Prostaglandin E2 (PGE2) is the predominant eicosanoid product released by macrophages at the site of inflammation. Binding of PGE2 to its cognate 7 transmembrane-spanning G protein–coupled receptors (GPCRs) activates signaling pathways, leading to the synthesis of the Fos transcription factor. Because the Ste20 serine/threonine protein kinase (S/TPK) is a critical signal transducer for the G protein–coupled pheromone receptor in Saccharomyces cerevisiae, we postulated that the PGE2 GPCRs may activate one of the Ste20 mammalian orthologs. We demonstrate here that the catalytic activity of a hematopoietic cell–restricted, Ste20-related S/TPK, HPK1, is positively regulated by exposure to physiological concentrations of PGE2. Furthermore, ectopic expression studies implicated HPK1 as a negative regulator of PGE2-induced transcription of the fos gene. Our data suggest that PGE2-induced activation of HPK1 may represent a novel negative regulatory pathway capable of modulating PGE2-mediated gene transcription.
Introduction
Signals generated by the prostaglandin E2 receptors (PGE2Rs) are thought to be propagated via the classical, well-characterized G-protein signal transduction pathways, utilizing phospholipase B and adenylyl cyclase as effector molecules.1 We postulate that the PGE2Rs may activate other pathways, perhaps, engaging one of the Ste20 family members, and utilize it to transmit or regulate the PGE2-induced fos gene transcription. Data presented here support our contention that exposure to PGE2 activates HPK1 kinase activity, which, in turn, negatively regulates PGE2-induced fos gene transcription.
Study design
Cells, antibodies, and other reagents
All cell lines used in this study are available from ATCC (Manassas, VA). The antihemagglutinin (HA) epitope antibody (12CA5) was purified from hybridoma supernatants in our laboratory. The anti-HPK1 polyclonal antibody no. 7,2 a generous gift from Dr F. Kiefer (Max-Planck Institute for Physiological and Clinical Research, Bad Nauheim, Germany) was used in Western blot analyses. Affinity-purified rabbit polyclonal antibody against the amino acid residues 341 to 366 of human HPK1 used for immunoprecipitations was custom produced for our laboratory by Cocalico Biologicals (Reamstown, PA). γ-[32P]adenosine triphosphate (γ-[32P]ATP) was obtained from Perkin Elmer Life Science (Boston, MA). Histone H2A, the exogenous substrate used in in vitro kinase reactions, was purchased from Roche Applied Science (Indianapolis, IN). PGE2 was purchased from Calbiochem-Novabiochem (San Diego, CA).
Molecular constructs
The pcDNA3 vector carrying mouse cDNAs that encode the HA-tagged wild-type and kinase-inactive (K46E) mutant of murine HPK1 was a generous gift from Dr F. Kiefer. A construct encoding the firefly luciferase reporter gene under the control of human fospromoter (Hu-fos-Luc) was described.3
Transient transfections and in vitro kinase assays
Jurkat T cells (1.5 × 107) were transfected as previously described.4 Whole cell lysates derived from resting or stimulated transfectants were subjected to immunoprecipitations by the indicated antibodies and were subjected to in vitro kinase reactions as described.2 Anti-HPK1 polyclonal antibody no. 7 2 was used in immunoblot assays to confirm the amounts of immunoprecipitated HPK1.
Luciferase assays
Jurkat cells were cotransfected with 2.5 μg of the Hu-fos-Luc reporter construct and 100 ng of pNullRenilla luciferase reporter construct along with 10 μg of the indicated experimental constructs. Dual luciferase assays were performed according to the manufacturer's direction (Promega, Madison, WI), and the relative luciferase activity was measured by the Monolight 2010 luminometer (Analytical Luminescence Laboratory, Ann Arbor, MI).
Results and discussion
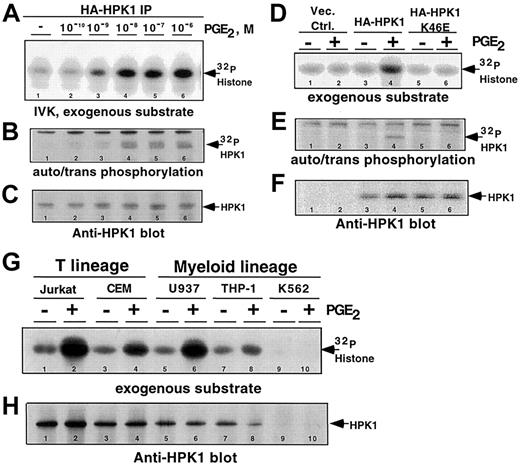
We hypothesized that the binding of PGE2 to its cognate receptors would generate signals that activate the catalytic activities of mammalian Ste20 family members. As observed in many cell types,5 6 our in vitro kinase activity screen of Ste20 orthologs revealed that most are constitutively active, and are not responsive to PGE2 stimulation, in Jurkat T cells (data not shown). However, HPK1, a hematopoietic cell–specific Ste20 family member, is robustly activated upon in vivo exposure to PGE2, in a concentration-dependent manner (Figure 1A-B). Exposure to 1 nM PGE2induced HPK1 kinase activity. Maximum levels of kinase activity were achieved when cells were stimulated with at least 10 nM PGE2 (Figure 1A-B, lanes 4-6). The 2 lowest PGE2 concentrations that activated HPK1, 1 and 10 nM, are at physiological levels found at the sites of inflammation.
It is possible that the kinase activity detected in our assay was due to a PGE2-responsive kinase that coprecipitated with HPK1. To address this concern, we transfected either the wild-type or a kinase-defective HA-HPK1 K46E mutant into Jurkat cells and stimulated the transfectants with 10 nM PGE2. The lack of kinase activity from the HA-HPK1 K46E mutant (Figure 1D-E, lanes 5-6) strengthened our conclusion that the observed kinase activities in Figure 1A-B were catalyzed by HPK1.
Furthermore, using anti-HPK1 antibody, we demonstrated that endogenous HPK1 responded to PGE2 stimulation in all hematopoietic cell lines tested (Figure 1G, lanes 1-8). Kinase activity was not detected in the anti-HPK1 immunoprecipitates from the K562 cells, a chronic myelogenous line that does not express HPK1 (Figure 1G, lanes 9-10). Western blot analysis using anti-HPK1 antibody confirmed that comparable amounts of HPK1 were present in all kinase reactions (Figure1C,F,H). Taken together, these observations established PGE2 as a potent activator of HPK1 kinase activity in hematopoietic cells. This is the first example of G protein–coupled receptor (GPCR) regulation of the catalytic activity of a Ste20 ortholog.
Physiological concentrations of PGE2-induced HPK1 kinase activity.
HA-HPK1 was immunoprecipitated from lysates derived from Jurkat T cells transfected with HA-HPK1. Cells were exposed to varying concentrations of PGE2 for 2 minutes at 37°C prior to lysis and immunoprecipitation. In vitro immune complex HPK1 kinase assays (IVK) were performed in the presence of exogenous substrate, 5 μg histone H2A. (A) 32P-incorporated histone H2A. (B)32P-incorporated auto-transphosphorylated HA-HPK1. (C) Western blot of immunoprecipitated HPK1 from panel B, using the anti-HPK1 no. 7 antibody. (D) In vitro immune complex HPK1 kinase assays were performed on lysates of PGE2-stimulated (10 nM) transfectants expressing wild-type or a catalytically inactive mutant (K46E) of HPK1. The autoradiographic bands depict32P-incorporated histone H2A. (E)32P-incorporated autophosphorylated/transphosphorylated wild-type and (K46E) mutant HPK1. (F) Western blot of immunoprecipitated HPK1 from panel E, using the anti-HPK1 no. 7 antibody. (G) In vitro immune complex kinase assays performed with endogenous HPK1 immunoprecipitated from nontreated cells or from cells treated with 10 nM PGE2. (H) Western blot of immunoprecipitated HPK1 from panel G, using the anti-HPK1 no. 7 antibody. Lane numbers are indicated at the bottom of all lanes. Data in all figures are representative of at least 3 independent experiments.
Physiological concentrations of PGE2-induced HPK1 kinase activity.
HA-HPK1 was immunoprecipitated from lysates derived from Jurkat T cells transfected with HA-HPK1. Cells were exposed to varying concentrations of PGE2 for 2 minutes at 37°C prior to lysis and immunoprecipitation. In vitro immune complex HPK1 kinase assays (IVK) were performed in the presence of exogenous substrate, 5 μg histone H2A. (A) 32P-incorporated histone H2A. (B)32P-incorporated auto-transphosphorylated HA-HPK1. (C) Western blot of immunoprecipitated HPK1 from panel B, using the anti-HPK1 no. 7 antibody. (D) In vitro immune complex HPK1 kinase assays were performed on lysates of PGE2-stimulated (10 nM) transfectants expressing wild-type or a catalytically inactive mutant (K46E) of HPK1. The autoradiographic bands depict32P-incorporated histone H2A. (E)32P-incorporated autophosphorylated/transphosphorylated wild-type and (K46E) mutant HPK1. (F) Western blot of immunoprecipitated HPK1 from panel E, using the anti-HPK1 no. 7 antibody. (G) In vitro immune complex kinase assays performed with endogenous HPK1 immunoprecipitated from nontreated cells or from cells treated with 10 nM PGE2. (H) Western blot of immunoprecipitated HPK1 from panel G, using the anti-HPK1 no. 7 antibody. Lane numbers are indicated at the bottom of all lanes. Data in all figures are representative of at least 3 independent experiments.
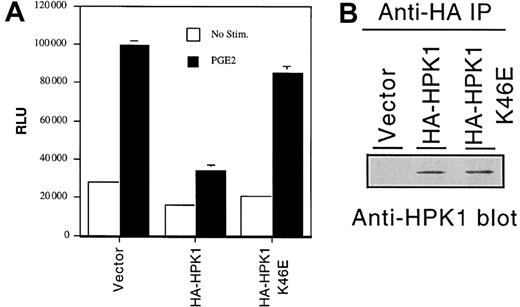
PGE2 stimulation induces c-fos gene transcription in many cell types7,8 via a poorly characterized cyclic adenosine monophosphate (cAMP)–independent mechanism.9 Because Ste20 orthologs have been implicated as critical signaling molecules in many receptor systems,10 we postulated that HPK1 might function as a facilitator or a regulator of the PGE2-induced Fos activation signal. To assess the effect of HPK1 on fos gene transcription, we cotransfected either a wild-type HPK1 or the catalytically inactive K46E mutant construct, along with afos promoter–regulated luciferase reporter construct, into the Jurkat T-cell line. Consistent with findings observed in other cell types, PGE2 treatment increased fos promoter activity by approximately 3-fold in the Jurkat T-cell line (Figure2A). Ectopic expression of wild-type HA-HPK1 inhibited the PGE2-induced fos promoter activity by approximately 60% to 70%. The K46E mutant failed to inhibit fos transcription, suggesting that the HPK1 kinase activity is required for the inhibition of fostranscription. This conclusion is consistent with a recent finding that, despite its ability to activate Jun N-terminal kinase (JNK) and c-Jun, HPK1 functions as a negative regulator of T-cell antigen receptor (TCR)– and B-cell antigen receptor (BCR)–mediated AP-1–dependent gene transcription.11 However, one must be cognizant of the possibility that the observed inhibition offos transcription may be due to kinase activity–dependent sequestration of critical signal transduction components by the overexpressed HPK1. It has recently been demonstrated that sequestration of SLP-76 family member(s) represents a possible mechanism underlying the HPK1-mediated inhibition of TCR and BCR signals.12 A better understanding of HPK1's role in biologic processes awaits the development of an HPK1-deficient animal model.
HPK1 inhibits PGE2-induced
fos promoter activity. (A) Fospromoter–regulated relative luciferase activity from cells transfected with empty vector or HPK1 constructs. Transfectants were left untreated (■) or were stimulated with 10 nM PGE2 (▪) for 6 hours prior to cell harvest. Duplicate transfectants were lysed, and the lysates were tested for luciferase activity. Relative light unit (RLU) values were normalized with the Renilla luciferase values. Error bars indicate standard deviations. This histogram depicts an experiment that represents the trend observed in 4 of 5 experiments. (B) Western blot analysis revealing the expression levels of the transfected HA-HPK1.
HPK1 inhibits PGE2-induced
fos promoter activity. (A) Fospromoter–regulated relative luciferase activity from cells transfected with empty vector or HPK1 constructs. Transfectants were left untreated (■) or were stimulated with 10 nM PGE2 (▪) for 6 hours prior to cell harvest. Duplicate transfectants were lysed, and the lysates were tested for luciferase activity. Relative light unit (RLU) values were normalized with the Renilla luciferase values. Error bars indicate standard deviations. This histogram depicts an experiment that represents the trend observed in 4 of 5 experiments. (B) Western blot analysis revealing the expression levels of the transfected HA-HPK1.
The mechanism by which PGE2Rs couple activation signals to HPK1 is not known. While 4 PGE2R genes have been identified (EP1 to EP4), the Gs-linked EP4 receptor is the predominant PGE2R expressed in Jurkat T cells.13 Reverse transcriptase–polymerase chain reaction (RT-PCR) analysis revealed that all cell lines used in this study also express a high level of EP4 receptor (data not shown). The EP4 receptor links activation signals to Gsα, which, in turn, activates adenylyl cyclase, leading to elevation of cAMP concentrations. Although it is enticing to implicate cAMP as the second messenger that activates HPK1, administration of cell-permeable forms of cAMP (N6, O2′-dibutyryl-cAMP, 8-bromo-cAMP, and 8-4-chlorophenylthio cAMP) to Jurkat cells failed to activate HPK1 (data not shown). These cAMP agents are biologically active as shown by their abilities to activate cAMP-dependent serine/threonine protein kinase A (PKA; data not shown). The inability of exogenous cAMP to activate HPK1 revealed that both the negative regulatory signal and the positive transcriptional signal9are not downstream of the cAMP signaling pathway. Further characterization of this pathway may reveal an insight into a novel mechanism for negative regulation.
We are grateful to Dr Joanne Pratt (Olin College, Needham, MA) for thorough reading of the manuscript. We thank Dr J. Pratt, Dr S. Baksh, Dr S. Pyarajan, and Dr Y.-J. Jin for thoughtful discussions. We are grateful to Dr F. Kiefer for providing the reagents.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/ blood-2002-07-2316.
Supported by National Institutes of Health, National Cancer Institute grant no. CA70758 (S.J.B.). S.S. is supported by a grant from the Association for International Cancer Research, St Andrews, Scotland.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sansana Sawasdikosol, New York University Medical Center, Skirball Institute of Biomolecular Medicine, Department of Pathology, 540 1st Ave, 5th floor, Laboratory 1, New York, NY 10016; e-mail: sawasdik@saturn.med.nyu.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal