Abstract
Imatinib-treated chronic myeloid leukemia (CML) patients with acquired resistance commonly have detectable BCR-ABL kinase domain mutations. It is unclear whether patients who remain sensitive to imatinib also have a significant incidence of mutations. We evaluated 144 patients treated with imatinib for BCR-ABL kinase domain mutations by direct sequencing of 40 accelerated phase (AP), 64 late chronic phase (≥ 12 months from diagnosis, late-CP), and 40 early-CP patients. Mutations were detected in 27 patients at 17 different residues, 13 (33%) of 40 in AP, 14 (22%) of 64 in late-CP, and 0 of 40 in early-CP. Acquired resistance was evident in 24 (89%) of 27 patients with mutations. Twelve (92%) of 13 patients with mutations in the adenosine triphosphate (ATP) binding loop (P-loop) died (median survival of 4.5 months after the mutation was detected). In contrast, only 3 (21%) of 14 patients with mutations outside the P-loop died (median follow-up of 11 months). As the detection of mutations was strongly associated with imatinib resistance, we analyzed features that predicted for their detection. Patients who commenced imatinib more than 4 years from diagnosis had a significantly higher incidence of mutations (18 [41%] of 44) compared with those treated within 4 years (9 [9%] of 100), P < .0001. Lack of a major cytogenetic response (MCR) was also associated with a higher likelihood of detecting a mutation; 19 (38%) of 50 patients without a MCR had mutations compared with 8 (8.5%) of 94 with an MCR, P < .0001. In conclusion, the detection of kinase domain mutations using a direct sequencing technique was almost always associated with imatinib resistance, and patients with mutations in the P-loop had a particularly poor prognosis. (Blood. 2003; 102:276-283)
Introduction
Positive results from trials of imatinib mesylate in patients with chronic myeloid leukemia (CML) have established it as the new standard of care in the chronic phase (CP) and accelerated phase (AP) of the disease.1-3 However, resistance to imatinib develops in both phases. The most common mechanism of acquired imatinib resistance is the reactivation of BCR-ABL kinase activity within the leukemic cell despite the presence of imatinib, by either gene amplification4-6 or point mutation.6-11 Point mutations within the ABL kinase domain of the BCR-ABL gene are emerging as the most frequent mechanism for reactivation of kinase activity.7,11 Structural data suggest that imatinib induces the BCR-ABL protein into the inactive conformation by binding to amino acids in the ABL kinase domain and blocking adenosine triphosphate (ATP) binding.12,13 This prevents the transfer of phosphate from ATP and blocks the downstream signal transduction pathways, leading to growth arrest or apoptosis.14-16 It is postulated that mutations within the ATP binding site can prevent imatinib from binding by either interrupting critical contact points between imatinib and the protein or inducing a conformation to which imatinib is unable to bind. A number of mutations have been well characterized in terms of their ability and degree to which they induce resistance.6,9,11,17-20
Previous studies of the detection of mutations have been predominantly conducted in patients in whom imatinib resistance had developed. A comprehensive study has not been performed to determine the frequency of mutations in patients in AP and CP, regardless of whether imatinib resistance had already developed. Furthermore, it has not been established if mutations are detectable in patients with CML treated with imatinib as first-line therapy nor whether the clinical consequences of mutations differ in the various disease phases. Varying levels of resistance have been demonstrated for some mutations,6,9,11,17-20 but this resistance has not been correlated with clinical outcomes. It has been reported that imatinib response decreases with the time from initial diagnosis in patients in CP and that those who fail to achieve a major cytogenetic response (MCR) are at a higher risk of progression to blast phase than those who achieve an MCR.2 It is not clear, however, if the association is due to a higher incidence of mutations in these patients. Answers to these issues may identify patients at highest risk for imatinib resistance because of kinase domain mutations and may guide therapeutic options to reduce this risk.
In this study we have performed an evaluation of the BCR-ABL kinase domain mutation status of 144 patients with CML treated with imatinib in Australia who had RNA available for testing. We determined the frequency of mutations in the different phases of the disease and correlated this frequency to the development of resistance, disease outcome, interval from diagnosis to the start of imatinib therapy, and cytogenetic response.
Patients and methods
Patients
More than 300 patients received imatinib at multiple centers in Australia over the time of the study. Our intent was to test all patients who had received at least 6 months of imatinib and who had available RNA for analysis. The samples were initially assessed for the level of BCR-ABL and only preceded to mutation analysis if the stored RNA contained a measurable level of BCR-ABL and the control gene level indicated nondegraded RNA. Samples from 156 patients were available, but 12 patients could not be tested, 4 because of low levels of BCR-ABL and 8 because of inadequate RNA quality, leaving 144 patients. Not all centers were able to collect samples from all patients treated with imatinib for various reasons; thus, the subset studied may not be fully representative of the entire patient group of 300. Patients tested had either received 6 or more months of imatinib therapy or had developed resistance and ceased therapy before 6 months (n = 2). The patients were grouped according to the disease stage at start of imatinib; 40 in AP, 64 in late-CP (defined as < 12 months from diagnosis), and 40 in early-CP (defined as < 12 months from diagnosis). Sixteen of the patients in early-CP had failed previous interferon (IFN)–α therapy, and 24 had only received hydroxyurea prior to imatinib therapy.
Accelerated phase was defined by the presence of one or more of the following: blasts in blood or bone marrow 10% or more but less than 30%, blasts plus promyelocytes in blood or bone marrow 20% or more, peripheral basophils 20% or more, thrombocytopenia less than 100 × 109/L unrelated to therapy, progressive splenomegaly, or clonal evolution (chromosomal abnormalities in addition to a single Philadelphia chromosome). Fifteen patients were classified as AP on the basis of clonal evolution as the sole criterion. Chronic phase was defined as less than 10% blasts in peripheral blood or bone marrow, less than 20% blasts plus promyelocytes in the peripheral blood or bone marrow, less than 20% basophils, and no extramedullary involvement apart from liver and spleen. Patients in AP were initially treated with 600 mg/d of imatinib and patients in CP with 400 mg/d.
We based our definition of imatinib resistance in patients in CP on the criteria used for the phase 2 study of CP CML after IFN-α treatment failure.2 Acquired resistance was defined as a sustained complete hematologic remission (CHR) that was followed by transformation to accelerated or blastic phase, loss of a sustained CHR without transformation, loss of an MCR (defined as at least 65% Philadelphia chromosome–positive cells or an increase of at least 30%), or loss of a complete cytogenetic response (CCR) with a corresponding increase in BCR-ABL levels of at least 1 log. Primary resistance was defined as failure to achieve and sustain a CHR for at least 4 weeks during the first 6 months of imatinib therapy. In some of these patients a hematologic response was achieved, but our criterion for a sustained CHR was not fulfilled. Transformation to accelerated or blastic phase in these patients was defined as acquired resistance.
We based our definition of imatinib resistance in patients in AP on the criteria used for the phase 2 study of AP CML.3 Primary resistance was defined as failure to achieve or sustain a hematologic response (defined as (1) CHR, (2) marrow response, or (3) return to CP) for at least 4 weeks during the first 6 months of imatinib therapy. Acquired resistance was defined as a sustained hematologic response that was followed by transformation to blastic phase, loss of a sustained hematologic response, loss of a MCR, or loss of a CCR with a corresponding increase in BCR-ABL levels of at least 1 log.
Depending on available RNA, patients were tested for mutations at between 1 and 18 different time points (median = 2). RNA samples (353) were analyzed. The median duration of imatinib therapy was calculated from commencement to the last time mutation analysis was performed. Patients in AP had received a median of 9 months of imatinib therapy (range, 4-24 months); late-CP, 10 months (range, 6-24 months); and early-CP, 14 months (range, 5-24 months). Approval for these studies was obtained from the Royal Adelaide Hospital institutional review board. Informed consent was provided according to the Declaration of Helsinki.
Sequencing of the ABL kinase domain of BCR-ABL
The complete ABL kinase domain of the BCR-ABL allele was analyzed using seminested reverse transcriptase–polymerase chain reaction (RTPCR) followed by direct sequencing as previously described.7 Briefly, the BCR-ABL allele was amplified using a forward primer that annealed in BCR exon b2 and a reverse primer that annealed at the junction of ABL exons 9 and 10. An 863 base pair (bp)–fragment containing the BCR-ABL kinase domain was amplified in a seminested PCR and sequenced in the forward and reverse directions using Dye Terminator Chemistry and a 3700 DNA Sequencer (Applied Biosystems, Foster City, CA).
The ability to detect mutations was very dependent on the quality and integrity of the RNA. To minimize RNA degradation, samples were processed into Trizol RNA stabilization solution (Invitrogen Life Technologies, Carlsbad, CA) within hours of peripheral blood collection and frozen at -80°C before delivery to the laboratory. The quality of the RNA was assessed prior to sequencing by measurement of the level of BCR-ABL and normal BCR using real-time quantitative PCR.21 A cutoff level based on the number of detectable BCR gene transcripts in the sample was established as an indicator of degraded RNA. If samples that did not comply with these cutoff values were used for mutation analysis, then unreliable results were obtained as evidenced by conflicting results between repeated analysis. This situation was especially important in patients in CCR who had lower levels of BCR-ABL/BCR (ratio of 1:100). The indicators used in this study were BCR values of more than 100 000 transcripts if the BCR-ABL/BCR ratio was less than 1:1000, BCR values of more than 50 000 transcripts if the BCR-ABL/BCR ratio was 1:1000 to 1:100, and BCR values more than 7000 transcripts if the BCR-ABL/BCR ratio was more than 1:100. In addition, for patients in CCR, the amount of cDNA added to the first-round PCR was increased to 150 ng compared with 100 ng for patients who were not in CCR. The first-round PCR products from patients in CCR were purified using PCR purification columns (Mo Bio, Carlsbad, CA) prior to the seminested PCR. Using the above measure of RNA quality and with compensating amounts of RNA for patients with low BCR-ABL levels, we were able to sequence samples with BCR-ABL values spanning 5 logs.
When mutations were detected in patients for the first time, the whole procedure was repeated using a second RNA sample from the same time point if available or from the same RNA if a second sample was not available. Detection of the mutation was only considered confirmed if the same mutation was detected in the repeat analysis.
To determine the sensitivity of the assay, a BCR-ABL cell line containing the Tyr253Phe mutation was mixed with wild-type BCR-ABL cells from 10% to 90% in 10% increments. RNA was extracted from each sample using the Trizol procedure, and mutation analysis was performed in duplicate. The BCR-ABL mutant was clearly detected at a dilution ratio of 20% (data not shown).
The amino acid substitutions were determined using GenBank accession no. M14752. However, this sequence has a number of nucleotides that differed to other ABL sequences in GenBank, including mRNA sequence X16416 and the DNA sequence U07563, as well as the sequence found in patients. This situation is relevant to the current analysis because we detected a mutation at a conflicting site. The discrepant nucleotides were 894A>G, 1062A>G, 1334G>T, 1335T>G, 1375A>G (the first nucleotides are those published in GenBank; accession no. M14752).
Statistical analysis
The Mann-Whitney test was used to compare the number of years from diagnosis to the start of imatinib therapy and the duration of imatinib therapy. The chi-square statistic was used to determine the association between mutation detection and disease phase, clonal evolution, cytogenetic response, and the time from diagnosis to imatinib start. A Kaplan-Meier curve was constructed for time to death from progressive disease and survival and compared using the log-rank test.
Results
BCR-ABL kinase domain mutations
In total, 27 of the 144 patients evaluated had detectable mutations at a median of 8 months (range, 3-18 months) after the start of imatinib therapy. Seventeen different mutations in the BCR-ABL kinase domain were found in these 27 patients. Details of the mutations are given in Table 1. These were all point mutations and were located within a sequence of 728 nucleotides, involving amino acids 244 to 486. Seven patients had 2 to 4 mutations, and one patient had 2 different mutations at the same nucleotide that both altered the amino acid at position 252 from Gln to His. The mutations, Leu248Val at the N-terminal and Ser417Tyr, Glu459Lys, and Phe486Ser at the C-terminal of the kinase domain have not previously been described.
BCR-ABL kinase domain mutations
Mutation . | Nucleotide substitution (GenBank no. M14752) . | No. of patients with the mutation* . |
|---|---|---|
| Met244Val | 730A>G | 1 |
| Leu248Val | 742C>G | 1 |
| Gly250Glu | 749G>A | 2 |
| Gln252His | 756G>C | 4 |
| Gln252His | 756G>T | 1 |
| Tyr253Phe | 758A>T | 2 |
| Glu255Lys | 763G>A | 5 |
| Glu255Val | 764A>T | 1 |
| Thr315lle | 944C>T | 2 |
| Phe317Leu | 951C>G | 2 |
| Met351Thr | 1052T>C | 8 |
| Glu355GLy | 1064A>G | 3 |
| Phe359Val | 1075T>G | 2 |
| His396Arg | 1187A>G | 1 |
| Ser417Tyr | 1250C>A | 1 |
| Glu459Lys | 1375G>A | 1 |
| Phe486Ser | 1457T>C | 1 |
Mutation . | Nucleotide substitution (GenBank no. M14752) . | No. of patients with the mutation* . |
|---|---|---|
| Met244Val | 730A>G | 1 |
| Leu248Val | 742C>G | 1 |
| Gly250Glu | 749G>A | 2 |
| Gln252His | 756G>C | 4 |
| Gln252His | 756G>T | 1 |
| Tyr253Phe | 758A>T | 2 |
| Glu255Lys | 763G>A | 5 |
| Glu255Val | 764A>T | 1 |
| Thr315lle | 944C>T | 2 |
| Phe317Leu | 951C>G | 2 |
| Met351Thr | 1052T>C | 8 |
| Glu355GLy | 1064A>G | 3 |
| Phe359Val | 1075T>G | 2 |
| His396Arg | 1187A>G | 1 |
| Ser417Tyr | 1250C>A | 1 |
| Glu459Lys | 1375G>A | 1 |
| Phe486Ser | 1457T>C | 1 |
Seven patients had multiple mutations.
Association of mutations with imatinib resistance
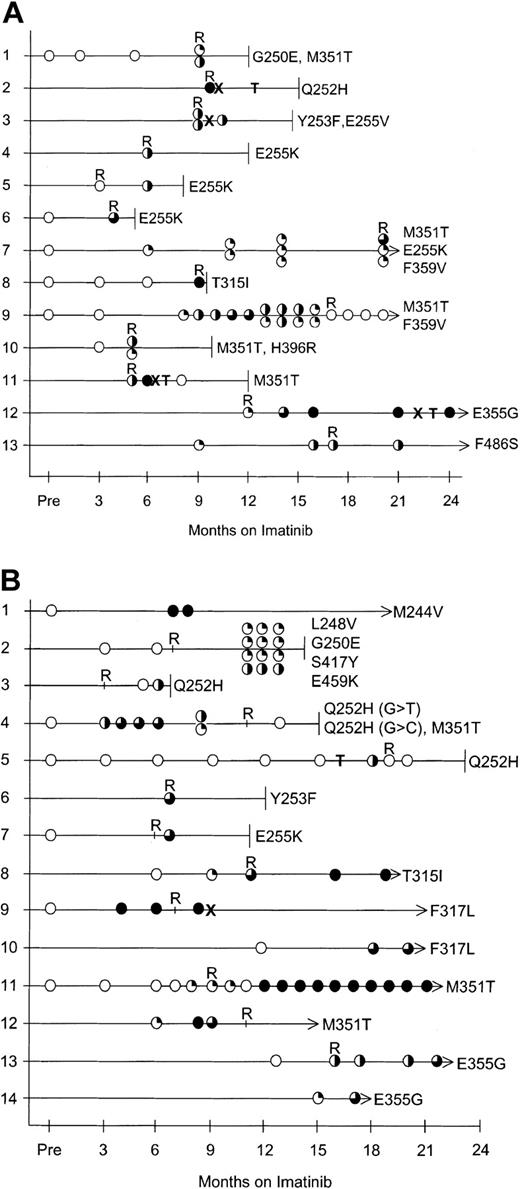
Patients (100%) in AP with mutations acquired imatinib resistance. Mutations were detected in 13 of 40 patients in AP. All 13 developed imatinib resistance that was evident in 10 patients at the time the mutation was detected and at 8, 9, and 14 months after the mutation was detected in the remaining 3 patients. Of the 27 patients without detectable mutations, only 2 acquired imatinib resistance. Therefore, 13 (87%) of 15 patients in AP with acquired imatinib resistance had detectable mutations. The 15 patients with acquired resistance included 5 with transformation to blast phase, 9 with recurrence of AP, and 1 with loss of a CCR and a corresponding 1-log increase in BCR-ABL. The disease outcome of the patients in AP with mutations is listed in Table 2, and Figure 1A tracks the emergence of mutations.
Disease outcome of the 13 patients in AP with mutations
Patient no. . | Age, y/sex . | Y from diagnosis . | Mutation . | Mo mutation first detected . | Best response by 6 mo . | Status when mutation first detected . | Disease status (mo after imatinib start) . |
|---|---|---|---|---|---|---|---|
| 1 (AP-CE) | 71/F | 4.5 | Gly250Glu | 9 | CHR | PHR | Died from disease progression (12), acquired imatinib resistance |
| Met351Thr | |||||||
| 2 | 41/F | 4.3 | Gln252His (G→C) | 10 | CHR | MBC | Imatinib withdrawn (10), died in MBC after MUD transplantation (15), acquired imatinib resistance |
| 3 (AP-CE) | 62/F | 5.8 | Tyr253Phe | 9 | CHR | CP | Imatinib withdrawn because of loss of CHR (10), died from disease progression (14), acquired imatinib resistance |
| Glu255Val | |||||||
| 4 (AP-CE) | 41/M | 5.3 | Glu255Lys | 6 | CHR | AP | Imatinib withdrawn (6) because of loss of response, died from disease progression (12), acquired imatinib resistance |
| 5 | 60/M | 3.0 | Glu255Lys | 6 | CHR | CP | Lost CP, died from disease progression (8), acquired imatinib resistance |
| 6 (AP-CE) | 44/M | 1.8 | Glu255Lys | 4 | CHR | CP | Died from disease progression (5), acquired imatinib resistance |
| 7 | 60/M | 6.3 | Met351Thr | 6 | CP | CP | Dose increased to 800 mg because of increased WCC (7), dose decreased to 400 mg because of side effects (9), Hydroxyurea commenced because of increased WBC (10), AP (20), acquired imatinib resistance |
| Glu255Lys | 11 | CP | |||||
| Phe359Val | 14 | CP | |||||
| 8 | 61/F | 7.8 | Thr315lle | 9 | CCR | LBC | Died in LBC (9.5), acquired imatinib resistance |
| 9 (AP-CE) | 64/M | 7.8 | Met351Thr | 8 | CHR | CHR | Dose increased to 800 mg (11), MBC (17), mutations not detected at onset of MBC when double Ph-chromosome was present, alive in MBC (20), acquired imatinib resistance |
| Phe359Val | 13 | CHR | |||||
| 10 | 58/F | 1.7 | Met351Thr | 5 | CP | MBC | Died in MBC (10), Acquired imatinib resistance |
| His396Arg | |||||||
| 11 | 48/M | 5.4 | Met351Thr | 5 | CP | AP | Imatinib withdrawn (7) because of MBC, acquired imatinib resistance, died in MBC after autograft (12) |
| 12 | 54/M | 7.5 | Glu355Gly | 12 | CHR | CP | AP (17), proceeded to allograft (23), relapsed into CP, mutation detected after allograft (24), acquired imatinib resistance |
| 13 (AP-CE) | 47/F | 7.7 | Phe486Ser | 9 | CCR | CCR | Lost CCR (17), dose increased to 800 mg, lost MCR (24), acquired imatinib resistance |
Patient no. . | Age, y/sex . | Y from diagnosis . | Mutation . | Mo mutation first detected . | Best response by 6 mo . | Status when mutation first detected . | Disease status (mo after imatinib start) . |
|---|---|---|---|---|---|---|---|
| 1 (AP-CE) | 71/F | 4.5 | Gly250Glu | 9 | CHR | PHR | Died from disease progression (12), acquired imatinib resistance |
| Met351Thr | |||||||
| 2 | 41/F | 4.3 | Gln252His (G→C) | 10 | CHR | MBC | Imatinib withdrawn (10), died in MBC after MUD transplantation (15), acquired imatinib resistance |
| 3 (AP-CE) | 62/F | 5.8 | Tyr253Phe | 9 | CHR | CP | Imatinib withdrawn because of loss of CHR (10), died from disease progression (14), acquired imatinib resistance |
| Glu255Val | |||||||
| 4 (AP-CE) | 41/M | 5.3 | Glu255Lys | 6 | CHR | AP | Imatinib withdrawn (6) because of loss of response, died from disease progression (12), acquired imatinib resistance |
| 5 | 60/M | 3.0 | Glu255Lys | 6 | CHR | CP | Lost CP, died from disease progression (8), acquired imatinib resistance |
| 6 (AP-CE) | 44/M | 1.8 | Glu255Lys | 4 | CHR | CP | Died from disease progression (5), acquired imatinib resistance |
| 7 | 60/M | 6.3 | Met351Thr | 6 | CP | CP | Dose increased to 800 mg because of increased WCC (7), dose decreased to 400 mg because of side effects (9), Hydroxyurea commenced because of increased WBC (10), AP (20), acquired imatinib resistance |
| Glu255Lys | 11 | CP | |||||
| Phe359Val | 14 | CP | |||||
| 8 | 61/F | 7.8 | Thr315lle | 9 | CCR | LBC | Died in LBC (9.5), acquired imatinib resistance |
| 9 (AP-CE) | 64/M | 7.8 | Met351Thr | 8 | CHR | CHR | Dose increased to 800 mg (11), MBC (17), mutations not detected at onset of MBC when double Ph-chromosome was present, alive in MBC (20), acquired imatinib resistance |
| Phe359Val | 13 | CHR | |||||
| 10 | 58/F | 1.7 | Met351Thr | 5 | CP | MBC | Died in MBC (10), Acquired imatinib resistance |
| His396Arg | |||||||
| 11 | 48/M | 5.4 | Met351Thr | 5 | CP | AP | Imatinib withdrawn (7) because of MBC, acquired imatinib resistance, died in MBC after autograft (12) |
| 12 | 54/M | 7.5 | Glu355Gly | 12 | CHR | CP | AP (17), proceeded to allograft (23), relapsed into CP, mutation detected after allograft (24), acquired imatinib resistance |
| 13 (AP-CE) | 47/F | 7.7 | Phe486Ser | 9 | CCR | CCR | Lost CCR (17), dose increased to 800 mg, lost MCR (24), acquired imatinib resistance |
AP-CE indicates the patients with clonal evolution as the sole criterion for AP; MBC, myeloid blast crisis; LBC, lymphoid blast crisis; CHR, complete hematologic response (white blood count [WBC] < 10.0, platelets < 450, no blasts, myelocytes + metamyelocytes. < 5%, no promyelocytes, no disease-related symptoms or extramedullary disease).
Mutation analysis in patients with mutations. (A) Patients in AP. (B) Patients in late-CP. Open circles represent wild-type BCR-ABL sequence. The degree of shading indicates the relative percentage of mutant compared with wild type. T indicates transplantation (refer to Tables 2 and 3 for transplantation details), X indicates that imatinib was withdrawn, terminated lines indicate the month the patient died, and arrows indicate that the patient is alive. R indicates when acquired imatinib resistance was evident. For space considerations, the mutations are shown in the figure with single-letter codes rather than 3-letter codes. For patients with multiple mutations the top circle corresponds to the amino acid with the lowest number, eg, in panel A, patient 1, the top circle corresponds to Gly250Glu and the lower circle Met351Thr. The double shading in panel B, patient 4, represents 2 mutations at the same nucleotide; no wild type was detected.
Mutation analysis in patients with mutations. (A) Patients in AP. (B) Patients in late-CP. Open circles represent wild-type BCR-ABL sequence. The degree of shading indicates the relative percentage of mutant compared with wild type. T indicates transplantation (refer to Tables 2 and 3 for transplantation details), X indicates that imatinib was withdrawn, terminated lines indicate the month the patient died, and arrows indicate that the patient is alive. R indicates when acquired imatinib resistance was evident. For space considerations, the mutations are shown in the figure with single-letter codes rather than 3-letter codes. For patients with multiple mutations the top circle corresponds to the amino acid with the lowest number, eg, in panel A, patient 1, the top circle corresponds to Gly250Glu and the lower circle Met351Thr. The double shading in panel B, patient 4, represents 2 mutations at the same nucleotide; no wild type was detected.
Clonal evolution was the only criterion for classification as AP in 15 of the 40 patients in AP. Seven of these 15 patients acquired imatinib resistance, and mutations were detected in 6 (86%) of 7. There was no significant difference in the frequency of mutations in patients with clonal evolution as the sole criterion for AP compared with the patients with other criteria for classification as AP.
Only 1 of 40 patients had primary hematologic resistance. This patient was treated for AP at diagnosis, and there was no evidence that the initial lack of hematologic response was due to the presence of a mutation, as only wild-type BCR-ABL was detected at 3.5 months of imatinib.
Patients (79%) in late-CP with mutations acquired imatinib resistance. Mutations were detected in 14 of 64 patients in late-CP. Eleven of the 14 patients acquired imatinib resistance. However, 2 of the 14 patients have only been followed for 2 months since the mutation was detected, which may not be long enough for resistance to be clinically evident. Of the 11 patients with mutations and acquired resistance, 6 had evidence of resistance at the time the mutations were detected, and resistance was acquired within 8 months in the other 5 patients. The disease outcome of the patients with mutations in late-CP is listed in Table 3, and Figure 1B tracks the emergence of mutations.
Disease outcome of the 14 patients in late-CP with mutations
Patient no. . | Age, y/sex . | Y from diagnosis . | Mutation . | Mo mutation first detected . | Best response by 6 mo . | Status when mutation first detected . | Disease status (mo after imatinib start) . |
|---|---|---|---|---|---|---|---|
| 1 | 48/M | 3.4 | Met244Val | 7 | PHR | PHR | Primary imatinib resistance at 400 mg, dose increased to 600 mg at 6 mo because of lack of response, MCR achieved, 25% Ph (18) |
| 2 | 51/F | 6.9 | Leu248Val | 11 (all mutations) | CHR | PHR | Died in AP (14), acquired imatinib resistance |
| Gly250Glu | |||||||
| Ser417Tyr | |||||||
| Glu459Lys | |||||||
| 3 | 65/F | 5.3 | Gln252His (G>C) | 6 | PHR | MBC | Died in MBC (6.5), acquired imatinib resistance |
| 4 | 71/M | 2.4 | Gln252His (G>T) | 3 | PHR | PHR(3) | WCC decreased to 4.9 but was not sustained; WCC increased (3), MBC (11); died in MBC (15); the mutations were not detected when tested at 13 mo in MBC when double Ph-chromosome was present; acquired imatinib resistance |
| Gln252His (G>C) | 3 | ||||||
| Met351Thr | 8 | PHR (8) | |||||
| 5 | 65/M | 4.2 | Gln252His (G>C) | 18 | CHR | CHR | Autograft (16); recommenced imatinib 1 mo after autograft (17); mutation detected at 18 mo but not subsequently in MBC (19) when double Ph-chromosome was present; died in MBC (23); acquired imatinib resistance |
| 6 | 52/M | 4.3 | Tyr253Phe | 7 | CHR | PHR | Dose increased to 600 mg at 7 mo because of increased WCC, died in AP (12), acquired imatinib resistance |
| 7 | 27/M | 1.7 | Glu255Lys | 7 | CHR | PHR | Died from disease progression (11), acquired imatinib resistance |
| 8 | 60/M | 9.3 | Thr315lle | 8.5 | MCR | MCR | Ph percentage increased (12), dose increased to 600 mg because of 100% Ph positive (19), acquired imatinib resistance |
| 9 | 61/M | 1.1 | Phe317Leu | 4 | CHR | CHR | MBC 3 mo after mutation detected, acquired imatinib resistance, imatinib withdrawn (11), in remission after chemotherapy (19) |
| 10 | 59/F | 2.3 | Phe317Leu | 18 | CCR | CCR | Remains in CCR (20) |
| 11 | 53/F | 5.8 | Met351Thr | 8 | CCR | CCR | AP* at 9 mo, acquired imatinib resistance, dose increased to 600 mg (9), MCR maintained (21) |
| 12 | 67/M | 3.6 | Met351Thr | 6 | MCR | MCR | PHR (11), CP (15), dose increased to 500 mg, acquired imatinib resistance |
| 13 | 51/F | 6.3 | Glu355Gly | 16 | CCR | AP* | Acquired imatinib resistance, dose increased to 600 mg (17), MCR (22) |
| 14 | 51/M | 11.0 | Glu355Gly | 15 | MCR | MCR | MCR (17) |
Patient no. . | Age, y/sex . | Y from diagnosis . | Mutation . | Mo mutation first detected . | Best response by 6 mo . | Status when mutation first detected . | Disease status (mo after imatinib start) . |
|---|---|---|---|---|---|---|---|
| 1 | 48/M | 3.4 | Met244Val | 7 | PHR | PHR | Primary imatinib resistance at 400 mg, dose increased to 600 mg at 6 mo because of lack of response, MCR achieved, 25% Ph (18) |
| 2 | 51/F | 6.9 | Leu248Val | 11 (all mutations) | CHR | PHR | Died in AP (14), acquired imatinib resistance |
| Gly250Glu | |||||||
| Ser417Tyr | |||||||
| Glu459Lys | |||||||
| 3 | 65/F | 5.3 | Gln252His (G>C) | 6 | PHR | MBC | Died in MBC (6.5), acquired imatinib resistance |
| 4 | 71/M | 2.4 | Gln252His (G>T) | 3 | PHR | PHR(3) | WCC decreased to 4.9 but was not sustained; WCC increased (3), MBC (11); died in MBC (15); the mutations were not detected when tested at 13 mo in MBC when double Ph-chromosome was present; acquired imatinib resistance |
| Gln252His (G>C) | 3 | ||||||
| Met351Thr | 8 | PHR (8) | |||||
| 5 | 65/M | 4.2 | Gln252His (G>C) | 18 | CHR | CHR | Autograft (16); recommenced imatinib 1 mo after autograft (17); mutation detected at 18 mo but not subsequently in MBC (19) when double Ph-chromosome was present; died in MBC (23); acquired imatinib resistance |
| 6 | 52/M | 4.3 | Tyr253Phe | 7 | CHR | PHR | Dose increased to 600 mg at 7 mo because of increased WCC, died in AP (12), acquired imatinib resistance |
| 7 | 27/M | 1.7 | Glu255Lys | 7 | CHR | PHR | Died from disease progression (11), acquired imatinib resistance |
| 8 | 60/M | 9.3 | Thr315lle | 8.5 | MCR | MCR | Ph percentage increased (12), dose increased to 600 mg because of 100% Ph positive (19), acquired imatinib resistance |
| 9 | 61/M | 1.1 | Phe317Leu | 4 | CHR | CHR | MBC 3 mo after mutation detected, acquired imatinib resistance, imatinib withdrawn (11), in remission after chemotherapy (19) |
| 10 | 59/F | 2.3 | Phe317Leu | 18 | CCR | CCR | Remains in CCR (20) |
| 11 | 53/F | 5.8 | Met351Thr | 8 | CCR | CCR | AP* at 9 mo, acquired imatinib resistance, dose increased to 600 mg (9), MCR maintained (21) |
| 12 | 67/M | 3.6 | Met351Thr | 6 | MCR | MCR | PHR (11), CP (15), dose increased to 500 mg, acquired imatinib resistance |
| 13 | 51/F | 6.3 | Glu355Gly | 16 | CCR | AP* | Acquired imatinib resistance, dose increased to 600 mg (17), MCR (22) |
| 14 | 51/M | 11.0 | Glu355Gly | 15 | MCR | MCR | MCR (17) |
MBC indicates myeloid blast crisis; LBC, lymphoid blast crisis; PHR, partial hematologic response (blasts in blood + bone marrow < 15%); CHR, complete hematologic response.
Double Philadelphia chromosome.
In total 3 of the 64 patients in late-CP had primary hematologic resistance, and all 3 were among the 14 patients with detectable mutations. One of the 3 had a mutation (Met244Val) without evidence of acquired resistance and has achieved an MCR after receiving a dose increase from 400 to 600 mg imatinib. There was no evidence that the initial lack of hematologic response was due to the presence of mutations because only wild-type BCR-ABL was detected within the first 6 months of imatinib therapy (n = 1) or prior to commencing therapy (n = 2).
Two of the 50 patients without detectable mutations acquired imatinib resistance. Therefore, of the 64 patients in late-CP, 13 acquired imatinib resistance and 11 (85%) of the 13 had detectable mutations. The 13 patients with acquired resistance included 5 with transformation to blast phase, 6 to AP, and 2 with a loss of an MCR. The remaining 48 patients continued to be responsive to imatinib.
No mutations were detected in patients in early-CP. Six of the 40 patients in early-CP acquired imatinib resistance. The 6 resistant patients included 1 with transformation to blast crisis, 2 to AP, 1 with loss of a MCR, and 2 with loss of a CCR and a corresponding significant increase in BCR-ABL levels of at least 1 log. In this small subset of patients with resistance in early-CP, we have not seen evidence of detectable mutations; however, the follow-up was relatively short (median 14 months of imatinib; range, 5-24 months). Only 1 of the 6 patients who acquired resistance had evidence of gene amplification as determined by the presence of an extra Philadelphia chromosome by cytogenetic analysis. Of the 16 patients who failed previous IFN-α therapy, only 1 acquired imatinib resistance.
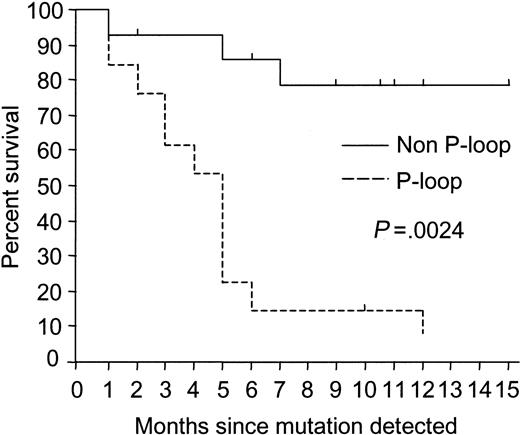
P-loop mutations were associated with poor outcomes
In 13 patients, mutations were detected within the ATP phosphate-binding loop (P-loop) at the N-terminal of the kinase domain at amino acids 250, 252, 253, or 255. Twelve (92%) of the 13 patients (6 of 7 AP and 6 of 6 late-CP) died at a median of 4.5 months (range, 0.5-12 months) after the mutations were detected. In contrast, only 3 of 14 patients with mutations outside the P-loop died at a median of 5 months (range, 0.2-7 months) after the mutation was detected. The follow-up of the remaining 11 patients with non–P-loop mutations was a median of 10.5 months after the first detection of mutations (range, 2-15 months; Figure 2).
Kaplan-Meier survival curves for patients with mutations. There was a significant difference in the survival rate of patients in AP and late-CP with P-loop and non–P-loop mutations. With one exception, patients with P-loop mutations died.
Kaplan-Meier survival curves for patients with mutations. There was a significant difference in the survival rate of patients in AP and late-CP with P-loop and non–P-loop mutations. With one exception, patients with P-loop mutations died.
Although the overall survival was better for those with non–P-loop mutations, survival for patients in AP was not statistically significant (P = .14) because 3 of 6 patients with non–P-loop mutations also died. However, the difference between the outcome of the 14 patients in late-CP with and without P-loop mutations was highly significant as only the 6 patients with P-loop mutations died (P = .0013). In contrast, of the 5 patients in late-CP with acquired resistance and mutations outside the P-loop, 3 have maintained CP (Thr315Ile, Phe317Leu, and Met351Thr) with an increased imatinib dose or change of therapy, and 2 have achieved a MCR (Met351Thr and Glu355Gly) with an increased imatinib dose (Table 3). The median follow-up was 10.5 months (range, 6-15 months) in these patients.
Factors predicting for mutation detection
Variables were evaluated to identify factors that may be associated with the detection of mutations and in particular with P-loop mutations because these factors were associated with a poor outcome. The baseline factors tested were the disease phase, time from diagnosis, and the presence of clonal evolution. The 6-month cytogenetic response was also evaluated. These factors were assessed by univariate analysis, and the results are listed in Table 4. The factors with a significant association overall with mutation detection were the time from diagnosis to imatinib start, failure to achieve a MCR by 6 months of imatinib therapy, and the disease phase. Other baseline variables known to be associated with response and survival such as blast percentage, hemoglobin, and previous response to IFN-α could not adequately be evaluated by univariate or multivariate analysis because of the low frequency of mutations and small patient numbers in some categories.
Significance of factors in the detection of mutations
Factor . | No. of patients . | Total no. with mutations (%) . | P . |
|---|---|---|---|
| Time from diagnosis to imatinib start | < .0001 | ||
| Less than 1 y | 46 | 0 | |
| 1-2 y | 26 | 4 (15) | |
| More than 2 y to 4 y | 28 | 5 (18) | |
| More than 4 y | 44 | 18 (41) | |
| Disease phase at imatinib start | |||
| Early-CP. | 40 | 0 | .0039 |
| Late-CP | 64 | 14 (22) | |
| Early-CP | 40 | 0 | |
| AP | 40 | 13 (33) | .0003 |
| Late-CP | 64 | 14 (22) | |
| AP | 40 | 13 (33) | .3308 |
| Clonal evolution in AP patients | |||
| No clonal evolution | 19 | 5 (26) | |
| Clonal evolution | 21 | 8 (38) | .6482 |
| Cytogenetic response at 6 mo | |||
| MCR | 94 | 8 (8.5) | <.0001 |
| No MCR | 50 | 19 (38) |
Factor . | No. of patients . | Total no. with mutations (%) . | P . |
|---|---|---|---|
| Time from diagnosis to imatinib start | < .0001 | ||
| Less than 1 y | 46 | 0 | |
| 1-2 y | 26 | 4 (15) | |
| More than 2 y to 4 y | 28 | 5 (18) | |
| More than 4 y | 44 | 18 (41) | |
| Disease phase at imatinib start | |||
| Early-CP. | 40 | 0 | .0039 |
| Late-CP | 64 | 14 (22) | |
| Early-CP | 40 | 0 | |
| AP | 40 | 13 (33) | .0003 |
| Late-CP | 64 | 14 (22) | |
| AP | 40 | 13 (33) | .3308 |
| Clonal evolution in AP patients | |||
| No clonal evolution | 19 | 5 (26) | |
| Clonal evolution | 21 | 8 (38) | .6482 |
| Cytogenetic response at 6 mo | |||
| MCR | 94 | 8 (8.5) | <.0001 |
| No MCR | 50 | 19 (38) |
Time from diagnosis. Of the 144 patients evaluated, those who commenced imatinib therapy more than 4 years from diagnosis had a higher incidence of mutations, in which 18 (41%) of 44 patients had mutations (Table 4). Seventeen of the 18 patients with mutations acquired imatinib resistance. Only 1 patient with a mutation in this group has not acquired resistance, but the patient has only had 2 months' follow-up since the detection of the mutation. Of the 26 patients without detectable mutations who were treated more than 4 years from diagnosis, 24 remained responsive to imatinib with a median follow-up of 15 months (range, 10-20 months) since imatinib start, whereas the other 2 patients acquired resistance.
In contrast, only 9 (9%) of 100 patients treated with imatinib within 4 years of diagnosis had detectable mutations. When these 100 patients were further divided, no mutations were detected in any of the 46 patients treated with imatinib within 1 year of diagnosis (40 early-CP and 6 AP). Seven of the 9 patients with mutations acquired imatinib resistance. Of the remaining 91 patients treated within 4 years of diagnosis who did not have detectable mutations, 83 continued to be responsive to imatinib with a median follow-up of 16 months (range, 6-24 months) since imatinib start, whereas 8 acquired resistance, including 6 patients in early-CP.
When patients in AP were further divided using a stricter criteria for classification of AP by excluding the patients with only clonal evolution, there was still a higher incidence of mutations and resistance in those patients with a time from diagnosis of more than 4 years (5 [45%] of 11 patients) compared with those less than 4 years (2 [14%] of 14 patients).
Failure to achieve a MCR within 6 months. Fifty of the 144 patients evaluated failed to achieve a MCR by 6 months of imatinib therapy. A higher incidence of mutations was detected in this group with 19 (38%) of 50 patients having detectable mutations at a median of 7 months (range, 3-18 months) of imatinib therapy (Table 4). Eighteen of these 19 patients acquired imatinib resistance, which was evident at a median of 9 months (range, 4-20 months) of imatinib therapy. Only 6 of the 50 patients who failed to achieve a MCR by 6 months acquired resistance without detectable mutations.
Of the 94 patients who achieved a MCR by 6 months of imatinib therapy, only 8 patients (8.5%) had mutations detected at a median of 9 months (range, 6-18 months) of imatinib therapy. Six of these patients acquired resistance that was evident at a median of 11.5 months (range, 9-17 months) of imatinib therapy. Two of the 8 patients with mutations have only had 2 months' follow-up since the mutation was detected. Of the 86 remaining patients without detectable mutations, only 4 patients in early-CP acquired imatinib resistance.
Disease phase. The difference in the frequency of mutations in the various disease phases was significant when patients in early-CP were compared with patients in AP and late-CP. Although there was a higher incidence of mutations in patients in AP compared with patients in late-CP, the difference was not significant (Table 4).
P-loop mutations. There was no significant association with baseline variables and the detection of P-loop mutations. The 13 patients with P-loop mutations had a median time from diagnosis to commencing imatinib of 4.3 years (range, 1.7-6.9 years) compared with 6.3 years (range, 1.1-11 years) for the 14 patients with non–P-loop mutations, P = .123. The frequency of P-loop mutations in patients in AP compared with late-CP was not significant, P = .306. However, all 13 patients with P-loop mutations failed to achieve a MCR within 6 months of commencing imatinib, whereas only 6 of the 14 patients with non–P-loop mutations failed to achieve a MCR.
Discussion
In this study we found that virtually every CML patient treated with imatinib who had a BCR-ABL kinase domain mutation detected by direct sequencing already had evidence of or subsequently developed evidence of imatinib resistance. The 144 patients evaluated were divided by their disease phase at the start of imatinib therapy as either AP, late-CP, or early-CP. Our aims were to determine the frequency of mutations in the various disease phases and to correlate mutation detection with imatinib resistance. The disease outcomes in patients with various mutations and imatinib resistance were assessed to determine if some mutations had consistent patterns of response to therapy and disease progression. Available baseline variables and the cytogenetic response to imatinib were evaluated to determine an association with mutation detection.
Mutations were detected in 27 patients at 17 different residues. Twenty-five of the 27 patients were followed for more than 2 months after the mutation was detected, and acquired resistance was evident in all but 1 patient. Mutations at amino acids 248, 417, 459, and 486 have not been previously reported. Amino acid 248 is predicted to contact imatinib through van der Waals interactions,12,13 and mutagenesis at this site to alanine rendered the ABL protein inactive.18 This finding suggested that residue 248 was also critical for ATP binding. Mutation to the slightly larger valine instead of alanine as seen in our patient may decrease sensitivity to imatinib while retaining kinase activity. Amino acids 417, 459, and 486 are located at the C-terminal of the kinase domain distant to the imatinib binding site. It is possible that mutation at these sites causes conformational changes to the BCR-ABL protein that alters the equilibrium to the active conformation and impairs imatinib binding. To our knowledge, other investigators have not sequenced beyond the activation loop in the C-terminal lobe and, thus, would not have detected these changes, even if they were present. Our data suggest that mutations beyond the activation loop are also implicated in imatinib resistance, and patients with these mutations may benefit from therapeutic intervention.
Mutations were only detected in patients classified as AP or late-CP. We did not detect mutations in any of the patients in early-CP, indicating that different mechanisms of resistance may be acting in this setting. Amplification of the BCR-ABL gene is a documented mechanism of imatinib resistance,6 but only 1 of the 6 patients in early-CP with acquired resistance had cytogenetic evidence of BCR-ABL amplification as demonstrated by a double Philadelphia chromosome. Further study of this cohort over time will determine if mutations eventually emerge or whether the inactivation of the BCR-ABL kinase and the associated reduction in leukemic cell mass make the development of mutations less likely.
The P-loop is a highly conserved region of the kinase domain involved in ATP binding and is a frequent site of mutations.7-9,11,17,19 Thirteen of 27 patients in our study had mutations within this site. Twelve of the 13 patients died of disease progression by 12 months of the mutations being detected, including all 6 patients in late-CP. In contrast, of the 14 patients with non–P-loop mutations, only 3 have died of disease progression. Point mutations in the P-loop of other proteins have been reported. Mutation in the P-loop of the ras protein p21 interferes with phosphate transfer and renders the protein oncogenic.22-24 A point mutation in the ATP binding pocket of the c-erbB tyrosine kinase increases its kinase activity and transforming potential.25,26 Interestingly, the P-loop mutation Tyr253Phe demonstrated intermediate resistance to imatinib both in vitro and in vivo when compared with Thr315Ile.17 However, Tyr253Phe in cABL demonstrated oncogenic activity27 and has been shown to induce significantly more phosphotyrosine in vivo than wild-type BCR-ABL.17 This finding supports the possibility that at least some P-loop mutations may increase the oncogenicity of BCR-ABL. Although the correlation of death and P-loop mutations in patients in late-CP was statistically significant, it did not reach significance in the patients in AP. This finding was most likely because AP is a more advanced disease, and other transforming events cooperate with BCR-ABL activity, leading to disease progression. The number of patients was small, however, and additional evaluation is required. Further study on the biologic activity of mutations within the P-loop may provide data on whether our finding in patients in late-CP is consistent with the P-loop mutations actually conferring enhanced transforming capacity and contributing to disease progression. The Thr315Ile mutation is one of the most resistant mutations in vitro6,11,17-19 ; however, only 2 patients in our series had this mutation, one of whom died in blast crisis. Thus, our data are not sufficient to draw any conclusions about the prognosis for patients with this mutation or whether they should be grouped with the P-loop mutations.
A number of patient variables were identified that were associated with a higher incidence of detectable mutations and imatinib resistance. These variables were a longer time from diagnosis to imatinib start and failure to achieve a MCR by 6 months. Patients who started treatment with imatinib more than 4 years after diagnosis had the highest incidence of mutations. This finding supports the concept of the gradual accumulation of a pool of BCR-ABL mutants over time, which, if they have a lower affinity for imatinib, are capable of expanding under the selective pressure of imatinib therapy. This concept is supported by the recent finding of kinase domain mutations in samples tested prior to therapy in 2 patients in blast crisis with primary hematologic resistance to imatinib.11 Another recent study reported baseline variables that independently predicted the probability of achieving a MCR in patients in CP treated with imatinib. These variables included the number of blasts, hemoglobin level, a time from diagnosis to imatinib start of less than 1 year, and a history of cytogenetic relapse during IFN-α therapy.2 A further study identified a long chronic phase at baseline as an adverse factor.28 We could not appropriately evaluate all of these factors by multivariate analysis to determine those that may be independently associated with the detection of mutations because there was a low frequency of mutations for a number of these categories. Analysis of a larger cohort of patients may identify independent predictors for mutation detection.
Several studies have reported that different kinase domain mutations retain varying sensitivities to higher concentrations of imatinib,11,18,20 which suggests that higher doses could prevent the selection and expansion of some resistant mutant clones. Patients in our series were treated initially with 400 mg for late-CP or 600 mg for AP. Seven of the 17 different mutations detected were common to both treatment arms, including the Met351Thr and Glu355Gly mutations. These mutations were found to retain biochemical sensitivity to imatinib at increased concentrations.11 Our data do not support a significant difference in the selection of resistant mutations at an imatinib dose of 400 or 600 mg. A comprehensive evaluation of patients treated with 800 mg is required to determine if a limited subset of mutations occur with higher doses. It has recently been reported that the Met244Val mutation has a low cellular IC50 (concentration that inhibits 50%) compared with previously analyzed mutants and that clinical response may be regained with higher imatinib doses if this mutation is the sole abnormality.20 This finding is supported by the clinical response of a patient in our series with the Met244Val mutation who achieved a MCR with an increased dose to 600 mg of imatinib after failure to obtain a response to the initial dose of 400 mg.
The incidence of mutations in our study was high and raises the question whether our data might be biased toward patients with mutations and/or resistance. Our intent was to collect all samples from patients who had received imatinib for at least 6 months. However, not all centers were able to collect samples from all patients, and those patients responding well may have been less likely to have blood collected during the study period. If this were the case, we may have received a disproportionate number of samples from patients with poor response to imatinib. However, we found the rates of MCR and CCR for patients in late-CP were similar to those reported in a study of patients with CML in CP in whom IFN-α had failed.2 The rates of MCR for patients in AP could not accurately be compared with the rates of other trials of imatinib in AP3,29 because the entry criteria for these trials was more restrictive and excluded patients with clonal evolution as the sole criterion for AP. However, the estimated 12-month death rate was similar to our study.3 Interestingly, 7 (47%) of 15 patients with AP with clonal evolution as the sole criterion of disease acceleration acquired resistance, and 6 of the 7 patients had mutations. This incidence of resistance was higher than a recent study that found no treatment failure in 15 similar patients.30
The direct sequencing method for mutation analysis of the complete BCR-ABL kinase domain enabled the detection of single or multiple mutations in patients with disease status ranging from CCR to overt relapse. The reliability of mutation analysis was highly dependent on the quality of the RNA. Therefore, RNA of high quality was required, and poor quality samples, as determined by quantitative PCR of a control gene, were excluded from analysis. This selection of samples for analysis may well be a significant difference between our study and others that reported a lower incidence of mutations in patients with resistance to imatinib.10,31,32 The direct sequencing method is less cumbersome than the subcloning techniques used in other studies6,11 and, thus, is more suitable for widespread testing. The sensitivity of the method at 20% is the same as the subcloning technique. Sequencing of the complete kinase domain is required because the mutations were detected throughout the domain and additional mutations became detectable over time in some patients.
In summary, most patients with detectable BCR-ABL kinase domain mutations acquired imatinib resistance. Mutations that have no clinical significance seem to be quite rare. Mutations in the P-loop were frequent and were particularly associated with a poor prognosis. Those patients with longer duration of CML prior to initiation of imatinib therapy had a higher incidence of detectable mutations and resistance. Failure to achieve cytogenetic disease control in the first 6 months of therapy often reflects the presence of mutations or a high probability that mutations will subsequently be detected. These results suggest that imatinib therapy may be most effective when initiated early in the disease course and that molecular monitoring in the first months of therapy may play a crucial role in detecting patients at high risk of resistance. This monitoring may identify candidates suitable for more aggressive therapy.
Prepublished online as Blood First Edition Paper, March 6, 2003; DOI 10.1182/blood-2002-09-2896.
Supported in part by grants from Novartis Pharmaceuticals Australia.
K.L. is employed by Novartis Pharmaceuticals, whose product (imatinib mesylate) was used in this study.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Bernadette Miller and Nancy Lerda for technical support; the data managers and laboratory staff of the participating centers, in particular Alison Harper, Rosie Hoyt, Liz O'Flaherty, Melinda Higgins, Michael Kersten, Sonja Downey, Jenny Bourne, April Josephsen, Deb Taylor, Paula Ambrosoli, Andrea Moore, and Pat Plenge; and Dr Petranel Ferrao for the donation of the Tyr253Phe-containing cells.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal