Abstract
Fanconi anemia (FA) is an autosomal recessive syndrome characterized by progressive bone marrow failure and cancer predisposition. Eight FA complementation groups have been identified. The FANCA, FANCC, FANCE, FANCF, and FANCG proteins form a nuclear complex required for the monoubiquination of the FANCD2 protein. To investigate the architecture of the FA protein complex, the yeast 2-hybrid system was used to map contact points of the FANCA/FANCG, FANCC/FANCE, and FANCF/FANCG interactions. FANCG was shown to interact with both the amino-terminus of FANCA and the carboxyl-terminal region of FANCF. A FANCG mutant truncated at the carboxyl-terminus retained the ability to interact with FANCA. The interaction between FANCG and FANCF was ablated by a Leu71Pro mutant of FANCG. A central region of FANCE was sufficient for FANCC binding. A Leu554Pro mutant of FANCC failed to interact with FANCE. To further examine complex assembly, the yeast 3-hybrid system was used to investigate the ability of FANCG to act as a molecular bridge in mediating interaction between other FA proteins. FANCG was able to mediate interaction between FANCA and FANCF, as well as between monomers of FANCA. Direct interaction between FANCE and FANCD2 was also demonstrated in the yeast 2-hybrid system. This interaction involving an amino-terminal region of FANCD2 may provide a link between the FA protein complex and its downstream targets. (Blood. 2003;102:136-141)
Introduction
Fanconi anemia (FA) is a rare autosomal recessive syndrome characterized by pancytopenia, predisposition to cancer, and a wide range of congenital malformations.1 Cells from patients with FA exhibit elevated spontaneous chromosome breakage and are hyper-sensitive to DNA cross-linking agents, including mitomycin C and diepoxybutane.2 FA is a genetically heterogeneous disorder comprising at least 8 complementation groups (FA-A, -B, -C, -D1, -D2, -E, -F, -G),3,4 the genes for 6 of which (FANCA,5,6 FANCC,7 FANCD2,4 FANCE,8 FANCF,9 and FANCG10 ) have been cloned. Most recently biallelic inactivation of BRCA2 was identified in FA cell lines from complementation groups FA-B and FA-D1.11
With the exception of FANCD2, which has conserved sequences in Arabidopsis thaliana, Drosophila melanogaster, and Caenorhabditis elegans, FANC protein orthologs have not been identified in nonvertebrates.12 In addition, despite the similar clinical phenotype generated by their individual absence, the FANC proteins do not bear any sequence similarity to each other. This phenomenon has given rise to the question of how, when, and where these orphan proteins might interact to maintain genomic integrity.
Database searches have revealed few identifiable protein-interaction motifs among the FANC proteins. Neither FANCC nor FANCF contain any such motifs. FANCE has a putative nuclear localization signal (NLS) in the central region of its sequence.8 FANCA has overlapping bipartite nuclear localization signals at the amino-terminus of the protein and a more carboxyl-terminal leucine zipper motif 5,6 that are conserved in the mouse homolog.13,14 The human FANCG has a putative leucine zipper motif, but it is not conserved in the mammalian homologs.15
FANCC has a predominantly cytoplasmic localization,16,17 with a smaller fraction present in the nucleus.18 The FANCA, FANCF, and FANCG proteins are found predominantly in the nucleus, with some cytoplasmic localization.19-23 FANCE has recently been shown to localize to the nucleus, concentrated in foci.24 FANCD2 is localized entirely to the nucleus, predominantly in a diffuse pattern, but also with a subset of cells containing foci.25 In addition to these variations in cellular compartmentalization, some FANC proteins appear to be involved in the localization of others, such as the ability of FANCE to promote the nuclear accumulation of FANCC.24
The FANCA, FANCC, and FANCG proteins immunoprecipitate as a complex,21,26 which has been shown to localize to chromatin and the nuclear matrix.27 The FANCF and FANCE proteins have also been identified in this nuclear complex,23,24 the formation of which is disrupted in cell lines from FA complementation groups FA-A, -B, -C, -E, -F, and -G.23,28,29 The FANCD2 protein, believed to function downstream of the FA protein complex, has been immunoprecipitated only with FANCE.24 In the absence of complex formation FANCD2 is not monoubiquinated or targeted to nuclear foci in response to DNA-damaging agents.25
Several direct interactions have been documented between the known members of the FA nuclear protein complex. Direct binding of the N-terminal region of FANCA with FANCG22,26,30 has been well documented. Interactions of FANCG with FANCF, as well as FANCE with FANCC, have also been demonstrated.31 The possibility of weak or transient interaction between other FANC protein pairs, including FANCA/FANCA, FANCG/FANCG,32 FANCA/FANCC,33 FANCA/FANCE, and FANCG/FANCE31 has been suggested by some yeast 2-hybrid studies.
To gain a better understanding of the architecture of the FA protein complex, as well as its connection to FANCD2, a GAL4 yeast 2-hybrid system34 was used to identify direct interactions between full-length FANC proteins and to map the contact points of these interactions. The effect of patient-derived putative missense mutants on protein interactions was also investigated. As FANCG had been demonstrated to interact directly with both FANCA and FANCF, the yeast 3-hybrid system35 was used to investigate the ability of FANCG to mediate interactions between the other FANC proteins, uncovering a possible role for FANCG at multiple stages of complex assembly.
Materials and methods
Bacterial and yeast strains
Escherichia coli strains DH10B (Invitrogen, Carlsbad, CA) or XL1-Blue (Stratagene, La Jolla, CA) were used in the construction and propagation of all plasmid constructs. E coli was propagated in Luria-Bertani (LB) media at 30°C for FANCD2 and FANCE and 37°C for other expression constructs. Saccharomyces cerevisiae strain PJ69-4A36 (American Type Culture Collection, Manassas, VA) was used in the yeast 2-hybrid assay. S cerevisiae strains AH109 and Y187 (Clontech, Palo Alto, CA) were used in the 3-hybrid assay. Yeast was grown at 30°C in YPD or synthetic dropout minimal media (SC).
Vectors and expression constructs
All constructs were made using vectors supplied in the Matchmaker GAL4 2-hybrid systems (Clontech). Two-hybrid DNA-binding domain (BD) constructs were made in the pAS2-1 vector. Three-hybrid BD constructs were made in the pBridge vector. Both pAS2-1 and pBridge contain the TRP1 gene for selection on tryptophan-deficient media. All activation domain (AD) constructs were made in the pGADGH or pGADT7 vectors containing the LEU2 gene for selection on leucine-deficient media. Full-length cDNAs were subcloned from the pREP4 (Invitrogen) vectors in which they were originally cloned, with the exception of FANCD2, which was subcloned from pMMPD2.4 Deletion constructs were designed on the basis of the presence of restriction sites or were engineered to express particular fragments. In cases in which polymerase chain reaction (PCR) was required to generate an in-frame fusion with the GAL4 domain, PCR was performed using Pwo polymerase (Roche, Laval, QC). All PCR products and linker regions were verified by DNA sequencing. The Leu71Pro and Gly546Arg mutants of FANCG37,38 were made using the QuikChange XL site-directed mutagenesis system (Stratagene, Cedar Creek, TX). The Leu554Pro mutation of FANCC7 had been generated previously.39 All point mutations were verified in the expression constructs by DNA sequencing.
Immunoblotting of GAL4 fusion proteins to confirm expression
Only constructs shown to express GAL4 fusion proteins by Western blot (data not shown) were used in 2-hybrid and 3-hybrid analyses. Cell lysates were prepared by the method of Langlands et al.40 Lysate (25 μL per lane) was loaded on a 10% Tris (tris(hydroxymethyl)aminomethane)–glycine gel (Invitrogen) for sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene diflouride (PVDF) membrane (Millipore, Bedford, MA). Membranes were probed with GAL4-BD or GAL4-AD mouse monoclonal antibodies (Santa Cruz Biotechnology, Santa Cruz, CA), followed by antimouse immunoglobulin G (IgG)–peroxidase conjugate and chemiluminescent detection (Amersham, Piscataway, NJ).
Yeast 2-hybrid assay
PJ69-4A was cotransformed simultaneously with 2 plasmids by the lithium acetate method as described in the Yeast Protocols Handbook of the Matchmaker system (Clontech). Cotransformants growing on both -Ade and -His selective media and controls were assayed for β-galactosidase activity by the liquid ONPG (o-nitrophenol-β-D-galactoside) sarkosyl permeabilization method of Kippert.41
Yeast 3-hybrid assay
Plasmid constructs were transformed into AH109 or Y187 by the lithium acetate method (Clontech), and transformants were mated according to the standard protocol given in the Clontech Yeast Protocols Handbook. Diploids were plated to selective media without methionine or with 4 mM methionine. HA-FANCG expression at 4 mM methionine was not detectable by Western blot using anti-HA monoclonal antibody (Roche) (data not shown). Cotransformants were assayed for β-galactosidase activity by the method of Kippert.41
Results
Yeast 2-hybrid system
In the PJ-694A strain of S cerevisiae, 3 reporters are used to assess interaction in the GAL4-based yeast 2-hybrid system: GAL2p-ADE2, GAL1p-HIS3, and GAL7p-LacZ.36 Cotransformants expressing full-length FANC proteins fused to the GAL4 activation domain (AD) or DNA-binding domain (BD) were assayed pairwise for activation of the 2 nutritional markers (Table 1). Interaction was observed between the AD-FANCC and BD-FANCE combination, as well as the reciprocal pairing of AD-FANCE and BD-FANCC. AD-FANCE also interacted with BD-FANCD2. Interactions were also observed between AD-FANCG and both BD-FANCA and BD-FANCF. The BD-FANCG construct autoactivated all 3 reporters strongly. Nonetheless, measurement of β-galactosidase activity of the pairings by liquid ONPG assay revealed that the LacZ reporter activation differed markedly from the basal activity of the BD-FANCG construct when it was paired with AD-FANCA or AD-FANCF (Table 2), consistent with the reciprocal assays. When cotransformed with AD-FANCA, the LacZ activation was considerably above background, in agreement with the interaction seen between the reciprocal pairing of BD-FANCA and AD-FANCG. The basal LacZ autoactivation was actually lowered for the BD-FANCG/AD-FANCF pairing, suggesting that FANCF may interfere with the strong ability of FANCG to autoactivate. The AD-FANCD2 construct also showed mild autoactivation of reporters and was not used in further analysis.
Pairwise analysis of interaction between full-length FANC proteins
. | BD-FANCA . | BD-FANCC . | BD-FANCD2 . | BD-FANCE . | BD-FANCF . |
|---|---|---|---|---|---|
| AD-FANCA | − | − | − | − | − |
| AD-FANCC | − | − | − | + | − |
| AD-FANCE | − | + | + | − | − |
| AD-FANCF | − | − | − | − | − |
| AD-FANCG | + | − | − | − | + |
. | BD-FANCA . | BD-FANCC . | BD-FANCD2 . | BD-FANCE . | BD-FANCF . |
|---|---|---|---|---|---|
| AD-FANCA | − | − | − | − | − |
| AD-FANCC | − | − | − | + | − |
| AD-FANCE | − | + | + | − | − |
| AD-FANCF | − | − | − | − | − |
| AD-FANCG | + | − | − | − | + |
Growth (+) or (−) of PJ69-4A cotransformants on -Ade -His media. The AD-FANCD2 and BD-FANCG constructs are not included because they autoactivated the reporters.
LacZ reporter activation in cotransformants of BD-FANCG with AD-FANC constructs
Full-length protein fused to GAL4-AD . | β-Galactosidase activity* . |
|---|---|
| No insert | 41.2 ± 1.5 |
| FANCA | 100.1 ± 5.2 |
| FANCC | 42.8 ± 3.3 |
| FANCD2 | 41.0 ± 2.6 |
| FANCE | 44.5 ± 6.9 |
| FANCF | 23.2 ± 1.6 |
| FANCG | 42.7 ± 1.4 |
Full-length protein fused to GAL4-AD . | β-Galactosidase activity* . |
|---|---|
| No insert | 41.2 ± 1.5 |
| FANCA | 100.1 ± 5.2 |
| FANCC | 42.8 ± 3.3 |
| FANCD2 | 41.0 ± 2.6 |
| FANCE | 44.5 ± 6.9 |
| FANCF | 23.2 ± 1.6 |
| FANCG | 42.7 ± 1.4 |
Units of β-galactosidase activity in the liquid ONPG assay were calculated as 1000 × optical density at 420 nm (OD420) divided by the product of the volume (milliliter) × time (minutes) × OD600.
To better understand the nature of these interactions among the FA proteins, deletion constructs were used to map contact sites for the binding of FANCA/FANCG, FANCF/FANCG, FANCC/FANCE, and FANCE/FANCD2 demonstrated in the yeast 2-hybrid system. The amino-terminal 40 amino acids of FANCA encompassing the NLS motifs were sufficient for interaction with full-length FANCG. As a polypeptide lacking only the first 35 amino-terminal amino acids of FANCA could not sustain this interaction, no secondary independent sites for FANCG binding were detected (Figure 1A). Of the truncated AD-FANCG polypeptides tested, only an AD-FANCG lacking the 142 C-terminal amino acids retained any ability to interact with FANCA (Figure 1B). The carboxyl-terminal region of FANCF was sufficient for interaction with FANCG (Figure 1C). None of a series of truncated polypeptides spanning the length of FANCG were able to mediate interaction with FANCF (Figure 1D). The effect of patient-derived Leu71Pro and Gly546Arg missense mutations of FANCG on interaction with both FANCA and FANCF was examined. ADFANCGLeu71Pro was unable to interact with BD-FANCF, and the reporter activation in the AD-FANCGLeu71Pro/BD-FANCA cotransformants was much reduced compared with that observed for wild-type AD-FANCG/BD-FANCA. AD-FANCGGly546Arg retained the ability to interact with both BD-FANCA and BD-FANCF. LacZ reporter activation was lower for AD-FANCGGly546Arg/BD-FANCA cotransformants than for wild-type AD-FANCG/BD-FANCA.
Mapping binding sites for the interactions of FANCG with FANCA and FANCF. (A) Mapping the FANCG binding region on FANCA. (B) Mapping the FANCA binding region on FANCG. (C) Mapping the FANCG binding region on FANCF. (D) Mapping the FANCF binding region on FANCG. Yeast strain PJ69-4A was cotransformed with the expression constructs indicated. Constructs activating the reporter genes above background levels are shaded. The dotted lines represent internal deletions. (++), (+), and (-) indicate good, weak, and absent growth, respectively. β-Galactosidase activity units in the liquid ONPG assay were calculated as 1000 × OD420 divided by the product of the volume (milliliter) × time (minutes) × OD600. The values shown indicate the mean and SE for 4 assays, each done in triplicate. Nd indicates that no β-galactosidase assay was performed because of nonactivation of the ADE2 and HIS3 reporters. NLS indicates nuclear localization signal; LZ, leucine zipper.
Mapping binding sites for the interactions of FANCG with FANCA and FANCF. (A) Mapping the FANCG binding region on FANCA. (B) Mapping the FANCA binding region on FANCG. (C) Mapping the FANCG binding region on FANCF. (D) Mapping the FANCF binding region on FANCG. Yeast strain PJ69-4A was cotransformed with the expression constructs indicated. Constructs activating the reporter genes above background levels are shaded. The dotted lines represent internal deletions. (++), (+), and (-) indicate good, weak, and absent growth, respectively. β-Galactosidase activity units in the liquid ONPG assay were calculated as 1000 × OD420 divided by the product of the volume (milliliter) × time (minutes) × OD600. The values shown indicate the mean and SE for 4 assays, each done in triplicate. Nd indicates that no β-galactosidase assay was performed because of nonactivation of the ADE2 and HIS3 reporters. NLS indicates nuclear localization signal; LZ, leucine zipper.
The central portion of FANCE encompassing amino acids 150 to 371 was sufficient for interaction with FANCC, although the use of constructs encoding additional amino acids on either side activated the reporters more strongly (Figure 2A). None of the amino- or carboxyl-terminal–truncated FANCC polypeptides tested were capable of interaction with FANCE. The AD-FANCCLeu554Pro mutant also failed to interact with BD-FANCE (Figure 2B). This result was confirmed by assaying cotransformants of the BDFANCCLeu554Pro/AD-FANCE combination (data not shown).
Mapping binding sites for the interactions of FANCE with FANCC and FANCD2. (A) Mapping the FANCC binding region on FANCE. (B) Mapping the FANCE binding region on FANCC. (C) Mapping the FANCE binding region on FANCD2. Yeast strain PJ69-4A was cotransformed with the expression constructs indicated. Constructs activating the reporter genes above background levels are shaded. (++), (+), and (-) indicate good, weak, and absent growth, respectively. β-Galactosidase activity units were calculated as in Figure 1. The values shown indicate the mean and SE for 4 assays, each done in triplicate. Nd indicates that no β-galactosidase assay was performed because of nonactivation of the ADE2 and HIS3 reporters. NLS indicates nuclear localization signal; Leu554Pro, missense mutation in FANCC polypeptide.
Mapping binding sites for the interactions of FANCE with FANCC and FANCD2. (A) Mapping the FANCC binding region on FANCE. (B) Mapping the FANCE binding region on FANCC. (C) Mapping the FANCE binding region on FANCD2. Yeast strain PJ69-4A was cotransformed with the expression constructs indicated. Constructs activating the reporter genes above background levels are shaded. (++), (+), and (-) indicate good, weak, and absent growth, respectively. β-Galactosidase activity units were calculated as in Figure 1. The values shown indicate the mean and SE for 4 assays, each done in triplicate. Nd indicates that no β-galactosidase assay was performed because of nonactivation of the ADE2 and HIS3 reporters. NLS indicates nuclear localization signal; Leu554Pro, missense mutation in FANCC polypeptide.
The amino-terminal 291 amino acids of FANCD2 were sufficient for interaction with FANCE, although this region alone activated the reporters less strongly than larger segments of the amino-terminus. Further truncation resulted in loss of activation (Figure 2C). The more carboxyl-terminal regions of FANCD2 were not eliminated as possible secondary sites of interaction.
Yeast 3-hybrid system
As FANCG was identified as able to bind more than one other FA protein in the 2-hybrid system, the yeast 3-hybrid system35 was used to investigate the ability of FANCG to act as a molecular “bridge” in mediating interaction between the FANC proteins. The BrAG construct encodes a constitutively expressed BD-FANCA together with an HA-tagged FANCG under the control of the methionine repressible Met25 promoter.35,42 BrAG autoactivated all 3 reporters only when HA-FANCG was coexpressed in the absence of methionine. As FANCA and FANCG have been shown to interact, and BD-FANCG to autoactivate, this autoactivation was not surprising as the DNA-binding domain would be brought into contact with FANCG. However, elevated β-galactosidase activity on conditional expression of HA-FANCG in the presence of AD-FANCA revealed the ability of HA-FANCG to mediate an interaction between monomers of BD-FANCA and AD-FANCA (Figure 3). The presence of AD-FANCF reduced the activation of the BrAG construct, just as AD-FANCF reduced the autoactivation of BD-FANCG in the 2-hybrid assay. BrAG paired with an AD-FANCG construct greatly elevated β-galactosidase activity in the presence of methionine (absence of HA-FANCG), as this was essentially a duplication of the FANCA/FANCG 2-hybrid result. Introduction of the HA-FANCG in the absence of methionine reduced this level of activation somewhat, as HA-FANCG was present to compete with AD-FANCG for binding to BD-FANCA (Figure 3). The 3-hybrid β-galactosidase values are not comparable to those seen in the 2-hybrid assay as the LacZ gene is under different GAL4 responsive promoters in the different strains, AH109/Y187 versus PJ69-4A. However, it is the relative increase in reporter activation seen within the 3-hybrid system that demonstrates a previously unknown ability of FANCG to mediate the multimerization of FANCA.
Mediation of BD-FANCA interactions by conditionally expressed HA-FANCG in the 3-hybrid system. The BrAG construct was paired with constructs expressing the indicated AD-fusion proteins by mating haploid yeast strains. HA-FANCG is expressed from the BrAG construct in the absence of methionine (□) but repressed by the presence of 4 mM methionine in the media (▦). The BD-FANCA fusion protein is constitutively expressed, as are all AD fusions. Activation of the LacZ reporter gene was measured in AH109/Y187 diploids by liquid β-galactosidase assay. β-Galactosidase activity units were calculated as in Figure 1. The results shown indicate the mean and SE for 3 (+Met) or 4 (-Met) assays, each done in triplicate.
Mediation of BD-FANCA interactions by conditionally expressed HA-FANCG in the 3-hybrid system. The BrAG construct was paired with constructs expressing the indicated AD-fusion proteins by mating haploid yeast strains. HA-FANCG is expressed from the BrAG construct in the absence of methionine (□) but repressed by the presence of 4 mM methionine in the media (▦). The BD-FANCA fusion protein is constitutively expressed, as are all AD fusions. Activation of the LacZ reporter gene was measured in AH109/Y187 diploids by liquid β-galactosidase assay. β-Galactosidase activity units were calculated as in Figure 1. The results shown indicate the mean and SE for 3 (+Met) or 4 (-Met) assays, each done in triplicate.
A second 3-hybrid construct, BrFG, featured constitutively expressed BD-FANCF paired with the methionine-repressible HA-FANCG. This construct also autoactivated slightly, possibly because the previously demonstrated FANCF/FANCG interaction is expected to create an indirect BD-FANCG combination. Analysis of β-galactosidase reporter activity revealed elevated levels for cotransformants of BrFG and AD-FANCA (Figure 4), implying the ability of HA-FANCG to mediate an interaction between BDFANCF and AD-FANCA. BrFG paired with an AD-FANCG construct elevated β-galactosidase activity in the absence of HA-FANCG, a duplication of the BD-FANCF/AD-FANCG 2-hybrid result. HA-FANCG expression in the absence of methionine reduced this activation as HA-FANCG was present to compete with AD-FANCG for binding to BD-FANCF. Altogether, 3-hybrid experiments with the BrFG construct both affirmed the direct binding of FANCF with FANCG seen in the 2-hybrid system, and demonstrated the ability of FANCG to mediate an interaction between FANCA and FANCF.
Mediation of BD-FANCF interactions by conditionally expressed HA-FANCG in the 3-hybrid system. The BrFG construct was paired with constructs expressing the indicated AD-fusion proteins by mating haploid yeast strains. HA-FANCG is expressed from the BrFG construct in the absence of methionine (□) but repressed by the presence of 4 mM methionine in the media (▦). The BD-FANCF fusion protein is constitutively expressed, as are all AD fusions. Activation of the LacZ reporter gene was measured in AH109/Y187 diploids by liquid β-galactosidase assay. β-Galactosidase activity units were calculated as in Figure 1. The results shown indicate the mean and SE for 3 (+Met) or 4 (-Met) assays, each done in triplicate.
Mediation of BD-FANCF interactions by conditionally expressed HA-FANCG in the 3-hybrid system. The BrFG construct was paired with constructs expressing the indicated AD-fusion proteins by mating haploid yeast strains. HA-FANCG is expressed from the BrFG construct in the absence of methionine (□) but repressed by the presence of 4 mM methionine in the media (▦). The BD-FANCF fusion protein is constitutively expressed, as are all AD fusions. Activation of the LacZ reporter gene was measured in AH109/Y187 diploids by liquid β-galactosidase assay. β-Galactosidase activity units were calculated as in Figure 1. The results shown indicate the mean and SE for 3 (+Met) or 4 (-Met) assays, each done in triplicate.
Discussion
The PJ69-4A strain of S cerevisiae was designed with 3 reporter genes under the control of different GAL4-responsive promoters to decrease the incidence of false positives while remaining a highly sensitive method of detecting protein-protein interaction.36 Pairwise analysis of FANC proteins in this version of the 2-hybrid system revealed 4 interactions (FANCA/FANCG, FANCF/FANCG, FANCC/FANCE, and FANCD2/FANCE). Comparison of reporter activation between these pairings of FANC proteins may not be a reliable indicator of relative interaction strength, as differences in the expression level and toxicity of proteins will affect reporter activation.43 However, all interactions were adequate to permit the use of deletion studies to map contact points involved in binding.
Identification of the NLS region of the FANCA protein as a binding site for FANCG concurred with reports from site-directed mutagenesis studies that amino acids within the NLS are important for FANCG binding.29,30 Failure of the amino-terminal–truncated FANCA lacking only amino acids 1 to 35 also agreed with an earlier study demonstrating that an epitope-tagged FANCA missing only the most amino-terminal NLS region failed to immunoprecipitate with FANCG.29 The ability of a carboxyl-terminal–truncated FANCG to sustain interaction with FANCA was consistent with the reported immunoprecipitation of FANCG amino acids 1 to 428 with full-length FANCA.26 Retention of the FANCA/FANCG interaction in FA cell lines in which the larger FA nuclear complex is absent, together with failure of an NLS mutant of FANCA to immunoprecipitate with FANCC, suggests this interaction may be an early requirement for complex formation.22,44 Direct binding of FANCG to the NLS of FANCA may explain why FANCG has been shown to promote the nuclear accumulation of FANCA.21,29
The detrimental effect of the Leu71Pro mutation on FANCG protein interactions is not surprising, given it occurs in a region described as having similarity to a leucine zipper,37 and substitution of a proline could be expected to disrupt α-helical secondary structure. In addition to the disruption of FANCF binding, reduced activation of all 3 reporters suggested that the FANCG/FANCA interaction could also be partially affected. Reduced activation of the nutritional reporters is particularly telling, as they exert strong selective pressure for interaction. The ability of the Gly546Arg mutant to bind both FANCA and FANCF is interesting, given that, when previously expressed in FA-G cells, this mutant cDNA was unable to correct mitomycin C (MMC) sensitivity.38 It is difficult to assess the relevance of the 2-fold decrease in LacZ reporter activation for AD-FANCGG546R/BD-FANCA cotransformants, particularly in the absence of effects on the nutritional reporters, as differences in the expression level or toxicity of mutants could affect reporter activation. One study reported that in vivo, the base change predicted to cause this glycine to arginine substitution actually results in an in-frame deletion,37 presumably due to altered RNA processing. Nevertheless, the inability of a cDNA-based expression construct to correct MMC sensitivity suggests that FANCGGly546Arg is deficient in some aspect of FANCG function required to maintain genomic integrity.
The 3-hybrid studies suggested a role for FANCG in mediating an interaction between FANCA and FANCF. FANCG may form a physical bridge complexing FANCA and FANCF together, or it could modify one or both of FANCA and FANCF in a manner required for the promotion of a direct FANCA/FANCF interaction. As FANCG was demonstrated to directly interact with both FANCA and FANCF in the 2-hybrid system, the physical bridge concept is a strong possibility. This likelihood is further strengthened by the ability of FANCA to immunoprecipitate with FANCF from normal cells, but not from a patient-derived cell line homozygous for a mutation resulting in the truncation of the carboxyl-terminus of FANCF.23 This mutation is in the same region of FANCF that was found in this study to interact with FANCG. FANCF has also been shown to stabilize the interaction between FANCG and FANCA.23
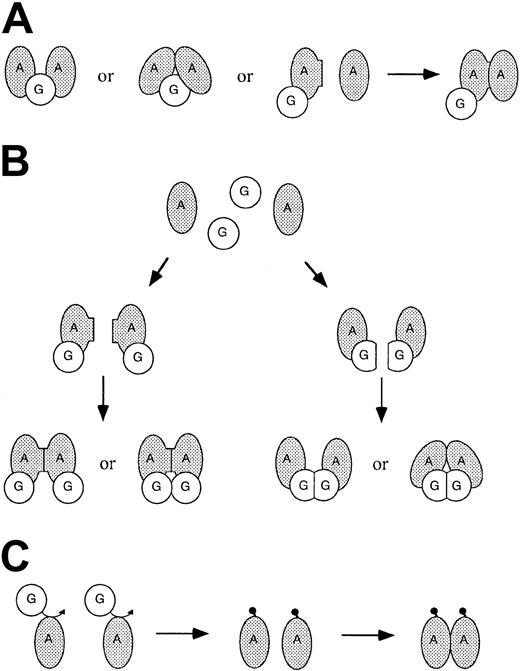
The 3-hybrid experiments also demonstrated that FANCG is able to mediate interaction between FANCA monomers. The stoichiometry of the FANCG protein in this interaction is unclear. Some studies have suggested that FANCA may be able to bind to noncontiguous regions of the FANCG protein,30,32 which would enable a single FANCG to interact with more than one monomer of FANCA (Figure 5A). Alternatively, multiple FANCG molecules could be involved. Although no evidence for FANCG dimerization was observed in our 2-hybrid experiments, FANCA binding could potentially alter the structure of FANCG to favor FANCG self-interaction (Figure 5B). Alternatively, FANCG could modify one or more FANCA molecules to allow direct FANCA dimerization (Figure 5C). Whether the binding of FANCG with FANCA always involves more than one monomer of FANCA, or whether the stoichiometry of the interaction varies under different circumstances, remains to be determined. In any event, the ability of FANCG to bind FANCA, in what appears to be an early event essential for complex formation,22,29 as well as to mediate interaction of FANCA with FANCF, suggests a key role for FANCG in the architecture of the FA complex.
Models for FANCG-mediated interaction between monomers of FANCA. (A) A single molecule of FANCG mediates interaction between FANCA monomers. This interaction may involve physical bridging, with or without contact between the FANCA monomers. Models involving direct FANCA/FANCA contact could include the stabilization of a preexisting weak or transient interaction between FANCA monomers not detectable in the 2-hybrid system. Alternatively, FANCG may bind to only one FANCA monomer, causing structural change that allows for interaction with a second monomer. (B) Multiple molecules of FANCG are involved in the interaction. Simple examples of 2:2 FANCA/FANCG stoichiometry are illustrated. Such interactions could involve FANCA promoting or stabilizing FANCG/FANCG interactions not detectable in the 2-hybrid system. (C) FANCG modifies one or more molecules of FANCA, allowing for multimerization of FANCA without direct involvement of FANCG in the complex. Although this is possible, the strong direct interaction of FANCA and FANCG seen in the 2-hybrid system favors models involving FANCG complexed with FANCA.
Models for FANCG-mediated interaction between monomers of FANCA. (A) A single molecule of FANCG mediates interaction between FANCA monomers. This interaction may involve physical bridging, with or without contact between the FANCA monomers. Models involving direct FANCA/FANCA contact could include the stabilization of a preexisting weak or transient interaction between FANCA monomers not detectable in the 2-hybrid system. Alternatively, FANCG may bind to only one FANCA monomer, causing structural change that allows for interaction with a second monomer. (B) Multiple molecules of FANCG are involved in the interaction. Simple examples of 2:2 FANCA/FANCG stoichiometry are illustrated. Such interactions could involve FANCA promoting or stabilizing FANCG/FANCG interactions not detectable in the 2-hybrid system. (C) FANCG modifies one or more molecules of FANCA, allowing for multimerization of FANCA without direct involvement of FANCG in the complex. Although this is possible, the strong direct interaction of FANCA and FANCG seen in the 2-hybrid system favors models involving FANCG complexed with FANCA.
The failure of the FANCCLeu554Pro mutant to interact with FANCE is interesting, given that overexpression of this mutant in 293 cells has been shown to induce MMC sensitivity.45 One possible explanation is that the failure of FANCCLeu554Pro to bind FANCE may result in the sequestering of other FANCC-binding proteins away from FANCE and possibly the FA protein complex. FANCCLeu554Pro fails to localize to the nucleus in HeLa cells,46 and recently FANCE was found to promote the nuclear accumulation of wild-type FANCC but not FANCCLeu554Pro.24 Thus, FANCE binding may be required for nuclear import or retention of FANCC, consistent with the failure of FANCC to form a nuclear complex in FANCE cell lines.28 The smallest region of FANCE mapped as sufficient for FANCC binding included the putative NLS. However, unlike the interaction of FANCG with a minimal NLS region of FANCA, a smaller construct still fully encompassing the putative NLS of FANCE was not sufficient to interact with FANCC. Thus, either the NLS region does not interact with FANCC, or, although this construct was expressed in yeast, it could not assume the correct conformation for FANCC binding. The failure of any of a series of truncated FANCC polypeptides spanning the entire FANCC protein to interact with FANCE suggests that noncontiguous regions of FANCC may be involved in this interaction.
The direct interaction of FANCE with FANCD2 provides a possible link between the FA nuclear protein complex and the downstream activity of the FANCD2 protein. The amino-terminal fragments of FANCD2 able to maintain FANCE binding are known to contain biologically significant residues. These residues include the site of monoubiquination (Lys561)25 as well as the site of ionizing radiation-inducible ataxia telangiectasia mutated (ATM)–dependent phosphorylation (Ser222),47 which is located in the most truncated polypeptide able to sustain interaction. However, these posttranslational protein modifications may not occur in yeast, and the possibility of a role for these residues in the FANCD2/FANCE interaction requires further investigation. It also remains to be determined whether FANCE is capable of binding both FANCC and FANCD2 simultaneously, or if the interactions occur sequentially as part of a cascade, as FANCE is the only FANC protein demonstrated to immunoprecipitate with FANCD2.24 Elucidation of the FANCD2 binding region of FANCE may provide further insight into this question. The recent identification of BRCA2 mutations in FA-D1 cell lines11 provides another interesting candidate for FANC protein interaction studies, as FA-D1 cells have intact FA nuclear complex formation and FANCD2 monoubiquination pathways.11,23,25
In summary, this study mapped the binding sites involved in 3 protein-protein interactions between members of the FA nuclear protein complex (FANCA/FANCG, FANCF/FANCG, and FANCC/FANCE). A fourth demonstrated interaction, that of FANCE with the amino-terminal region of FANCD2, provides a possible link between the FANCA/C/E/F/G complex and the downstream action of the FANCD2 protein. The ability of FANCG to mediate interaction between monomers of FANCA as well as between FANCA and FANCF suggests that the FANCG protein may play an important structural role in the architecture of the FA protein complex with involvement at multiple stages of complex assembly. A model for the architecture of the nuclear FA complex based on these observations is presented in Figure 6. Taken together, these results demonstrate an increased level of complexity in the web of interactions among proteins underlying the molecular pathogenesis of Fanconi anemia.
A model for the architecture of the nuclear FA protein complex. Letters indicate the FANC proteins investigated in this study. FANCG can directly bind to both FANCA and FANCF as well as mediate an interaction between them. The FANCG-mediated multimerization of FANCA is presented in one of the simpler possible forms. This study demonstrated direct interaction of FANCC only with FANCE, although previous immunoprecipitation experiments have suggested that both FANCC and FANCE are part of the nuclear FA complex (dotted lines). FANCE is currently the only FANC protein demonstrated to interact either directly or indirectly with FANCD2. Thus, in the absence of evidence otherwise, FANCE is shown as exiting the core FA complex to bind to FANCD2. With the exception of the FANCG-mediated FANCA multimerization demonstrated in this study, all interactions among the FANC proteins are presented as occurring with 1:1 stoichiometry for simplicity.
A model for the architecture of the nuclear FA protein complex. Letters indicate the FANC proteins investigated in this study. FANCG can directly bind to both FANCA and FANCF as well as mediate an interaction between them. The FANCG-mediated multimerization of FANCA is presented in one of the simpler possible forms. This study demonstrated direct interaction of FANCC only with FANCE, although previous immunoprecipitation experiments have suggested that both FANCC and FANCE are part of the nuclear FA complex (dotted lines). FANCE is currently the only FANC protein demonstrated to interact either directly or indirectly with FANCD2. Thus, in the absence of evidence otherwise, FANCE is shown as exiting the core FA complex to bind to FANCD2. With the exception of the FANCG-mediated FANCA multimerization demonstrated in this study, all interactions among the FANC proteins are presented as occurring with 1:1 stoichiometry for simplicity.
Prepublished online as Blood First Edition Paper, March 20, 2003; DOI 10.1182/blood-2002-11-3517.
Supported by the Canadian Institutes of Health Research (CIHR), the Hospital for Sick Children (HSC) Foundation, and the Lombard Chair in Pediatrics Research (M.B.). S.M.G. held a Natural Sciences and Engineering Research Council of Canada (NSERC) Postgraduate Scholarship and a Doctoral Research Award from Canadian Institutes of Health (CIHR).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr M. Grompe for the kind gift of the FANCD2 cDNA and Dr J. Lightfoot and Dr J. Rommens for helpful discussions and comments on the manuscript.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal