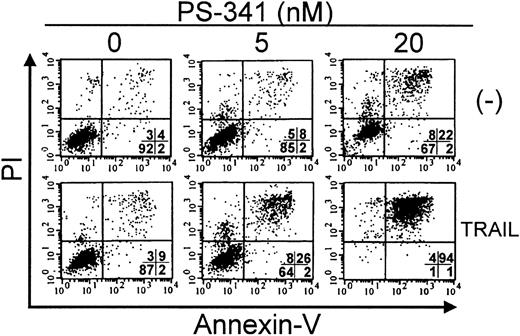
Apoptosis of C1498 cells in response to PS-341 plus TRAIL. C1498 cells were incubated with PS-341 at 0, 5 nM, or 20 nM for 2 hours prior to the addition of TRAIL (500 ng/mL). After a further 16 hours, cell death was estimated by FACS analysis after Annexin V and propidium iodide staining as described in “Materials and methods.” Percentage of cells in each quadrant is shown. Viable cells are in lower left quadrant.
Apoptosis of C1498 cells in response to PS-341 plus TRAIL. C1498 cells were incubated with PS-341 at 0, 5 nM, or 20 nM for 2 hours prior to the addition of TRAIL (500 ng/mL). After a further 16 hours, cell death was estimated by FACS analysis after Annexin V and propidium iodide staining as described in “Materials and methods.” Percentage of cells in each quadrant is shown. Viable cells are in lower left quadrant.
Blocking of tumor cell apoptosis by caspase inhibitors. C1498 cells were labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (A) zFA-FMK (50 μM) or (B) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” Renca cells were incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (C) zFA-FMK (50 μM) or (D) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.”
Blocking of tumor cell apoptosis by caspase inhibitors. C1498 cells were labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (A) zFA-FMK (50 μM) or (B) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” Renca cells were incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (C) zFA-FMK (50 μM) or (D) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.”
Although these short-term assays did show a synergistic interaction between PS-341 and TRAIL (isobologram analysis, data not shown), we had previously noted that high doses (> 100 nM) of PS-341 alone could cause significant death of these tumor cells usually 48 hours after exposure of tumor cells to the proteasome inhibitor. In order to determine whether the combination of PS-341 plus TRAIL was indeed superior to PS-341 alone, we compared the effects in 18 hours of exposure of tumor cells with these agents, alone and in combination, on the long-term clonal survival of C1498 cells in liquid culture. Therefore, after 18 hours of exposure, cells were washed and cultured in fresh media. As seen in Table 1, TRAIL alone slowed the growth of C1498. Overnight exposure of C1498 cells to PS-341 at 20 nM resulted in strong antiproliferative effects that produced a reduction of tumor cells obtained on day 7 by approximately 3 logs. A comparison of PS-341 alone with PS-341 plus TRAIL treatments reveals that the combination further decreases tumor cell number by more than 2 logs when comparing numbers of cells obtained 11 days after the respective treatments.
The effect of PS-341 and TRAIL treatment on long-term growth of C1498
Treatment* . | . | Cell number, × 106† . | . | . | |||
|---|---|---|---|---|---|---|---|
| Inhibitor (nM) . | TRAIL . | 2 days . | 7 days . | 11 days . | |||
| 0 | 0 | 7.8 ± 0.9 | (7988) | — | |||
| 0 | + | 2.7 ± 0.5 | — | — | |||
| PS-341 (1) | 0 | 7.2 ± 0.9 | — | — | |||
| PS-341 (1) | + | 2.7 ± 0.8 | — | — | |||
| PS-341 (5) | 0 | 8.8 ± 1.0 | — | — | |||
| PS-341 (5) | + | 4.2 ± 1.2 | — | — | |||
| PS-341 (20) | 0 | BD | 5.9 ± 1.2 | (1510) | |||
| PS-341 (20) | + | BD | BD | 7.1 ± 0.4 | |||
Treatment* . | . | Cell number, × 106† . | . | . | |||
|---|---|---|---|---|---|---|---|
| Inhibitor (nM) . | TRAIL . | 2 days . | 7 days . | 11 days . | |||
| 0 | 0 | 7.8 ± 0.9 | (7988) | — | |||
| 0 | + | 2.7 ± 0.5 | — | — | |||
| PS-341 (1) | 0 | 7.2 ± 0.9 | — | — | |||
| PS-341 (1) | + | 2.7 ± 0.8 | — | — | |||
| PS-341 (5) | 0 | 8.8 ± 1.0 | — | — | |||
| PS-341 (5) | + | 4.2 ± 1.2 | — | — | |||
| PS-341 (20) | 0 | BD | 5.9 ± 1.2 | (1510) | |||
| PS-341 (20) | + | BD | BD | 7.1 ± 0.4 | |||
— indicates not determined; BD, below detection level of the MTS assay.
C1498 cells (1 × 105) were incubated overnight in the presence or absence of PS-341 and TRAIL (1 μg/mL). Cells were then washed thoroughly and resuspended in 1 mL fresh media. Various dilutions of cells were transferred to microtiter plates.
Viable cell numbers were estimated at various time points using an MTS assay based on a standard curve with fixed numbers of C1498. The number in parentheses indicates a theoretical number assuming a 4-fold increase in cell number per day.
Proteasome inhibition in tumor cells
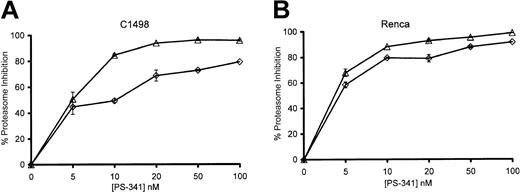
We determined the extent of proteasome inhibition that occurred when PS-341 sensitized C1498 and Renca tumor cells to TRAIL-mediated cytolysis. As seen in Figure 4, treatment of C1498 and Renca cells with PS-341 for 2 hours could result in up to 90% inhibition of proteasome activity depending on the concentration of PS-341. Prolonged incubation up to 18 hours enhanced the inhibition of enzyme activity seen at the lower doses of PS-341. Interestingly, if the PS-341 was washed off after 2 hours and cells were incubated for a further 16 hours, proteasome enzyme activity recovered to a significant degree (data not shown). In 18-hour incubations, the concentrations of PS-341 necessary to sensitize tumor cells to TRAIL-mediated lysis generally inhibited proteasome activity of C1498 cells by 60% to 80%. Therefore, the sensitization of cells to TRAIL-mediated apoptosis by PS-341 required that the level of proteasome inhibition exceeded a critical threshold level. Proteasome inhibition of less than 50% does not sensitize these tumor cells to TRAIL.
Proteasome inhibition of C1498 and Renca by PS-341. C1498 (A) and Renca (B) cells were incubated for 2 hours (⋄) or 18 hours (▵) in the presence or absence of various concentrations of PS-341. Cells were washed, and proteasome enzymatic activity (± SEM) was determined as described in “Materials and methods.”
Proteasome inhibition of C1498 and Renca by PS-341. C1498 (A) and Renca (B) cells were incubated for 2 hours (⋄) or 18 hours (▵) in the presence or absence of various concentrations of PS-341. Cells were washed, and proteasome enzymatic activity (± SEM) was determined as described in “Materials and methods.”
PS-341 effects on NF-κB
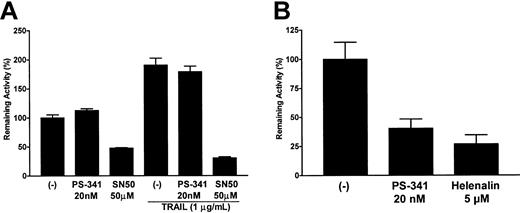
One major effect of proteasome inhibition in many cells is the inhibition of NF-κB translocation to the nucleus, due to blocking of the cytosolic degradation of Iκ-B. Indeed, this is thought to be the major mechanism whereby PS-341 sensitizes tumor cells to apoptosis. In order to assess the effects of PS-341 on NF-κB activation, we transfected C1498 cells with an NF-κB reporter construct coupled to luciferase to estimate levels of activated NF-κB. After a rest period of 36 hours the transfectants were pretreated for 2 hours with PS-341 followed by a 4-hour incubation in the presence or absence of TRAIL. As seen in Figure 5A, endogenous levels of NF-κB activity in C1498 cells were surprisingly not inhibited to any significant degree by PS-341 at concentrations up to 20 nM, even when incubations with PS-341 were extended to 18 hours (data not shown). In contrast, SN50 (a peptide inhibitor of NF-κB translocation) did block NF-κB activation in C1498 cells by 60% to 80%, and Helenalin (a chemical blocker of NF-κB DNA binding) inhibited the activity of the NF-κB reporter construct by up to 60% (data not shown). PS-341, SN50, and Helenalin did not have any significant effects in control C1498 cells transfected with the pTATA-luc minimal promoter (data not shown). TRAIL treatment of C1498 cells induced an increase in NF-κB reporter activity, but while SN50 still inhibited activity very significantly, PS-341 was without effect. Similar experiments using the A20 leukemia cell line did show significant effects of PS-341 on NF-κB reporter activity (Figure 5B). Interestingly, in contrast to PS-341, neither the SN50 peptide nor Helenalin sensitized C1498 cells to TRAIL-mediated apoptosis (data not shown). This further suggests that the apoptosis-promoting effects of PS-341 in C1498 cells may be independent of any effects on NF-κB activation.
Effects of PS-341 on NF-κB activity. Cells were transfected with either the pTATA-luc or the pNF-κB-luc plasmids. After a recovery period of 36 hours, transfectants were treated with various concentrations of PS-341 (20 nM), SN50 (50 μg/mL), or Helenalin (5 μM) for 2 hours followed by a further 4 hours in the presence or absence of TRAIL (1 μg/mL). Relative levels of NF-κB—driven promoter activity in C1498 (A) or A20 (B) were then calculated (± SEM) as previously described in “Materials and methods.”
Effects of PS-341 on NF-κB activity. Cells were transfected with either the pTATA-luc or the pNF-κB-luc plasmids. After a recovery period of 36 hours, transfectants were treated with various concentrations of PS-341 (20 nM), SN50 (50 μg/mL), or Helenalin (5 μM) for 2 hours followed by a further 4 hours in the presence or absence of TRAIL (1 μg/mL). Relative levels of NF-κB—driven promoter activity in C1498 (A) or A20 (B) were then calculated (± SEM) as previously described in “Materials and methods.”
Effects of PS-341 on levels of antiapoptotic proteins
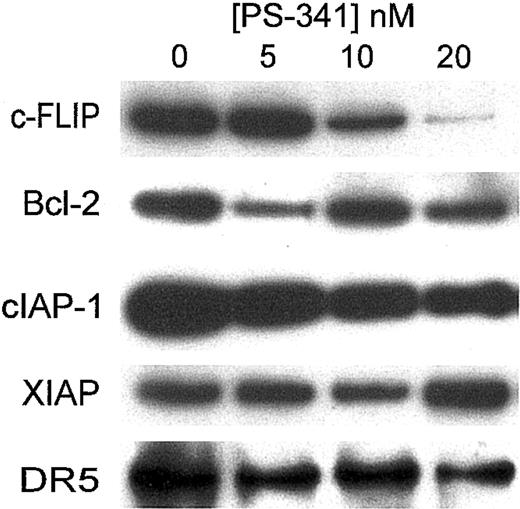
A number of proteins have been reported to block the apoptotic cascade.10 We therefore treated C1498 with PS-341 for various periods of time and then estimated levels of various antiapoptotic proteins by Western blotting. As shown in Figure 6 after 18 hours of treatment of C1498 cells, there were no major changes in levels of bcl-2 or XIAP. In a similar manner levels of bax or bcl-xL were unaltered (data not shown). Levels of cIAP-1 were somewhat reduced. Surprisingly there was a reduction in the expression of mouse DR-5 (the TRAIL receptor) on Western blotting, even when low doses of PS-341 were used. However, since we were unable to detect mouse DR-5 on fluorescence-activated cell sorter (FACS) analysis, we do not know how PS-341 affects the surface expression of DR-5.
Western blotting for antiapoptotic proteins following PS-341 treatment. C1498 cells were incubated for 18 hours in the presence or absence of various concentrations of PS-341. Cells were then washed and cell extracts were prepared. Western blotting was carried out using various antibodies as described in “Materials and methods.”
Western blotting for antiapoptotic proteins following PS-341 treatment. C1498 cells were incubated for 18 hours in the presence or absence of various concentrations of PS-341. Cells were then washed and cell extracts were prepared. Western blotting was carried out using various antibodies as described in “Materials and methods.”
The most dramatic change in antiapoptotic proteins occurred in levels of c-FLIP, which was clearly decreased in C1498 cells at concentrations corresponding to those that sensitize these cells to TRAIL-mediated apoptosis. This suggests that in C1498 cells, the reduction of c-FLIP levels may play some role in sensitizing these cells to apoptosis by TRAIL. Similar changes of expression of these proteins were observed in Renca cells (data not shown). Interestingly, C1498 cells transfected to overexpress c-FLIP (Figure 7A) were significantly more resistant to the apoptotic effects of PS-341 plus TRAIL (Figure 7B). However the c-FLIP transfectants were equally sensitive to the direct cytostatic effects of PS-341 alone (data not shown). This suggests that the reduction of c-FLIP levels following PS-341 treatment may play a major role in sensitizing these cells to TRAIL-mediated apoptosis.
Inhibition of TRAIL apoptosis by c-FLIP expression. (A) C1498 cells were transfected with a c-FLIP expression plasmid as described in “Materials and methods.” Cells were then cloned, and the levels of expression of the Flag-tagged c-FLIP and native c-FLIP in C1498 cells and representative clones 1, 2, and 3 were determined by Western blotting with an antibody to c-FLIP. (B) C1498 cells and clones 1, 2, and 3 were labeled with [111In] Ox. Cells were then incubated with media (□), PS-341 at 10 nM (▦), or PS-341 at 10 nM for 2 hours prior to the addition of 200 ng/mL TRAIL (▪). After a further 16 hours, cell lysis (± SEM) was estimated as described in “Materials and methods.” *P < .001 by Student t test when compared with C1498 cells.
Inhibition of TRAIL apoptosis by c-FLIP expression. (A) C1498 cells were transfected with a c-FLIP expression plasmid as described in “Materials and methods.” Cells were then cloned, and the levels of expression of the Flag-tagged c-FLIP and native c-FLIP in C1498 cells and representative clones 1, 2, and 3 were determined by Western blotting with an antibody to c-FLIP. (B) C1498 cells and clones 1, 2, and 3 were labeled with [111In] Ox. Cells were then incubated with media (□), PS-341 at 10 nM (▦), or PS-341 at 10 nM for 2 hours prior to the addition of 200 ng/mL TRAIL (▪). After a further 16 hours, cell lysis (± SEM) was estimated as described in “Materials and methods.” *P < .001 by Student t test when compared with C1498 cells.
PS-341 plus TRAIL treatment of bone marrow cultures containing tumor cells
Since the PS-341 plus TRAIL combination showed highly significant levels of toxicity on the C1498 leukemic cells, we attempted to determine if this combination could purge bone marrow of leukemic cells. Therefore, mixtures of bone marrow and tumor cells were incubated in the presence or absence of PS-341, TRAIL, or PS-341 plus TRAIL for 18 hours prior to their transfer to irradiated recipient mice. The effects of these agents on survival of leukemia cells could then be estimated. As anticipated from previous studies on tumor cells alone, PS-341 plus TRAIL resulted in a dramatic reduction in the number of viable C1498 cells within the bone marrow cultures, as assessed by tumor proliferation assays (Figure 8A). Furthermore, on transfer of these bone marrow—tumor cell mixtures to irradiated C57BL/6 mice, mice receiving bone marrow—tumor cell mixtures treated with TRAIL alone all succumbed to leukemia by about 25 to 35 days. Treatment of bone marrow—tumor cell mixtures with PS-341 alone did result in a significant increase in survival (P < .001) compared with untreated mixtures, with upto 50% of mice surviving more than 100 days (Figure 8B). In contrast, more than 90% of mice receiving the bone marrow—tumor cell mix treated with PS-341 plus TRAIL survived more than 100 days. Since the mice survived more than 100 days, this made it unlikely that PS-341 plus TRAIL had major detrimental effects on bone marrow cells in the inoculum. However, in order to examine the effects of PS-341 plus TRAIL on hematopoietic reconstitution in more detail, cellular blood counts (CBCs) were monitored in B6 mice at various time points after they received PS-341 plus TRAIL—treated bone marrow. There were some slight, but significant, changes in levels of red cells in mice receiving bone marrow that has been treated with either PS-341 alone or in combination with TRAIL at early stages (day 14) of hematopoietic recovery (Table 2). Also at day 14 there was a significant decrease in platelets, specifically in mice receiving marrow treated with PS-341 plus TRAIL. However, by day 60 CBCs had normalized in all groups. Therefore, the major bone marrow toxicity of PS-341 plus TRAIL was a mild, transient thrombocytopenia observed early after bone marrow transfer, which disappeared over time.
Effect of PS-341 plus TRAIL on purging tumor cells from the bone marrow. C1498 cells (104 cells/mL) were added to bone marrow cells (5 × 106 cells/mL), and these mixtures were incubated for 4 hours with PS-341 (20 nM) followed by a further 16 hours in the presence or absence of TRAIL (1000 ng/mL). (A) Dilutions of the cells from the various incubations were then put in media, and total tumor cell numbers (± SEM) were estimated at various time points using MTS staining as described in “Materials and methods.” ○ indicate C1498 cells alone; •, C1498 + bone marrow; ▵, C1498 + bone marrow + TRAIL (1 μg/mL); ▴, C1498 + bone marrow + PS-341 (20 nM); and ▪, C1498 + bone marrow + PS-341 (20 nM) + TRAIL (1 μg/mL). (B) Cells from the various incubations were transferred to irradiated C57BL/6 recipient mice as described in “Materials and methods.” □ indicate C1498 + bone marrow; ▵, C1498 + bone marrow + PS-341 (20 nM); ▿, C1498 + bone marrow + TRAIL (1 μg/mL); and ○, C1498 + bone marrow + PS-341 (20 nM) + TRAIL (1 μg/mL). Mice were continuously monitored, and survival was followed for 100 days after the bone marrow transplantations. **P < .001 comparing survival of mice receiving PS-341—treated bone marrow—tumor cell mixtures with controls; *P < .001 comparing PS-341 plus TRAIL—treated mixtures with PS-341 alone using the log-rank test. (C) At 95 days after bone marrow transfer, the percent of CD45.1+ donor-derived lymphoid cells in the spleen was determined by FACS analysis in (▪) mice receiving bone marrow but no tumor cells, (▴) surviving mice that had received bone marrow—tumor cell mixtures treated with PS-341 alone, and (•) surviving mice that had received bone marrow—tumor cell mixtures treated with PS-341 plus TRAIL. Values from individual mice from 3 independent experiments are shown with the bar on the right indicating the mean value.
Effect of PS-341 plus TRAIL on purging tumor cells from the bone marrow. C1498 cells (104 cells/mL) were added to bone marrow cells (5 × 106 cells/mL), and these mixtures were incubated for 4 hours with PS-341 (20 nM) followed by a further 16 hours in the presence or absence of TRAIL (1000 ng/mL). (A) Dilutions of the cells from the various incubations were then put in media, and total tumor cell numbers (± SEM) were estimated at various time points using MTS staining as described in “Materials and methods.” ○ indicate C1498 cells alone; •, C1498 + bone marrow; ▵, C1498 + bone marrow + TRAIL (1 μg/mL); ▴, C1498 + bone marrow + PS-341 (20 nM); and ▪, C1498 + bone marrow + PS-341 (20 nM) + TRAIL (1 μg/mL). (B) Cells from the various incubations were transferred to irradiated C57BL/6 recipient mice as described in “Materials and methods.” □ indicate C1498 + bone marrow; ▵, C1498 + bone marrow + PS-341 (20 nM); ▿, C1498 + bone marrow + TRAIL (1 μg/mL); and ○, C1498 + bone marrow + PS-341 (20 nM) + TRAIL (1 μg/mL). Mice were continuously monitored, and survival was followed for 100 days after the bone marrow transplantations. **P < .001 comparing survival of mice receiving PS-341—treated bone marrow—tumor cell mixtures with controls; *P < .001 comparing PS-341 plus TRAIL—treated mixtures with PS-341 alone using the log-rank test. (C) At 95 days after bone marrow transfer, the percent of CD45.1+ donor-derived lymphoid cells in the spleen was determined by FACS analysis in (▪) mice receiving bone marrow but no tumor cells, (▴) surviving mice that had received bone marrow—tumor cell mixtures treated with PS-341 alone, and (•) surviving mice that had received bone marrow—tumor cell mixtures treated with PS-341 plus TRAIL. Values from individual mice from 3 independent experiments are shown with the bar on the right indicating the mean value.
The effects of PS-341 and TRAIL on complete blood counts after BMT
. | Day 14 after BMT* . | . | . | . | . | Day 60 after BMT† . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment‡ . | BMCs . | BMCs + C1498 . | BMCs + C1498 + PS-341 . | BMCs + C1498 + TRAIL . | BMCs + C1498 + PS-341 + TRAIL . | BMCs . | BMCs + C1498 + PS-341 . | BMCs + C1498 + PS341 + TRAIL . | ||||||
| Lymphocytes,§ cells/mL × 106 | 1.16 ± 0.17 | 1.45 ± 0.16 | 1.12 ± 0.15 | 1.16 ± 0.10 | 1.74 ± 0.47 | 8.72 ± 1.07 | 8.01 ± 0.72 | 7.44 ± 0.42 | ||||||
| Neutrophils, cells/mL × 106 | 2.07 ± 0.29 | 1.55 ± 0.12 | 1.41 ± 0.21 | 1.76 ± 0.37 | 0.96 ± 0.30 | 2.87 ± 0.44 | 2.94 ± 0.45 | 2.53 ± 0.32 | ||||||
| RBCs, cells/mL × 109 | 8.80 ± 0.16 | 8.65 ± 0.13 | 7.78 ± 0.26∥ | 8.46 ± 0.10 | 6.93 ± 0.56∥ | 9.34 ± 0.33 | 9.85 ± 0.15 | 9.93 ± 0.30 | ||||||
| Platelets, cells/L × 109 | 548 000 ± 23 000 | 467 000 ± 33 000 | 459 000 ± 51 000 | 436 000 ± 23 000 | 323 000 ± 54 000∥ | 542 000 ± 54 000 | 661 000 ± 41 000 | 615 000 ± 23 000 | ||||||
. | Day 14 after BMT* . | . | . | . | . | Day 60 after BMT† . | . | . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Treatment‡ . | BMCs . | BMCs + C1498 . | BMCs + C1498 + PS-341 . | BMCs + C1498 + TRAIL . | BMCs + C1498 + PS-341 + TRAIL . | BMCs . | BMCs + C1498 + PS-341 . | BMCs + C1498 + PS341 + TRAIL . | ||||||
| Lymphocytes,§ cells/mL × 106 | 1.16 ± 0.17 | 1.45 ± 0.16 | 1.12 ± 0.15 | 1.16 ± 0.10 | 1.74 ± 0.47 | 8.72 ± 1.07 | 8.01 ± 0.72 | 7.44 ± 0.42 | ||||||
| Neutrophils, cells/mL × 106 | 2.07 ± 0.29 | 1.55 ± 0.12 | 1.41 ± 0.21 | 1.76 ± 0.37 | 0.96 ± 0.30 | 2.87 ± 0.44 | 2.94 ± 0.45 | 2.53 ± 0.32 | ||||||
| RBCs, cells/mL × 109 | 8.80 ± 0.16 | 8.65 ± 0.13 | 7.78 ± 0.26∥ | 8.46 ± 0.10 | 6.93 ± 0.56∥ | 9.34 ± 0.33 | 9.85 ± 0.15 | 9.93 ± 0.30 | ||||||
| Platelets, cells/L × 109 | 548 000 ± 23 000 | 467 000 ± 33 000 | 459 000 ± 51 000 | 436 000 ± 23 000 | 323 000 ± 54 000∥ | 542 000 ± 54 000 | 661 000 ± 41 000 | 615 000 ± 23 000 | ||||||
RBC indicates red blood cell.
Data from 3 independent experiments were pooled.
Data from 2 independent experiments were pooled.
Cells were cultured for 24 hours prior to infusion into irradiated C57BL/6 mice, as described in “Materials and methods.”
Peripheral blood samples were collected from 3 mice per group. The values for lymphocytes, neutrophils, red blood cells, and platelets show mean ± SEM.
A significant decrease (P < .05) compared with that of BMCs + C1498 control group, as determined by an unpaired Student t test.
It remained important to determine whether the reconstitution of the immune system of the irradiated recipient mice had occurred because of the transfer of donor hematopoietic cells. Therefore, the contribution of donor cells to the hematopoietic reconstitution of the host was analyzed in various lymphoid organs using FACS analysis. At day 95, mice that had just received bone marrow, or surviving mice whose bone marrow—tumor cell mixture had been treated with either PS-341 alone or PS-341 plus TRAIL, contained about 80% splenocytes of donor (CD45.1+) origin (Figure 8C). Similar high levels of reconstitution from donor cells were observed in other lymphoid organs (data not shown).
Discussion
Proteasome inhibition with specific inhibitors such as PS-341 is currently being used as a novel anticancer therapy in ongoing clinical trials.31 Furthermore, PS-341 has shown efficacy in a variety of tumor models both as a single agent1 or in combination with chemotherapy32,33 or radiation therapy.34 In this study we report that PS-341 can be successfully combined with the death ligand TRAIL to synergistically promote tumor cells apoptosis.
From our studies on the acute myeloid leukemia C1498, we could not conclude that the inhibition of NF-κB was involved in the sensitization of these cells to apoptosis mediated by TRAIL. On examination of levels of several antiapoptotic genes, we saw a substantial reduction in c-FLIP at concentrations of PS-341 that sensitize cells to TRAIL apoptosis. Interestingly, the transcription of c-FLIP is not regulated by NF-κB in all tumor cells.15 Furthermore, in some studies major reductions of c-FLIP protein seem to occur at the posttranslational level.35 It seems surprising that proteasome inhibition can cause a decrease in c-FLIP, particularly since c-FLIP is reported to be degraded by the proteasome in some cells.36 However, the relative importance of different pathways controlling levels of c-FLIP may vary in different cell types, and we are currently further investigating the molecular mechanisms controlling c-FLIP in C1498 and Renca cells. The concentrations of PS-341 that sensitize C1498 cells to TRAIL lysis also inhibited cell-cycle progression in C1498 cells, and this cell-cycle inhibition resulted in an accumulation of cells in S-G2/M (data not shown). Blockade at this stage of the cell cycle by PS-341 has been previously reported by others.1 Perturbation of cell cycle has been reported to sensitize cells to apoptosis, and it has also been reported that c-FLIP levels are modulated during the cell cycle in T cells, with peak levels occurring during G1 followed by a reduction in S-G2/M.37 Whether effects of PS-341 on the cell cycle are crucial in sensitizing C1498 cells to TRAIL-mediated apoptosis is unknown but is currently under investigation. Reductions of c-FLIP were also observed in Renca cells on PS-341 treatment, but the molecular basis of this is also unclear, and is also currently being investigated further. Overexpression of c-FLIP blocked the effects of PS-341 on TRAIL-mediated apoptosis of C1498 cells and Renca cells (data not shown). This suggests that levels of c-FLIP are critical in determining the sensitivity of certain cells to TRAIL, and the reductions of c-FLIP observed in response to PS-341 may sensitize cells to TRAIL-mediated apoptosis. Interestingly, fibroblasts from c-FLIP-/- mice are much more sensitive to the apoptotic effects of Fas ligand and TNF than are wild-type fibroblasts,38 indicating that a reduction of c-FLIP alone can be sufficient to sensitize some cells to death receptor—mediated apoptosis. Furthermore, reintroduction of c-FLIP to these c-FLIP-/- fibroblasts produces TRAIL resistance.39
In the bone marrow purging model used in this study, the combination of PS-341 and TRAIL could selectively deplete C1498 tumor cells from bone marrow—tumor cell mixtures. Although a slight thrombocytopenia was observed at early time points (day 14) following transfer of PS-341 plus TRAIL—treated bone marrow, all cellular blood counts had normalized by day 60. It has been reported that PS-341 as a single agent has only minor effects in bone marrow function at doses used in tumor therapy in mice or rats.1 In the clinical setting, PS-341 has been reported to have minimal effects on hematopoiesis, and elicits only a small transient thrombocytopenia. The mild thrombocytopenia seen here in mice did not appear to be problematic, and is similar to that seen in patients. In vitro studies indicate that TRAIL has either very minor or undetectable effects on hematopoesis in normal human bone marrow.40,41 We observed no effects of TRAIL as a single agent on hematopoesis in our studies.
Leukemia and lymphomas seem particularly sensitive to the effects of proteasome inhibition. Therefore, human multiple myeloma cells can be killed by PS-341 in vitro,4,42 and a number of clinical trials of PS-341 in multiple myeloma are ongoing. Indeed, additional beneficial cytotoxic affects of PS-341 plus TRAIL have been observed on multiple myeloma cells in vitro.43 Concerning AML, cells from different disease stages have shown differential sensitivity to TRAIL. Therefore, AML cells of stage M4 were reported to be sensitive to TRAIL-mediated apoptosis in vitro, whereas cells from patients with stages M1, M2, and M3 were not.44 It would be of great interest to determine if PS-341 could further enhance the sensitivity of human AML cells to TRAIL in an analogous manner to the effects on C1498. The study on sensitivity of human AML cells to TRAIL was performed using activated natural killer T (NKT) effector cells that expressed cell-surface TRAIL.44 Blocking antibodies to TRAIL inhibited the lysis of AML M4 cells by activated NKT cells. This also suggests that beneficial immunotherapeutic effects of NK cells, NKT cells, or graft-versus-leukemia responses to AML in vivo could have a TRAIL component. Whether such successful bone marrow purging by PS-341 plus TRAIL can be extended from leukemias to solid tumors is currently under investigation using the murine renal cancer Renca.
One important consideration is that not all tumors undergo apoptosis after exposure to TRAIL. Some solid tumors such as breast cancer and melanoma are reported to be sensitive to TRAIL-mediated apoptosis in vitro,17,45 whereas other tumor cells are resistant. In some cases (such as C1498) sensitivity can be increased by PS-341, whereas in other cases PS-341 has no effect (data not shown). Prior knowledge of the tumors' sensitivity to the PS-341 plus TRAIL treatment could therefore be a good advance predictor of the potential for the success or failure of treatment in a bone marrow transplantation setting. Our present studies have demonstrated substantial therapeutic benefits of PS-341 plus TRAIL in the BMT setting. It remains critical to determine whether PS-341 plus TRAIL can provide therapeutic benefit after systemic administration to mice bearing leukemias or solid tumors. TRAIL is reported to be far less toxic than other members of the TNF family on administration in vivo.46 However, there are reports that TRAIL does have significant toxicity to hepatocytes47 and neuronal cells.48 The concentrations of PS-341 required to sensitize C1498 or Renca cells to TRAIL-mediated lysis can be well tolerated in mice and humans.1,31 Indeed, the level of proteasome inhibition seen in the present study, and its reversal over time, mimics other data previously reported in vivo and from clinical data sets. This highlights the predictive nature of the 20S assay and its utility across in vitro, in vivo, and human studies.29 However, it remains crucially important to determine whether PS-341 plus TRAIL can directly promote tumor destruction after direct administration of both agents to mice, and what the limiting toxicities of this combination would be. Systemic administration of PS-341 plus TRAIL could be used to destroy certain tumor cells directly, or alternatively to destroy the residual tumor cells remaining after high-dose chemotherapy or radiation therapy regimens. In conclusion, these studies do indicate that the targeting of apoptotic pathways in tumor cells, using combinations of agents that synergistically interact to preferentially promote tumor cell apoptosis, could hold substantial promise for cancer therapy in the future.
Prepublished online as Blood First Edition Paper, March 13, 2003; DOI 10.1182/blood-2002-09-2975.
Supported in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number N01-C0-12400 and R01 CA72699.
P.J.E. was employed by Millennium Pharmaceuticals, whose product (PS-341) was studied in the present work.
By acceptance of this article, the publisher or recipient acknowledges the right of the US government to retain a nonexclusive, royalty-free license in and to any copyright covering the article.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to Ms Susan Charbonneau and Ms Connie Champion for assistance in preparing this manuscript. We thank Steven Stull for assistance with the animal studies; Drs John Ortaldo and Margaret Read for their critical reading of this manuscript; and Dr Gregory Alvord for consultations on isobologram analysis.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US government.

![Figure 1. Enhancement of tumor cell death by PS-341 plus TRAIL combinations. (A) C1498 cells labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), or 30 nM (□) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” (B) Renca cells were incubated with PS-341 at 0 (▪), 10 nM (○), 20 nM (♦), and 40 nM (▴) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.” *P < .01 and **P < .001 by Student t test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/1/10.1182_blood-2002-09-2975/5/m_h81334566001.jpeg?Expires=1769243859&Signature=k5EJmPGwERPCHTUDafyfX02At7Vc5IIUhflQzmmAGDGIE2qgRcDfwbI1SWS7blCQ-bJaepiolXDfuPFlwaZj9hk2QgKcCiOVq~RctiPl6AQc3v7sIGVrHE79oaO-whCts78vovMDwVjgDurDg~Z97GZannY0xQuw48yWc5PTyl7FZp91fze5MZ7YLOnyq9Eio2KF05vaVOs-0jfK8LqeSK8hyF4hK9fln-YMEBBdfLAKthRaPedq5KfxA6gO0PbhW9d18ric-83zQCOR1gV5-G6qOl8-Fqg1ybMDW02KDV6pdsCyiB~PBrVQg61ngAORGcGvuVjQ1XNK6bnPsJYrxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Blocking of tumor cell apoptosis by caspase inhibitors. C1498 cells were labeled with [111In] Ox and then incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (A) zFA-FMK (50 μM) or (B) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, lysis (± SEM) was estimated as described in “Materials and methods.” Renca cells were incubated with PS-341 at 0 (▪), 5 nM (▵), and 20 nM (♦) and (C) zFA-FMK (50 μM) or (D) zVAD-FMK (50 μM) for 2 hours prior to the addition of various concentrations of TRAIL. After a further 16 hours, cell viability (± SEM) was assessed by the uptake of the MTS dye as described in “Materials and methods.”](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/1/10.1182_blood-2002-09-2975/5/m_h81334566003.jpeg?Expires=1769243859&Signature=v6AA-hC6gDA-AEG7YwFB8qxKzOaKnDRB1YxRvUCAlRwE9DqGiTdz0nWpo1z2dIkQjPR05SnS021w4wSzwBAE7murDj8aua3Dh-ss-64S2moSvk3dbRh6Rta7aCMj4WNyIkth0edaEpoMws8F6HbyLY-Ob8QaLwtJrHzr2cqddgrr7wqb4zPdkVGsI1vE5nGEKf03JCznVO9kjdr9C7iDls1k~mc9AO1Dy97emvlbWMjkzXyP8iW5v0LOrj8sIQr7pXNEdHShF42-USMXXCkjGKXN27yOh4At5ItHUmLFHFGOh6QjZQp8IS~nONzUZrPLWNC9nhtvBe0sQs9XgeLC9Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 7. Inhibition of TRAIL apoptosis by c-FLIP expression. (A) C1498 cells were transfected with a c-FLIP expression plasmid as described in “Materials and methods.” Cells were then cloned, and the levels of expression of the Flag-tagged c-FLIP and native c-FLIP in C1498 cells and representative clones 1, 2, and 3 were determined by Western blotting with an antibody to c-FLIP. (B) C1498 cells and clones 1, 2, and 3 were labeled with [111In] Ox. Cells were then incubated with media (□), PS-341 at 10 nM (▦), or PS-341 at 10 nM for 2 hours prior to the addition of 200 ng/mL TRAIL (▪). After a further 16 hours, cell lysis (± SEM) was estimated as described in “Materials and methods.” *P < .001 by Student t test when compared with C1498 cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/102/1/10.1182_blood-2002-09-2975/5/m_h81334566007.jpeg?Expires=1769243859&Signature=TRqzCyUnNlLtk~vnB2pwFg19TpelnZiFPNkHeeRPb4gQNPVDssoUoUQFJNzChvexbZUtgQd9QX~evtXNo1H6l8s9PLAisHGK9n72a2xlFgJqVeg7FLvkEE3b0M4SrynDelcG5zoFE2QNSpvIGdRf67wGfXCxYc-VBT8FUTlTTSnC3vsTYI8~nTGJAszBS8icoeTYR-Z3P2r8cQNsDgq9LL8aZZ5Szeraiz6vRuJWKpeIqUc7dxzMFHnDK~nKraLKsEDkjBIGP64cOzYzNHJEj74zj33g1FLUkTS6zcj5VXY8cckha1UA7iN1BnM1AYjfJzhtdjHpURzSmD5H09QukQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal