Congenital afibrinogenemia is a rare autosomal recessive disorder characterized by the complete absence of detectable fibrinogen. We previously identified the first causative mutations for this disease, homozygous deletions of approximately 11 kb of the fibrinogen alpha chain gene (FGA). Subsequent analyses revealed that most afibrinogenemia alleles are truncating mutations of FGA, although mutations in all 3 fibrinogen genes, FGG, FGA andFGB have been identified. In this study, we performed the first prenatal diagnosis for afibrinogenemia. The causative mutation in a Palestinian family was a novel nonsense mutation in theFGB gene, Trp467Stop (W467X). Expression of the Trp467Stop mutant FGB cDNA in combination with wild-typeFGA and FGG cDNAs showed that fibrinogen molecules containing the mutant beta chain are not secreted into the media. The fetus was found to be heterozygous for the Trp467Stop mutation by direct sequencing and by linkage analysis, a result that was confirmed in the newborn by intermediate fibrinogen levels.
Introduction
Congenital afibrinogenemia (Mendelian Inheritance in Man, no. 202400; http://www.ncbi.nlm.nih.gov/omim), characterized by a complete absence of detectable fibrinogen, was originally described in 1920.1 To date some 150 families with this disorder have been reported,2 with approximately 50% of cases present in consanguineous families.3 Although functional assays of clot formation are infinitely prolonged in affected individuals, the coagulation defect is surprisingly no more severe than in severe hemophilias A and B.4 5 Umbilical cord hemorrhage is often the first sign of the disorder; gum bleeding, epistaxis, menorrhagia, gastrointestinal bleeding, and hemarthrosis occur with varying intensity, and spontaneous intracerebral bleeding and splenic rupture can occur throughout life.
Fibrinogen is synthesized in the hepatocyte from 3 homologous polypeptide chains, Aα, Bβ, and γ, which assemble to form the hexameric structure (AαBβγ)2. The 3 genes coding for fibrinogen gamma (FGG), alpha (FGA), and beta (FGB) are clustered in a region of approximately 50 kb on chromosome 4q28-q31.6 We previously identified the first causative mutations for this disorder.7 The genetic defect in a nonconsanguineous Swiss family was an apparently recurrent deletion of approximately 11 kb of DNA, which eliminates most of theFGA gene and so leads to a complete absence of functional fibrinogen. Since our identification of the disease locus, numerous causative mutations have been characterized in FGA (most of the cases)8 9 but also in FGG andFGB, allowing a precise molecular diagnosis for the patients, as well as prenatal diagnosis for the families concerned. In this study, the first prenatal diagnosis for afibrinogenemia was performed for a Palestinian family with 2 affected daughters.
Study design
Description of family
Informed consent was obtained from the adults participating in the study. The parents were a healthy consanguineous Palestinian couple with no history of bleeding tendency. Their first conception was a miscarriage during the first trimester. The second and third conceptions produced 2 daughters who were born at term with birth weights of 2.9 and 3 kg, respectively. The bleeding tendency in the 2 daughters was noted during midinfancy (between 4 and 5 months of age) in the form of serious intracranial bleeding after nonsignificant trauma. Fibrinogen levels were undetectably low (< 50 mg/dL), establishing the diagnosis of congenital afibrinogenemia. Following diagnosis, treatment consisted of infusions of fresh frozen plasma (FFP) following any significant bleeding, approximately once every 5-6 months. No infusion was required for the last 2 years because of increased parental awareness of accident and trauma prevention. Fibrinogen levels were determined for the parents and were found to be 170 and 150 mg/dL for the mother and the father, respectively.
Mutation screening in family members
Microsatellite analysis
The FGA intron 3 tetranucleotide repeat (FGAi3) was analyzed by PCR amplification with oligonucleotides FGA PCR2.1 (5′CCATAGGTTTTGAACTCACAG3′) and FGA PCR2.2 (5′CTTCTCAGATCCTCTGACAC3′) followed by denaturing polyacrylamide-urea gel electrophoresis according to standard procedures.
Amniocentesis
Amniocentesis was performed by the usual procedure at 16 weeks of gestation. Amniotic fluid cells were cultured and, after achieving adequate growth, DNA was prepared and analyzed for the mutation present in the affected daughters.
Expression and analysis of the FGB Trp467Stop mutation in COS-7 cells
The 3 human fibrinogen cDNAs, FGA, FGB,and FGG were obtained by reverse transcription–polymerase chain reaction (RT-PCR) on HepG2 cells and cloned into the pcDNA3.1/V5-His TOPO mammalian expression vector (Invitrogen, Groningen, The Netherlands). The Trp467Stop mutation was inserted in the FGB construct using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). Transient transfections of COS-7 cells were performed using LipofectAMINE Plus reagent (Invitrogen) according to the manufacturer's instructions. Transfections were performed in 100-mm dishes with 12 μg pcDNA3.1/V5-His TOPO vector (negative control) or with equal amounts (4 μg) of all expression vectors. In the case of the heterozygous mutant analysis, 2 μg of each Bβ construct (wild type and mutant) was used. Cells were washed with phosphate-buffered saline (PBS) 3 hours after transfection and incubated for 18 hours in media without serum but with protease inhibitors (Complete Mini, Roche, Basel, Switzerland). Conditioned medium was centrifuged to remove cell debris, harvested, and concentrated using Ultrafree-4 5K column (Millipore, Bedford, MA) before adding reducing or nonreducing Laemmli buffer. Cells were lysed in 2 × concentrated Laemmli buffer. After boiling at 95°C for 5 minutes, samples were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (7.5% or 10%). Western blot analysis was performed using rabbit anti–human fibrinogen antibodies (Dako, Zug, Switzerland) at a 1:2500 dilution. Immunoreactive bands were revealed with an enhanced chemiluminescence kit (ECL; Amersham Pharmacia Biotech, Dübendorf, Switzerland) according to manufacturer's instructions.
Results and discussion
A consanguineous Palestinian couple (first cousins) with 2 daughters with congenital afibrinogenemia requested a prenatal diagnosis for their ongoing pregnancy (Figure1A). In the first step, DNA was extracted from fresh EDTA (ethylenediaminetetraacetic acid) blood samples from parents and affected children for identification of the causative mutation transmitted in this family. Screening of all the coding regions and exon-intron junctions for FGA (α isoform, exons 1-5) and FGG by PCR and sequencing showed no mutation. Analysis of the FGB gene revealed a previously unreported nonsense mutation in the last exon, exon 8, Trp467Stop (TGG>TAG) (numbered from the first ATG codon according to guidelines for human mutation nomenclature11 12), which was found in heterozygosity in each of the parents and in homozygosity in each of the 2 affected daughters (Figure 1B).
Prenatal diagnosis for congenital afibrinogenemia.
(A) Family tree. SA: spontaneous abortion. The arrow indicates the fetus undergoing prenatal diagnosis (proband). (B) Partial sequence ofFGB exon 8 demonstrating the FGBTrp467Stop (W467X; TGG>TAG) mutation. Top panel, heterozygote; bottom panel, homozygote Trp467Stop. (C) Amino acid sequence of the fibrinogen beta chain. The Palestinian Trp467Stop mutation identified in this study is shown (bold) as well as the Trp470Stop (W470X) “fibrinogen Mount Eden” mutation19 and 3 missense mutations18 20: Leu383Arg, Gly430Asp, and Tyr467Gly. Amino acids encoded by odd-numbered exons 1, 3, 5, and 7 are in normal font; those encoded by even-numbered exons 2, 4, 6, and 8 are highlighted in gray. Amino acids encoded by codons separated in 2 consecutive exons are underlined. Amino acids are numbered from the initiator methionine; those included in the signal peptide are in bold.
Prenatal diagnosis for congenital afibrinogenemia.
(A) Family tree. SA: spontaneous abortion. The arrow indicates the fetus undergoing prenatal diagnosis (proband). (B) Partial sequence ofFGB exon 8 demonstrating the FGBTrp467Stop (W467X; TGG>TAG) mutation. Top panel, heterozygote; bottom panel, homozygote Trp467Stop. (C) Amino acid sequence of the fibrinogen beta chain. The Palestinian Trp467Stop mutation identified in this study is shown (bold) as well as the Trp470Stop (W470X) “fibrinogen Mount Eden” mutation19 and 3 missense mutations18 20: Leu383Arg, Gly430Asp, and Tyr467Gly. Amino acids encoded by odd-numbered exons 1, 3, 5, and 7 are in normal font; those encoded by even-numbered exons 2, 4, 6, and 8 are highlighted in gray. Amino acids encoded by codons separated in 2 consecutive exons are underlined. Amino acids are numbered from the initiator methionine; those included in the signal peptide are in bold.
Microsatellite analysis of the FGA intron 3 tetranucleotide repeat (FGAi3) was also performed. Both parents were found to be heterozygous for the FGAi3 microsatellite marker, whereas the 2 affected daughters were homozygous (data not shown).
Mutation analysis was performed on DNA isolated from cultured amniocytes, and the fetus was found to be heterozygous for the familial Trp467Stop mutation both by sequencing and FGAi3 microsatellite analysis. This result was confirmed in the newborn when the mother gave birth to a healthy baby boy weighing 2.9 kg at birth, and fibrinogen determination revealed a level of 120 mg/dL, consistent with heterozygosity for an afibrinogenemia mutation.
The Trp467Stop mutation was predicted to lead to the production of a truncated fibrinogen beta chain, with 25 amino acids missing from the C-terminus (Figure 1C). Alternatively, the truncated beta polypeptide might be unstable or the mutation might cause a defect in theFGB mRNA by aberrant splicing (nonsense-associated alternative splicing) or by affecting the stability of the mRNA through nonsense-mediated mRNA decay.13-15 However, because the Trp467Stop mutation lies within the last exon of FGB, these mechanisms are not thought to be activated.
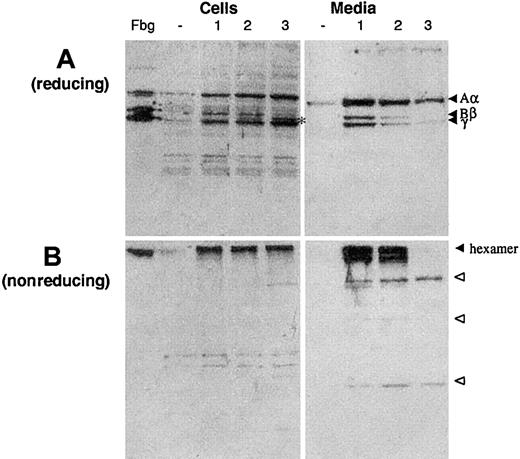
Previous studies in COS-1 cells expressing normal fibrinogen alpha and gamma chains in combination with beta-chain deletion mutants had led to the conclusion that the C-terminal portion of the beta chain, notably residues 238-491 (numbering from the initiator methionine), was not essential for fibrinogen assembly and secretion.16 In order to prove the causative nature of the mutation, Trp467Stop mutant and wild-type FGB cDNAs were transiently cotransfected with wild-type FGA and FGG cDNAs in COS-7 cells. Cells were lysed 18 hours after transfection and the conditioned media harvested. Individual fibrinogen chains and assembled hexamers were detected by Western blot analysis with a polyclonal antifibrinogen antibody (Figure 2).
Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs.
Samples of cell lysates and culture medium were subjected to (A) 10% SDS-PAGE under reducing conditions or (B) 7.5% SDS-PAGE under nonreducing conditions. The blots were incubated with a polyclonal anti–human fibrinogen antibody and cross-reacting bands revealed by chemiluminescence, as described in “Study design.” Fbg indicates purified fibrinogen control; −, COS cells transfected with an empty vector. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated; open arrowheads indicate probable α/γ intermediates (see “Results and discussion” and Hartwig and Danishefsky17). An asterisk marks the Trp467Stop mutant fibrinogen chain, which lacks 25 amino acid residues from the C-terminus. Transfections, lane 1: normal Aα, Bβ, and γ; lane 2: normal Aα, normal Bβ + mutant Trp467Stop Bβ, and normal γ; lane 3: normal Aα, mutant Trp467Stop Bβ (only), and normal γ.
Western blot analysis of cell extracts and conditioned media of COS-7 cells transfected with fibrinogen cDNAs.
Samples of cell lysates and culture medium were subjected to (A) 10% SDS-PAGE under reducing conditions or (B) 7.5% SDS-PAGE under nonreducing conditions. The blots were incubated with a polyclonal anti–human fibrinogen antibody and cross-reacting bands revealed by chemiluminescence, as described in “Study design.” Fbg indicates purified fibrinogen control; −, COS cells transfected with an empty vector. The positions of the hexameric complex and the normal Aα, Bβ, and γ chains are indicated; open arrowheads indicate probable α/γ intermediates (see “Results and discussion” and Hartwig and Danishefsky17). An asterisk marks the Trp467Stop mutant fibrinogen chain, which lacks 25 amino acid residues from the C-terminus. Transfections, lane 1: normal Aα, Bβ, and γ; lane 2: normal Aα, normal Bβ + mutant Trp467Stop Bβ, and normal γ; lane 3: normal Aα, mutant Trp467Stop Bβ (only), and normal γ.
When COS-7 cells are transfected with the 3 normal fibrinogen cDNAs, all 3 chains are correctly expressed and assembled inside the cell, and the fibrinogen hexamers are secreted into the media. When cells are transfected with normal FGA and FGGcDNAs and the Trp467Stop FGB mutant cDNA, again all 3 chains are correctly and stably expressed inside the cells. The mutant beta chain is incorporated into the fibrinogen hexamer inside the cell (Figure 2B, “Cells,” lane 3) but is not secreted: only incomplete forms containing α and/or γ chains and no fibrinogen hexamer are detectable in the supernatant (Figure 2A and B, “Media,” lane 3). These results closely resemble those previously described in COS cells transfected with different combinations of the 3 normal fibrinogen cDNAs.17
When cells are transfected with equal amounts of wild-type and mutantFGB cDNAs (and normal FGA and FGGcDNAs), imitating heterozygosity for the Trp467Stop mutation, both beta chains are clearly distinguished in the Western blot of cell lysates, with the normal chain being more abundant (Figure 2A, “Cells,” lane 2). By contrast, in the cell medium only the wild-type beta chain is found (Figure 2A, “Media,” lane 2).
The data demonstrate that truncation of the last 25 amino acid residues from the fibrinogen beta chain C-terminus does not inhibit hexamer assembly but eliminates its secretion, as previously reported for 2 missense mutations in FGB exons 7 and 8 identified in afibrinogenemia patients.18 These results are apparently in contradiction with the experimental observation that fibrinogen beta chains truncated at amino acid position 238 were able to assemble with alpha and gamma fibrinogen chains and were secreted into the media.16 However, the assembly and secretion of such a severely truncated polypeptide may not be physiologically relevant.
Interestingly, Homer et al19 reported a very similar mutation (Trp440Stop, amino acids numbered without the signal peptide, or Trp470Stop according to our nomenclature), which occurs only 3 codons downstream of the Trp467Stop mutation. The Trp470Stop mutation “fibrinogen Mount Eden” was identified in heterozygosity in a patient following laboratory investigations prior to a liver biopsy for hepatitis C. The patient had reduced fibrinogen levels (0.7 mg/mL) and a prolonged activated partial thromboplastin time. Other than mild epistaxis and gum bleeding, the patient was asymptomatic. The authors showed that the truncated fibrinogen beta chain was not found in the patient's plasma and suggested that removal of the C-terminal 22 residues does not allow incorporation of the mutant chain into mature fibrinogen hexamers. No fibrinogen inclusion bodies were detected in the liver biopsy, indicating that molecules containing the mutant chains do not accumulate in the patient's hepatocytes. We suggest that the molecular mechanism may be at the level of secretion, as with the Trp467Stop mutation we describe. The authors state that the mutation causes hypofibrinogenemia in heterozygosity; however, one can consider it an afibrinogenemia mutation because a homozygous individual for this mutation will most certainly have no circulating fibrinogen.
In conclusion, the Trp467Stop mutation identified in this study, along with the Trp470Stop mutation19 and 3 missense mutations18 20 identified in the same region in afibrinogenemia patients, confirms the necessity of intact C-terminal portions of the fibrinogen beta chain for the secretion of functional fibrinogen hexamers.
We are grateful to Dr Colette Rossier and Corinne Gumy for DNA sequencing and to Elena Barro for expert technical assistance.
Prepublished online as Blood First Edition Paper, January 2, 2003; DOI 10.1182/blood- 2002-10-3116.
Supported by Swiss National Science Foundation (SNF) grant no. 31-64987.01 and by the Dr Henri Dubois Ferrière-Dinu Lipatti Foundation. M.N.-A. is the recipient of an SNF professorship.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Marguerite Neerman-Arbez, Centre Médical Universitaire, 1 rue Michel Servet, CH-1211 Geneva, Switzerland; e-mail:marguerite.arbez@medecine.unige.ch.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal