In vitro models of granulopoiesis involving the inducible expression of either CCAAT enhancer binding protein alpha (C/EBPα) or C/EBPε in myeloid cells have been shown to lead to the induction of a granulocytic maturation program accompanied by the expression of myeloid-specific genes. Since members of the C/EBP family of transcription factors recognize and bind to similar DNA-binding motifs, it has been difficult to elucidate the specific role of each of the C/EBP family members in eliciting myeloid gene expression. In order to address this issue, we focused on the expression of the lactoferrin (LF) gene. LF expression is transcriptionally regulated in a C/EBP-dependent manner in myeloid cells. Using chromatin immunoprecipitation (ChIP) analysis we demonstrate that C/EBPα binds to the LF promoter in nonexpressing cells. Upon induction of maturation, C/EBPε binds to the LF promoter, which correlates with LF expression. Lack of LF expression in the acute promyelocytic leukemia cell line NB4, which harbors the t(15;17) translocation, cannot be correlated with aberrant binding at the C/EBP site in the LF promoter. It is, however, associated with the persistent binding of the silencer CCAAT displacement protein (CDP/cut) to the LF promoter in these cells. We conclude that C/EBPα, C/EBPε, and CDP/cut all play definitive roles in regulating late gene expression during normal myeloid development.
Introduction
Although a number of transcription factors have been implicated in myelopoiesis,1-4 several members of the CCAAT enhancer binding protein (C/EBP) family of transcription factors have been shown to be indispensable for normal development of the myeloid lineage. The C/EBP family of transcription factors is a class of basic region/leucine zipper (bZIP) factors that recognize the consensus DNA-binding sequence, 5′-ATTGCGCAAT-3′, as obligate dimers. Currently, 6 members of this family of transcription factors have been described, including C/EBP α, β, γ, δ, ε, and CHOP-10/GADD 153, all of which contain highly homologous dimerization (leucine-zipper) domains and DNA-binding (basic-region) motifs. These proteins have been implicated in regulating gene expression in a variety of cell types, including adipocytes (reviewed in Darlington et al5), constitutive and acute phase response genes in the liver (reviewed in Diehl6), and myelomonocytic cells (reviewed in Lekstrom-Himes and Xanthopoulos7).
Severe hematopoietic abnormalities have been reported for mice nullizygous for C/EBPα and ε. Although C/EBPα−/−mice die perinatally due to defects in gluconeogenesis,8,9they also demonstrate an early block in the differentiation of granulocytes. C/EBPα has been postulated to be a master regulator of the granulopoietic developmental program. Although C/EBPα is expressed in several tissues, its expression in the hematopoietic compartment is restricted to the granulocytic lineage, where it is expressed in the most primitive precursors. Recently, mutations within the C/EBPα gene have been demonstrated in patients with acute myeloid leukemia (AML) (French-American-British [FAB] classification M1 and M2).10 Additionally, the AML1/ETO fusion protein that results from the t(8;21) translocation in patients with M2 AML has been shown to down-regulate C/EBPα expression,11suggesting that loss of C/EBPα function in AML patients may contribute to the observed early block in the granulocytic maturation pathway.
C/EBPε is expressed relatively late in the granulocytic maturation pathway and has been shown to play a role in late myeloid gene expression.12-14 Absence of C/EBPε in mutant mice causes a late block in the differentiation of granulocytes. C/EBPε−/− mice produce hyposegmented granulocytes that are functionally defective. Mutant mice usually survive 2 to 5 months but eventually succumb to opportunistic bacterial infections.15 It has been demonstrated that C/EBPε−/− mice express absent or low levels of RNA for several genes, including the secondary granule protein (SGP) genes (murine lactoferrin [mLF], murine neutrophil gelatinase [mNG], and murine neutrophil collagenase [mNC]). Studies from our laboratory have demonstrated that expression of mLF and mNC in the developing neutrophil is dependent on intact C/EBP binding sites within their gene promoters (Khanna-Gupta et al16; and A.K.-G. and N.B., manuscript in preparation, 2003). Mutations within the C/EBPε gene have been described in patients with specific granule deficiency (SGD),17,18 a rare condition marked by defects in neutrophil function including atypical nuclear morphology, impaired bactericidal activity, and abnormalities in neutrophil migration, as well as a lack of both neutrophil and eosinophil secondary granule proteins.19 20
Previous studies using transient transfection analyses have demonstrated that both C/EBPα and C/EBPε, in concert with other factors such as PU.1, AML/CBFβ, and c-Myb, can regulate the promoters of several myeloid-specific genes.14,16,21,22 A number of primary granule protein genes including myeloperoxidase,23lysozyme, neutrophil elastase,24,25 and mim-1 are thought to be regulated by C/EBPα. Many of the same genes have also been shown to be regulated by C/EBPε.14 Interestingly, representational difference analysis (RDA), using fetal livers from wild-type and C/EBPα knock-out mice, showed that the expression of the late secondary granule protein (SGP) genes lactoferrin (LF), human neutrophil collagenase (HNC), and neutrophil gelatinase associated lipocalin (NGAL) were absent in C/EBPα−/−livers.26 In contrast, similar RDA studies using neutrophils and macrophages from C/EBPε−/− mice did not identify this group of late-expressing genes as targets of C/EBPε, even though transient transfection12-14 and C/EBPε−/− mice studies indicated otherwise.
In an attempt to further delineate the role of both C/EBPα and C/EBPε in myeloid maturation, several groups, including our own, have derived cell lines that inducibly express these factors in a myeloid setting.12,27,28 Granulocytic maturation has been reported following induced expression of C/EBPα as well as C/EBPε, suggesting that such in vitro overexpression models cannot be used to distinguish downstream targets of each factor. Additionally, a recent report has demonstrated that expression of C/EBPβ from theC/EBPα gene locus is sufficient to drive normal hematopoiesis in vivo.29 This apparent redundancy in C/EBP function is unlikely to be reflective of the in vivo role played by the individual C/EBP family members during granulopoiesis. Since the expression of C/EBPα in a number of myeloid cell lines induced toward the neutrophil lineage is biphasic, peaking in early and again in late myeloid cells,27 we hypothesized that C/EBPα and the late-expressing C/EBPε may together play a part in the in vivo regulation of late myeloid genes such as the SGP genes, which are hallmarks of terminal neutrophil differentiation.30 31
In this study, we used chromatin immunoprecipitation (ChIP) analysis to dissect the role of C/EBPα and C/EBPε in mediating the expression of the late myeloid gene lactoferrin (LF), which serves as a model for the coordinately regulated SGP group of genes during neutrophil maturation. We have previously demonstrated that high levels of LF expression are mediated in part via a C/EBP element within the LF gene promoter,16 while CCAAT displacement protein (CDP/cut), a highly conserved silencing factor that binds the LF promoter, coordinately represses expression of LF and all the SGP genes in early myeloid cells.32 33 Here we demonstrate that C/EBPα binds to the LF promoter in nonexpressing cells, whereas binding of C/EBPε occurs upon maturation and is correlated with LF expression. Expression also correlates with loss of CDP/cut binding to the promoter. We examined the modulation of these transcription factors in NB4 cells, an acute promyelocytic (APL) cell line that carries the t(15;17) translocation and fails to express LF upon induction with all-trans retinoic acid (ATRA). Lack of LF expression in these cells cannot be correlated with aberrant binding at the C/EBP site in the LF promoter, since binding of C/EBPα and C/EBPε appears to be intact. However, ATRA induction of NB4 cells is associated with persistent binding of the silencer CDP/cut to the LF promoter. We conclude that both C/EBPα and C/EBPε play definitive roles in regulating late gene expression during normal myeloid development, and that disruption of the interplay between the positive (C/EBP) and negative (CDP/cut) regulators of LF gene expression may contribute to the leukemic phenotype.
Materials and methods
Tissue culture, transient transfections, and luciferase assay
Human acute promyelocytic leukemia NB4 cells were obtained from Dr M. Lanotte (Institut National de la Santé et de la Recherche Médicale [INSERM], Paris, France), and were maintained and grown in RPMI 1640 medium (Gibco BRL, Grand Island, NY) supplemented with 10% heat-inactivated fetal calf serum (Gemini Bioproducts, Calabasas, CA), 0.2 mM glutamate, 50 units/mL penicillin, and 50 μg/mL streptomycin. 32Dwt18 cells (a gift from Dr Daniel Link, University of Washington, St Louis, MO) were grown in Iscoves modified Dulbecco medium (IMDM) supplemented with 10% fetal calf serum and 10% WEHI-conditioned medium, as a source of interleukin-3 (IL-3). MPRO cells were obtained from Dr Schickwann Tsai (Mt Sinai School of Medicine, NY) and were maintained in AIM-V medium (Gibco BRL) supplemented with 1% fetal calf serum and either with recombinant granulocyte-macrophage colony-stimulating factor (GM-CSF; Amgen, Thousand Oaks, CA) or 10% HM-5–conditioned medium as a source of GM-CSF. All cells were maintained at 37°C in a humidified 5% CO2 incubator. Differentiation of NB4 and MPRO cells was performed as described previously.34,35 Briefly, exponentially growing cells were washed twice with phosphate-buffered saline (PBS) and resuspended in growth medium containing 5 to 10 μM all-trans retinoic acid (ATRA; Sigma, St Louis, MO). Cells were incubated for the times stipulated. Differentiation of 32Dwt18 cells was performed as described previously.32 Exponentially growing cells were washed twice with phosphate-buffered saline (PBS) and resuspended in growth medium containing 1 U/mL erythropoietin (Epo; Amgen) in the absence of an IL-3 source. Maturation of all inductions was monitored by Wright-Giemsa staining.
Transient transfection experiments were performed as previously described.12 Briefly, approximately 1 × 10732Dwt18 cells were gently pelleted and washed twice with PBS and resuspended in 180 μL HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid)–buffered saline in electroporation cuvettes. Then, 10 to 20 μg of each reporter plasmid construct and 2 μg pCMVβgal (Clontech, Palo Alto, CA), an internal control plasmid used to monitor transfection efficiency, were added to each aliquot of cells. Following a 5-minute incubation period at room temperature, the DNA-cell samples were electroporated using a Biorad Gene Pulser (Biorad, Hercules, CA) at 400 V with a capacitance of 250 μF. Luciferase expression levels were normalized to the levels of β-galactosidase expression.32
Chromatin immunoprecpitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed using the protocol for the acetyl-histone H4 ChIP Assay Kit (Upstate Biotechnology, Lake Placid, NY). Briefly, 2 × 10732Dwt18 (uninduced and 4-day Epo-induced), MPRO (uninduced and 24-hour ATRA-induced), and NB4 (uninduced and 48-hour ATRA-induced) cells were cross-linked by addition of formaldehyde into the medium at a final concentration of 1% and incubated for 30 minutes at 37°C. Prior to cross-linking, an aliquot of the cells was removed for analysis of input chromatin DNA. Cells were then washed with ice-cold phosphate-buffered saline and resuspended in 200 μL of ChIP lysis buffer (1% SDS [sodium dodecyl sulfate]; 10 mM EDTA [ethylenediaminetetraacetic acid]; and 50 mM Tris [tris(hydroxymethyl)aminomethane]–HCl, pH 8.0) with protease inhibitors (Roche, Indianapolis, IN) and 1 μM diisopropyl fluorophosphate (DFP; Sigma), incubated on ice for 10 minutes, and sonicated at 28% power for 3 pulses of 10 seconds each in a Branson Sonifier 450 (Branson Ultrasonics, Danbury, CT). Sonicated lysates were then diluted to 2 mL with ChIP dilution buffer (0.01% SDS; 1.1% Triton X-100; 1.2 mM EDTA; 16.7 mM Tris-HCl, pH 8.0; and 167 mM NaCl), followed by preclearing with 80 μL protein A–agarose beads or protein G Plus–agarose beads for 60 minutes at 4°C with rotation. The precleared lysates were next immunoprecipitated using antibodies for either C/EBPε (4 μg; Santa Cruz Biotech, Santa Cruz, CA, sc-158), C/EBPα (8 μg; Santa Cruz Biotech, sc-61), or CDP (4 μg; Santa Cruz Biotech, sc-6327) at 4°C overnight with rotation. A no-antibody control was included with each experiment. Immune complexes were collected with 60 μL protein A–agarose or protein G Plus–agarose and washed once with 1 mL each of the following buffers: low salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 150 mM NaCl), high salt wash buffer (0.1% SDS; 1% Triton X-100; 2 mM EDTA; 20 mM Tris-HCl, pH 8.0; and 1500 mM NaCl), LiCl wash buffer (250 mM LiCl; 1% nonidet P-40 [NP-40]; 1% sodium deoxycholate; 1 mM EDTA; and 10 mM Tris-HCl, pH 8.0); and twice with 10 mM Tris-HCl, pH 8.0; and 1 mM EDTA. Immune complexes were next eluted using freshly prepared elution buffer (1% SDS and 0.1 M NaHCO3). Cross-links were reversed by heating at 65°C in the presence of NaCl followed by proteinase K treatment. The DNA was recovered by phenol/chloroform extraction followed by ethanol precipitation and resuspended in 40 μL distilled water. ChIP DNA (1 μL) was next used as a template for polymerase chain reaction (PCR) using the appropriate oligos (all oligos are oriented in the 5′ to 3′ direction). C/EBPε CDP/cut: FP (forward primer), GCTAACCGGAATATGCTAATCAG; RP (reverse primer), CCTTTCAGAGACACCTGCTC.12 LF CDP/cut: FP, GTTTAGTTTGCTTCCAACTG; RP, CCATCTCCTCCTTCTCTTT.32 Murine NE: FP, GACACCCCCACTGTCG; RP, TTATAGGTGGGAACCAGAG.25Murine LF C/EBP: FP, GTTTCCTGTACCAGCGCCT; RP, GTCTGTGGTCTTGGGAGA.36 Human LF C/EBP: FP, TGGCGGGGAGTGGGAGGGAA; RP, AAGCTTGTCGACCGACTTGGCAAACGAAG.16Gp91phox CDP/cut: FP, CCAATGATTATTAGCCAATTTCTG; RP, CATGGTGGCAGAGGTTGAATGT.37 HNP CDP/cut and C/EBP: FP, GTCAACTGTGTTAGGAGCCAT; RP, CGTGCACAAGTGGACTTC.38
All PCR products were subcloned into the pCRII vector (Invitrogen, Carlsbad, CA) and sequenced by standard dideoxy sequencing technology to confirm their identity.
Northern blot analysis
Northern blot analysis was performed as described previously.34 Briefly, 10 μg total RNA prepared using Trizol reagent (Gibco BRL) from uninduced and ATRA-induced NB4 cells was separated on a 1% denaturing agarose gel, transferred to nitrocellulose filters, and hybridized to 32P-labeled cDNA fragments at 68°C in Express-Hyb (Sigma). Filters were washed at high stringency in 0.1% sodium dodecyl sulfate (SDS) and 0.1 × SSC (1 × SSC, 3 M NaCl, 0.3 sodium citrate, pH 7.0) at 60°C and autoradiographed. Blots were probed sequentially with a previously described LF probe isolated in our laboratory,34 a cDNA probe for human gp91phox (provided by David Skalnik, Indiana University, Indianapolis, IN), and a previously described α defensin (HNP 1,3) probe.39 A γ-actin probe was used to confirm equal RNA loading in each lane.
Preparation of nuclear extracts and electrophoretic mobility shift analysis (EMSA)
Nuclear extracts were prepared as described previously12 from uninduced and 2-day ATRA-induced NB4 cells. Total protein concentration in the nuclear extract preparations was assayed using the Bradford assay (BioRad kit) per the manufacturer's instructions. In general, most preparations yielded 1.5 to 2 μg/μL protein. EMSA analysis was next carried out as previously described.12 EMSAs were performed by incubating 15 μg nuclear extracts with 20 000 cpm of double-stranded oligonucleotide harboring the CDP/cut site within the LF promoter32 in a 20-μL reaction mixture containing 10 mM HEPES-KOH buffer (pH 7.9), 50 mM KCl, 2.5 mM MgCl2, 1 mM dithiothreitol (DTT), 10% glycerol, 1 μg acetylated bovine serum albumin (New England Biolabs, Beverly, MA), and 0.5 μg poly(dI-dC) at 25°C for 20 minutes. For competition analysis, a 100-fold molar excess of unlabeled oligonucleotides was added to the nuclear extracts prior to the addition of the labeled probe. Oligos used for competition have been previously described.12 32 Binding reactions were resolved on a 4% nondenaturing polyacrylamide gel containing 1 × TBE (0.089 M Tris-borate, 0.089 M boric acid, and 0.002 M EDTA) and electrophoresed at 150 V for 3 hours at 4°C. Gels were exposed to x-ray film with an intensifying screen overnight at −80°C.
Western blot analysis
Approximately 40 μg nuclear extracts or total cell extracts prepared from 1 × 106 cells in lysis buffer (15 mM HEPES, pH 7.9; 0.4 MKCl; 0.1% NP-40; 4 mM NaF; 4 mM Na3VO4; 0.2 mM EDTA; 0.2 mM EGTA; 1 μM DFP; 1 mM DTT; 1 mM phenylmethylsulfonyl fluoride [PMSF]; and 10% glycerol) prepared from uninduced and 2-day ATRA-induced NB4 cells; uninduced and 24-hour ATRA-induced MPRO cells; and uninduced and 4-day Epo-induced 32Dwt18 cells were transferred to 2 × Laemelli loading buffer, boiled for 5 minutes, and loaded onto a 4% to 20% Tris/Glycine gel (Novex, San Diego, CA). Electrophoresis was carried out at 150 V for 2 hours at room temperature. Electrophoresed proteins were transferred to a polyvinylidene fluoride (PVDF) membrane (BioRad) and blocked with 5% nonfat dry milk in Tris-buffered saline containing 0.1% Tween-20 (TBS-T) for one hour at room temperature. The blocked membranes were next incubated with CDP, C/EBPε, and C/EBPα antibodies at a 1:3000 dilution in TBS-T at 4°C overnight. Each membrane was washed 3 times with TBS-T + 5% nonfat dry milk and incubated with either an antigoat horseradish peroxidase–conjugated secondary antibody (Santa Cruz Biotech) or an antirabbit horseradish peroxidase–conjugated secondary antibody for one hour at room temperature. The membrane was washed 5 times in TBS-T and chemiluminescent detection performed per the manufacturer's instructions (Amersham, Piscataway, NJ).
Results
C/EBPα C/EBPβ, and C/EBPε expression plasmids transactivate a C/EBP site containing lactoferrin promoter plasmid in myeloid cells
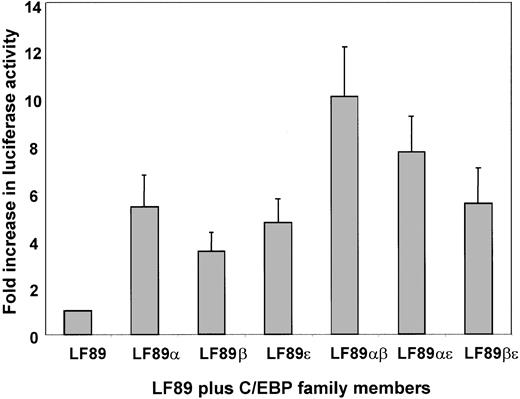
Transient cotransfection assays have been widely used to identify and characterize functional DNA-binding elements within the promoters of several genes. We have previously demonstrated that myeloid-specific expression of lactoferrin (LF), a model for secondary granule protein (SGP) gene expression, is C/EBP-dependent.16 In order to identify which family member of the C/EBP family of transcription factors is responsible for mediating high levels of LF expression during myeloid maturation, we performed a transient cotransfection assay using a previously identified 89-bp fragment (LF89) of the lactoferrin gene promoter harboring a C/EBP site cloned into the promoterless pGL3 basic reporter plasmid.16 The LF89 plasmid was cotransfected with C/EBP expression plasmids into 32Dwt18 cells. The 32Dwt18 cells are a subline of the murine myeloid 32Dcl3 cells that have been stably transfected with a chimeric receptor containing the extracellular domain of the erythropoietin receptor and the intracellular domain of the G-CSF receptor. These cells respond to erythropoietin (Epo) and undergo differentiation along the granulocytic lineage.32 As is evident in Figure1, no significant difference between transactivation effects of C/EBPα (5.8-fold) C/EBPβ (4-fold), or C/EBPε (5.2-fold) on LF89 was observed in 32Dwt18 cells. Since C/EBP family members bind DNA as dimers, we examined the possibility that cotransfection of 2 family members may cooperatively transactivate the LF89 plasmid, indicating a preference of a particular pair for the LF89 C/EBP site. Our data (Figure 1), however, indicate that pairwise cotransfections of C/EBP family members with LF89 result in an additive rather than a cooperative effect on LF89 expression. (For example, LF89α [5.8-fold] + LF89β [4-fold] = LF89αβ [10-fold].) The results from this experiment indicate that all 3 C/EBP family members can transactivate the LF89 plasmid equally well in this in vitro assay, and hence no definitive conclusion may be drawn as to the specific involvement of either individual or paired C/EBP family members in LF expression in a myeloid setting.
Transient cotransfection analysis of LF89 and expression plasmids for C/EBPα, β, and ε in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with an LF gene promoter fragment spanning −89 bp (LF89) harboring a C/EBP site cloned into the promoterless luciferase reporter pGL3-Basic plasmid (10 μg), and expression plasmids for C/EBPα, β, and ε (5 μg each), individually or pairwise. A pCMVβgal expression plasmid (2 μg) was included in each transfection to normalize for transfection efficiency. The total concentration of DNA per transfection was maintained at 22 μg by the addition of salmon sperm DNA where necessary. Transfected cells were harvested 24 hours after transfection and assayed for luciferase and β-galactosidase activity. Normalized luciferase values have been represented as a fold increase of luciferase activity over LF89 alone. Mean ± SE for 3 experiments performed in duplicate have been illustrated.
Transient cotransfection analysis of LF89 and expression plasmids for C/EBPα, β, and ε in 32Dwt18 cells.
32Dwt18 cells were transiently cotransfected with an LF gene promoter fragment spanning −89 bp (LF89) harboring a C/EBP site cloned into the promoterless luciferase reporter pGL3-Basic plasmid (10 μg), and expression plasmids for C/EBPα, β, and ε (5 μg each), individually or pairwise. A pCMVβgal expression plasmid (2 μg) was included in each transfection to normalize for transfection efficiency. The total concentration of DNA per transfection was maintained at 22 μg by the addition of salmon sperm DNA where necessary. Transfected cells were harvested 24 hours after transfection and assayed for luciferase and β-galactosidase activity. Normalized luciferase values have been represented as a fold increase of luciferase activity over LF89 alone. Mean ± SE for 3 experiments performed in duplicate have been illustrated.
Chromatin immunoprecipitation (ChIP) analysis of C/EBPα and C/EBPε in lactoferrin-expressing cells
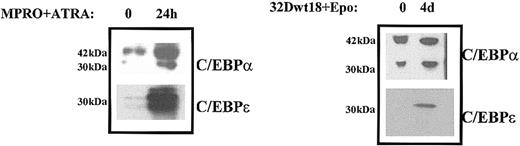
The use of ChIP analysis to gain an understanding of in vivo protein-DNA interactions in a number of cell systems has revolutionized the study of DNA-binding proteins and their role in mediating gene expression (eg, Boyd and Farnham40; Wells et al41; and Shang and Brown42). This technique provides a unique glimpse in real time of the binding activity of a given DNA-binding protein. In order to gain an understanding of the role of C/EBPα and C/EBPε in mediating high levels of LF expression during granulopoiesis, we performed ChIP analysis in 2 previously characterized murine myeloid cell lines, both of which are capable of expressing high levels of LF upon induction toward neutrophil maturation.32,43 MPRO cells are a promyelocytic cell line harboring a truncated dominant-negative RARα gene. These cells can be made to undergo neutrophil maturation upon the addition of pharmacologic levels of ATRA.35,43,44 Western blot analysis revealed that while the levels of C/EBPε are up-regulated upon induction in both ATRA-induced MPRO cells as well as Epo-induced 32Dwt18 cells, the levels of C/EBPα remained unchanged in the latter but were up-regulated in the former (Figure2). This apparent difference in expression of C/EBPα in the 2 myeloid cell lines probably reflects the observation that C/EBPα is expressed in a biphasic manner, peaking in early and again in late myeloid cells,27 and that the promyelocytic MPRO cells are not only further along the granulocytic maturation pathway than 32Dwt18 cells, but mature more rapidly as well. As is evident in Figure3A, ChIP analysis using anti-C/EBPα and anti-C/EBPε was carried out in MPRO cells before and after induction for 24 hours with ATRA. Isolated DNA was subjected to PCR both before (Figure 3A, lane 8, input) and after chromatin immunoprecipitation using primers designed to amplify the region within the LF promoter harboring the C/EBP binding site.16 The LF C/EBP site in uninduced MPRO cells appears to be bound to C/EBPα (Figure 3A, lane 2) but not to C/EBPε (Figure 3A, lane 4). Induction of MPRO cells toward neutrophil maturation with ATRA changes the dynamics of protein binding at the LF C/EBP site. C/EBPα remains bound (Figure 3A, lane 3), while C/EBPε becomes bound to the site (Figure 3A, lane 5). A no-antibody control (Figure 3A, lane 1) and preimmune serum controls (Figure 3A, lanes 6-7) yielded no PCR product corresponding to the LF C/EBP site, thus serving as negative controls. Additionally, irrelevant oligos did not yield a PCR product, thus emphasizing the specificity of the observations in this experiment (data not shown).
Western blot analysis of C/EBPα and C/EBPε expression during myeloid differentiation.
Left panel: Western blot analysis was performed using whole cell extracts prepared from uninduced (0) and ATRA-induced (24 h) MPRO cells. The blots were probed with C/EBPα and C/EBPε antibodies. Right panel: Western blot analyses were performed on whole cell extracts prepared from uninduced (0) or Epo-induced (4 d) 32Dwt18 cells. The blots were probed with C/EBPα and C/EBPε antibodies as outlined in “Materials and methods.” The molecular weights of the proteins are indicated in kilodaltons.
Western blot analysis of C/EBPα and C/EBPε expression during myeloid differentiation.
Left panel: Western blot analysis was performed using whole cell extracts prepared from uninduced (0) and ATRA-induced (24 h) MPRO cells. The blots were probed with C/EBPα and C/EBPε antibodies. Right panel: Western blot analyses were performed on whole cell extracts prepared from uninduced (0) or Epo-induced (4 d) 32Dwt18 cells. The blots were probed with C/EBPα and C/EBPε antibodies as outlined in “Materials and methods.” The molecular weights of the proteins are indicated in kilodaltons.
Chromatin immunoprecipitation (ChIP) analysis of C/EBPα and C/EBPε during myeloid differentiation.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (24 h) MPRO cells using antibodies specific for C/EBPα (lanes 2-3) and C/EBPε (lanes 4-5), a no-antibody control (−) (lanes 1,9), and preimmune serum controls (PIS) (lanes 6-7). The precipitated chromatin was analyzed using primers specific for the murine LF (mLF) C/EBP site. Input mLF chromatin (1:10 dilution) is represented in lane 8. (B) ChIP analysis was performed using uninduced (0) and 4-day Epo-induced (4d) 32Dwt18 cells and antibodies specific for C/EBPα (lanes 2-3) and C/EBPε (lanes 5-6), no-antibody controls (lanes 1, 4, and 10), and preimmune serum (PIS) controls (lanes 7-8). Precipitated chromatin was analyzed using primers for the murine LF (mLF) promoter. Input mLF chromatin (1:10 dilution) is represented in lane 9. M indicates molecular weight markers. (C) ChIP analysis of uninduced (0) and ATRA-induced (24 h) MPRO cells using C/EBPα antibodies (lanes 1-2), C/EBPε antibodies (lanes 3-4), and a no-antibody control (lane 5). Precipitated chromatin was analyzed by PCR using primers specific for murine neutrophil elastase (mNE). Each ChIP experiment was performed 2 to 3 times. All PCR products were subcloned and sequenced by dideoxy method to confirm their identities.
Chromatin immunoprecipitation (ChIP) analysis of C/EBPα and C/EBPε during myeloid differentiation.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (24 h) MPRO cells using antibodies specific for C/EBPα (lanes 2-3) and C/EBPε (lanes 4-5), a no-antibody control (−) (lanes 1,9), and preimmune serum controls (PIS) (lanes 6-7). The precipitated chromatin was analyzed using primers specific for the murine LF (mLF) C/EBP site. Input mLF chromatin (1:10 dilution) is represented in lane 8. (B) ChIP analysis was performed using uninduced (0) and 4-day Epo-induced (4d) 32Dwt18 cells and antibodies specific for C/EBPα (lanes 2-3) and C/EBPε (lanes 5-6), no-antibody controls (lanes 1, 4, and 10), and preimmune serum (PIS) controls (lanes 7-8). Precipitated chromatin was analyzed using primers for the murine LF (mLF) promoter. Input mLF chromatin (1:10 dilution) is represented in lane 9. M indicates molecular weight markers. (C) ChIP analysis of uninduced (0) and ATRA-induced (24 h) MPRO cells using C/EBPα antibodies (lanes 1-2), C/EBPε antibodies (lanes 3-4), and a no-antibody control (lane 5). Precipitated chromatin was analyzed by PCR using primers specific for murine neutrophil elastase (mNE). Each ChIP experiment was performed 2 to 3 times. All PCR products were subcloned and sequenced by dideoxy method to confirm their identities.
In order to determine if the pattern of C/EBPα and C/EBPε binding at the LF C/EBP site was unique to the MPRO cell system or whether it formed the basis of a common paradigm with respect to LF expression in the developing neutrophil, we performed ChIP analysis in a second myeloid cell line. We have previously shown that 32Dwt18 cells differentiate in response to Epo induction in the absence of IL-3, and express the LF gene at levels comparable with the 32Dcl3/G-CSF system, without encountering the 80% cell death associated with induction of 32Dcl3 with G-CSF.32 ChIP analysis of uninduced and Epo-induced 32Dwt18 cells demonstrated a dynamic pattern of C/EBPα and C/EBPε binding to the LF C/EBP site (Figure 3B), identical to that observed in the MPRO cell system (Figure 2A). Here again, C/EBPα alone was bound to the LF C/EBP site in uninduced cells (Figure 3B, lane 2), while both C/EBPα (Figure 3B, lane 3) and C/EBPε (Figure3B, lane 6) were bound to this site in the LF promoter in induced 32Dwt18 cells.
Our data thus far indicate a distinct pattern of C/EBPα binding and further suggest that binding of C/EBPε to the LF C/EBP site correlates with the up-regulation of the late SGP gene LF during neutrophil maturation. We next asked whether the binding dynamics of C/EBPα and C/EBPε were different for a C/EBP site in a primary granule protein in the MPRO cell system. As indicated in Figure 3C, ChIP analysis of the C/EBP binding site in the neutrophil elastase (NE) gene promoter25 indicates that C/EBPα is bound to the NE C/EBP site in uninduced cells (Figure 3C, lane 1). However, neither C/EBPα (Figure 3C, lane 2) nor C/EBPε (Figure 3C, lane 4) are bound to this site in induced MPRO cells (Figure 3C, lane 2). Thus the pattern of C/EBPα and C/EBPε engagement at the C/EBP sites of a primary (NE) and a secondary (LF) granule protein gene appears to be different. Our data reaffirm the previously held view that C/EBPε is involved in inducing expression of the secondary and not the primary granule protein genes in the maturing neutrophil.13
Chromatin immunoprecipitation (ChIP) analysis of C/EBPα and C/EBPε in lactoferrin nonexpressing cells
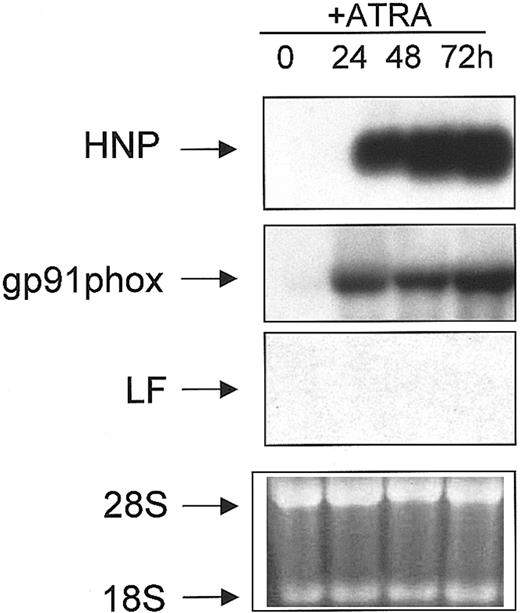
NB4 is a cell line derived from a patient with acute promyelocytic leukemia (APL) and harbors the t(15;17) cytogenetic abnormality.45 These cells undergo partial granulocytic maturation upon induction with ATRA, in that they undergo apparently normal phenotypic maturation but manifest a coordinate failure to express the SGP genes.34 To demonstrate the partial nature of the neutrophil maturation program induced by ATRA in NB4 cells, we performed Northern blot analysis. Total RNA isolated from uninduced (0 hours) and ATRA-induced (24, 48, and 72 hours) NB4 cells was subjected to Northern blot analysis and probed sequentially for the expression of myeloid-specific genes, including LF. As previously shown, the levels of the C/EBPε transcript were induced upon ATRA induction,46 as were the levels of α defensins (HNP) (Figure 4), a group of highly conserved low-molecular-weight cationic peptides that comprise 30% to 50% of the primary granule proteins,47 and gp91phox, a major component of the nicotinamide adenine dinucleotide phosphate (NADPH) oxidase complex in the mature neutrophil (reviewed in Skalnik4) (Figure 4). As previously shown, no LF mRNA was detected in either uninduced or ATRA-induced NB4 cells (Figure4).34
Northern blot analysis of NB4 cells treated with ATRA
. RNA (10 μg) isolated from uninduced (0) and ATRA-induced (24, 48, 72 h) NB4 cells were subjected to Northern blot analysis. The blot was sequentially probed with 32P-labeled cDNA probes for HNP, gp91phox, and LF as described in “Materials and methods.” Equal loading of RNA in each lane was determined by the presence of an equal concentration of both 18S and 28S ribosomal RNA in each lane in the ethidium bromide stained gel prior to blotting (bottom panel).
Northern blot analysis of NB4 cells treated with ATRA
. RNA (10 μg) isolated from uninduced (0) and ATRA-induced (24, 48, 72 h) NB4 cells were subjected to Northern blot analysis. The blot was sequentially probed with 32P-labeled cDNA probes for HNP, gp91phox, and LF as described in “Materials and methods.” Equal loading of RNA in each lane was determined by the presence of an equal concentration of both 18S and 28S ribosomal RNA in each lane in the ethidium bromide stained gel prior to blotting (bottom panel).
It has been previously hypothesized that in APL the PML/RARα fusion protein resulting from the t(15;17) translocation may interfere with the function of C/EBPα thereby leading to an arrest in these cells at the promyelocytic stage.48 In order to evaluate this hypothesis with respect to the nonexpression of the SGP gene LF in these cells, we performed ChIP analysis for both C/EBPα and C/EBPε at the LF C/EBP site in uninduced and induced NB4 cells. No apparent change was observed in the binding of C/EBPα or C/EBPε in uninduced or ATRA-induced NB4 cells (Figure 5A, top left panel). C/EBPα remained bound to the C/EBP LF site in both uninduced (Figure 5A, top left panel, lane 4) as well as induced (Figure 5A, top left panel, lane 5) NB4 cells. The pattern of C/EBPε binding at the LF C/EBP site was essentially identical to that for C/EBPα (Figure 5A, top left panel, lanes 2-3). In contrast, the binding of C/EBPα and C/EBPε to the recently described functional C/EBP site in the HNP gene promoter38showed significant changes, which likely correlate with the expression of this gene in ATRA-induced NB4 cells. As shown in Figure 5A, reciprocal binding of C/EBPα and C/EBPε was observed at the HNP C/EBP site in uninduced and ATRA-induced NB4 cells: C/EBPα bound the HNP C/EBP site in only the uninduced NB4 cells (Figure 5A, bottom left panel, lane 4), while C/EBPε associated with this site in induced NB4 cells (Figure 5A, bottom left panel, lane 3). The specificity of these observations was confirmed by the inability of either preimmune serum (Figure 5A, right panel, lanes 6-7) or a no-antibody control (Figure5A, lane 1) to yield the appropriate PCR products corresponding to the C/EBP cis elements in question. Our observations suggest that unlike neutrophil elastase (Figure 3C), this primary granule protein gene is regulated by C/EBPε binding activity. This is consonant with previous studies that have demonstrated that the defensins (HNP) are expressed later than most of the other primary granule proteins,49and that they share some regulatory features with secondary granule proteins. For example, the defensins are the only primary granule proteins that are absent from neutrophils in patients with neutrophil-specific granule deficiency (SGD).39
ChIP analysis of C/EBPα and C/EBPε in the leukemic NB4 cell line.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (48 h) NB4 cells using antibodies specific for C/EBPε (lanes 2-3) and C/EBPα (lanes 4-5), a no-antibody control (−) (lanes 1,9), and preimmune serum (PIS) controls (lanes 6-7). The precipitated chromatin was analyzed using primers specific for the human LF (LF) promoter (top panel) and the HNP promoter (bottom panel). Input HNP chromatin (1:10 dilution) is represented in lane 8. This experiment was repeated 3 times. The identities of PCR products obtained were confirmed by dideoxy sequencing. (B) Western blot analysis of whole cell extracts prepared from uninduced (0) and ATRA-induced (48 h) NB4 cells was performed and probed with C/EBPα and C/EBPε antibodies. The molecular weights of the proteins detected are shown in kilodaltons.
ChIP analysis of C/EBPα and C/EBPε in the leukemic NB4 cell line.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (48 h) NB4 cells using antibodies specific for C/EBPε (lanes 2-3) and C/EBPα (lanes 4-5), a no-antibody control (−) (lanes 1,9), and preimmune serum (PIS) controls (lanes 6-7). The precipitated chromatin was analyzed using primers specific for the human LF (LF) promoter (top panel) and the HNP promoter (bottom panel). Input HNP chromatin (1:10 dilution) is represented in lane 8. This experiment was repeated 3 times. The identities of PCR products obtained were confirmed by dideoxy sequencing. (B) Western blot analysis of whole cell extracts prepared from uninduced (0) and ATRA-induced (48 h) NB4 cells was performed and probed with C/EBPα and C/EBPε antibodies. The molecular weights of the proteins detected are shown in kilodaltons.
While protein levels of C/EBPε were markedly increased upon ATRA induction of NB4 cells (Figure 5B, top panel), levels of the larger 42-kDa C/EBPα isoform remained unchanged (Figure 5B, bottom panel). Interestingly, the levels of the shorter 30-kDa C/EBPα isoform appeared to decrease upon ATRA induction of NB4 cells. Previous studies have demonstrated that this isoform, when expressed at high levels relative to the 42-kDa isoform, acquires dominant-negative properties.10 The implications of this observation on the observed differential binding of C/EBPα to its cognate binding sites in the promoters of LF and HNP (Figure 5A) within the APL milieu remain to be elucidated.
Persistent binding of CDP/cut to the LF CDP binding site may account for nonexpression of LF in ATRA-induced NB4 cells
Even though the binding pattern of C/EBPα and C/EBPε to the LF C/EBP site correlates with increased LF expression in induced MPRO and 32Dwt18 cells (Figure 3A-B), it appears to be insufficient to up-regulate expression of LF in ATRA-induced NB4 cells (Figure 4). It seems likely that the defect associated with nonexpression of LF in NB4 cells induced with ATRA may not be associated with aberrant binding at the LF C/EBP site. Previous studies from our laboratory have shown that CCAAT displacement protein (CDP/cut), a highly conserved silencing factor that binds the LF promoter, coordinately represses expression of all SGP genes.32,33 CDP/cut is a homeodomain protein that contains 3 highly homologous domains of 70 amino acids called thecut repeats.50 CDP/cut is thought to play a role in determining cell-type specificity in both Drosophila melanogaster and mammals.51,52 In general, CDP/cut is thought to act as a repressor of developmentally regulated genes.52-56 In the myeloid compartment, CDP/cut has been implicated as a transcriptional repressor of the gp91phox gene in immature myeloid cells,54,57 the C/EBPε gene,12 and the SGP genes.32,33 A CDP/cut site has recently been described in the promoter of the primary granule HNP 1,3 genes,38 but its functional role in the expression of this family of genes has not been described.
We therefore assessed whether recruitment of CDP/cut to its cognate binding sites in the HNP, C/EBPε, gp91phox, and LF gene promoters could be correlated with expression of these genes in the NB4 cell system. ChIP analysis revealed that CDP/cut was recruited to the CDP/cut binding sites in uninduced NB4 cells in the promoters of all 4 genes examined (Figure 6A, lane 1). ATRA-induced differentiation resulted in loss of binding of CDP/cut in the HNP, C/EBPε, and gp91phox gene promoters (Figure 6A, lane 2). This loss of CDP/cut binding appears to correlate with expression of these 3 genes in ATRA-induced NB4 cells (Figure 4). CDP/cut binding, however, was not abolished in the LF promoter in ATRA-induced NB4 cells (Figure 6A, top panel, lane 2), an observation that correlates with the nonexpression of LF in the NB4 cell system. Loss of CDP/cut binding in the gp91phox, C/EBPε, and HNP promoters could not be explained by a decrease in protein levels of CDP/cut in ATRA-induced NB4 cells, as judged by Western blot analysis (Figure 6B), suggesting that the mechanism underlying loss of CDP/cut binding likely involves posttranslational modifications of the CDP/cut protein.
ChIP analysis of CDP/cut binding to myeloid promoters in NB4 cells.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (48 h) NB4 cells using an antibody specific for CDP/cut (lanes 1-2) and a no-antibody control (−) (lanes 3-4). The precipitated chromatin was analyzed using primers specific for the CDP/cut sites in the LF promoter, the HNP promoter, the C/EBPε promoter, and the gp91phox promoter. Input LF/CDP chromatin (1:10 dilution) is represented in lane 5. This experiment was repeated 3 times. The identities of all PCR products obtained were confirmed by dideoxy sequencing. (B) Western blot analysis was performed on 40-μg nuclear extracts prepared from uninduced (NB4; day 0), and 48-hour ATRA-induced (NB4; 48 h ATRA) NB4 cells. The blot was probed with a CDP/cut-specific antibody. The molecular-weight marker is indicated.
ChIP analysis of CDP/cut binding to myeloid promoters in NB4 cells.
(A) Chromatin immunoprecipitations were performed from uninduced (0) and ATRA-induced (48 h) NB4 cells using an antibody specific for CDP/cut (lanes 1-2) and a no-antibody control (−) (lanes 3-4). The precipitated chromatin was analyzed using primers specific for the CDP/cut sites in the LF promoter, the HNP promoter, the C/EBPε promoter, and the gp91phox promoter. Input LF/CDP chromatin (1:10 dilution) is represented in lane 5. This experiment was repeated 3 times. The identities of all PCR products obtained were confirmed by dideoxy sequencing. (B) Western blot analysis was performed on 40-μg nuclear extracts prepared from uninduced (NB4; day 0), and 48-hour ATRA-induced (NB4; 48 h ATRA) NB4 cells. The blot was probed with a CDP/cut-specific antibody. The molecular-weight marker is indicated.
The persistent binding of CDP/cut to the LF CDP/cut binding site in ATRA-induced NB4 cells was further confirmed by EMSA analysis. The LF CDP/cut binding site (DM2 probe) was used as a probe to compare the CDP/cut binding status of nuclear extracts prepared from NB4 and ATRA-induced NB4 cells. CDP/cut protein binding to the LF CDP/cut binding site was found to be unchanged in uninduced versus ATRA-induced NB4 cells (Figure 7A, lanes 2-3). The binding of CDP/cut protein from the NB4 nuclear extracts was found to be specific, as 3 of the DNA-protein complexes (Figure 7A, arrows) were specifically competed away only in the presence of a 100-fold molar excess of either the LF/CDP/cut (DM2) oligo itself (Figure 6A, lane 4), or the neural cell adhesion molecule (NCAM) oligos (Figure 7A, lane 5), which have previously been shown to bind to the CDP/cut protein.32 In a parallel EMSA experiment using the C/EBPε CDP/cut binding site as a probe, we were able to demonstrate a significant decrease in binding of CDP/cut to this site in ATRA-induced NB4 extracts when compared with uninduced NB4 extracts (Figure 7B, lanes 2-3). This finding further validates the observation that decreased binding of CDP/cut correlates with expression of C/EBPε in ATRA-induced NB4 cells (Figures 4,6A). Since persistent CDP/cut binding to the LF silencer element appears to be a hallmark of LF nonexpression, we conclude that the inability of ATRA-induced NB4 cells to express the LF gene correlates with persistent binding of the CDP/cut protein to the LF CDP/cut binding site.
Persistent binding of CDP/cut to CDP/cut site in the LF promoter.
(A) Electrophoretic mobility shift analysis was carried out using32P-labeled double-stranded oligos encoding the CDP/cut site in the LF promoter (DM2 probe, lane 1). Addition of nuclear extracts prepared from uninduced (NB4, U, lane 2) and 48-hour ATRA-induced (NB4, 2d ATRA, lane 3) NB4 cells resulted in the formation of specific protein-DNA complexes (arrows), which were specifically competed away by the addition of a 100-fold molar excess of unlabeled self (lane 4, 100 × DM2), or known CDP/cut binding oligos (ie, NCAM; lane 5). (B) Binding of CDP/cut to the C/EBPε promoter. EMSA analysis was carried out using 32P-labeled double-stranded oligos encoding the CDP/cut site in the C/EBPε promoter. Addition of nuclear extracts prepared from uninduced (NB4, U, lanes 2,5) and 48-hour ATRA-induced (NB4, 2d ATRA, lane 3) NB4 cells resulted in the formation of specific protein-DNA complex (arrow), which was specifically competed away by the addition of a 100-fold molar excess of unlabeled self (lane 4, × 100 DM2), or by the addition of increasing concentrations of a known CDP/cut binding site (ie, NCAM; lanes 6-8).
Persistent binding of CDP/cut to CDP/cut site in the LF promoter.
(A) Electrophoretic mobility shift analysis was carried out using32P-labeled double-stranded oligos encoding the CDP/cut site in the LF promoter (DM2 probe, lane 1). Addition of nuclear extracts prepared from uninduced (NB4, U, lane 2) and 48-hour ATRA-induced (NB4, 2d ATRA, lane 3) NB4 cells resulted in the formation of specific protein-DNA complexes (arrows), which were specifically competed away by the addition of a 100-fold molar excess of unlabeled self (lane 4, 100 × DM2), or known CDP/cut binding oligos (ie, NCAM; lane 5). (B) Binding of CDP/cut to the C/EBPε promoter. EMSA analysis was carried out using 32P-labeled double-stranded oligos encoding the CDP/cut site in the C/EBPε promoter. Addition of nuclear extracts prepared from uninduced (NB4, U, lanes 2,5) and 48-hour ATRA-induced (NB4, 2d ATRA, lane 3) NB4 cells resulted in the formation of specific protein-DNA complex (arrow), which was specifically competed away by the addition of a 100-fold molar excess of unlabeled self (lane 4, × 100 DM2), or by the addition of increasing concentrations of a known CDP/cut binding site (ie, NCAM; lanes 6-8).
Discussion
Gaining an understanding of the role played by closely related members of a transcription factor family such as the C/EBP family in a differentiation model has proved to be challenging. In vitro methods relying on overexpression models and transient transfection analyses have limitations, as closely related members such as C/EBPα and C/EBPε often produce identical results (Figure1).12-14,16,24,25,27 In this study, we have used chromatin immunoprecipitation (ChIP), a powerful method used to assess the status of transcription-factor binding to promoters of interest in live cells,58 to assess the role of 2 closely related members of the C/EBP family of transcription factors (α and ε) in myeloid cell differentiation. Our findings reveal that C/EBPα, which is expressed very early in the myeloid development program,27binds to both primary (HNP and NE gene) as well as secondary (LF, and HNC [data not shown]) granule protein gene promoters in uninduced cells in the 3 myeloid development cell systems used in this study. It is evident from our ChIP analysis that C/EBPα may act as a negative regulator by binding to cis elements either as a homodimer or as a heterodimer with a yet-to-be-defined binding partner in cells not expressing its target genes. It is unclear whether C/EBPα exerts this negative regulatory effect via its own previously described negative regulatory subdomain,59 or whether its putative binding partner has intrinsic negative regulatory properties. Recent studies have shown that disruption of the E2F repression domain within the C/EBPα gene is associated with abnormalities in both adipogenesis and granulopoiesis in transgenic mice.60
Our data confirm the previous observations that C/EBPε binds to and up-regulates the expression of the secondary granule protein genes (LF [Figure 2A-B] and HNC [data not shown]).12-14 In addition, we demonstrate that some primary granule protein genes are regulated by C/EBPε. Whereas murine neutrophil elastase (mNE) expression does not appear to be C/EBPε-dependent tin ATRA-induced MPRO cells (Figure 2C), the α defensins (HNP 1,3) do appear to require C/EBPε for high-level expression in ATRA-induced NB4 cells (Figure 4). As noted in “Results,” previous studies have demonstrated that defensins (HNP) are expressed later in the neutrophil maturation sequence than the other primary granule protein genes, and their synthesis overlaps the onset of SGP gene expression.49 Furthermore, the absent expression of defensins (HNP) in specific granule deficiency (SGD), in which the underlying defect is C/EBPε-associated, suggests that HNP expression, like the SGP genes, is C/EBPε-dependent.39
The presence of both C/EBPα and C/EBPε at the C/EBP binding site in the LF promoter in induced cells suggests that binding of both family members is necessary for high-level LF expression in the MPRO and 32Dwt18 cell systems (Figure 3A-B). A similar pattern of C/EBPα and C/EBPε binding at the LF C/EBP site was also observed in both uninduced and ATRA-induced NB4 cells (Figure 5). However, ATRA-induction of NB4 cells does not result in LF expression (Figure4). It therefore appears that the C/EBPα/ε heterodimer is not sufficient to mediate high expression levels of LF in the acute promyelocytic leukemic (APL) NB4 cell line harboring the t(15;17) translocation resulting in the PML/RARα fusion protein.45 Previous studies have suggested that the PML/RARα fusion protein may interfere with the function of C/EBPα by inhibiting the ability of C/EBPα to bind to cognate cis elements in the promoters of C/EBP-regulated genes, thereby blocking myeloid differentiation in APL cells (reviewed in Tenen48and references therein). The authors have further suggested a direct interaction of C/EBPα with the PML/RARα fusion protein, which is lost upon ATRA induction of APL cells resulting in increased binding of C/EBPα to its binding sites.48 Our ChIP data, however, show that C/EBPα is bound to the HNP C/EBP site in uninduced NB4 cells. Induction of HNP expression in these cells upon ATRA induction results in the loss of C/EBPα binding to its cis element (Figure 5). Based on our observations, it is likely that in APL cells the loss rather than the previously hypothesized gain48 of C/EBPα binding, in association with a concomitant gain of C/EBPε binding, is correlated with gene expression.
It seems likely that the defect associated with nonexpression of LF in NB4 cells induced with ATRA is not attributable to aberrant C/EBP binding since C/EBPα binding remains unimpaired at the LF C/EBP binding site. Previous studies from our laboratory have shown that CCAAT displacement protein (CDP/cut), a highly conserved silencing factor binds the LF promoter and modulates its activity during myeloid differentiation.32,33 ChIP analysis revealed that CDP/cut was recruited to the CDP/cut binding sites in uninduced NB4 cells in the promoters of all 4 myeloid-specific gene promoters examined (Figure6A). ATRA induction of NB4 cells resulted in loss of binding of CDP/cut in the HNP, C/EBPε, and gp91phox gene promoters. This loss of CDP/cut binding appears to correlate with expression of these 3 genes in ATRA-induced NB4 cells (Figure 4). CDP/cut binding, however, was not abolished in the LF promoter in ATRA-induced NB4 cells (Figure 6A, top panel, lane 2), an observation that correlates with the nonexpression of LF in the NB4 cell system. Changes in CDP/cut binding governing differential stage-specific gene expression are thought to be mediated by posttranslational modification of the CDP/cut protein rather than changes in the level of CDP/cut expression (reviewed in Nepveu61). Whereas this posttranslational activity remains normal at the HNP, C/EBPε, and gp91phox CDP/cut binding sites, we propose that the CDP/cut modifying activity associated with the LF CDP/cut site is defective in APL cells.
This observed differential repressive activity of CDP/cut may be inherent in the molecule itself. CDP/cut is a homeobox protein containing 3 highly conserved DNA-binding repeats referred to ascut repeats, each of which is capable of recognizing and binding specific DNA motifs in target genes (reviewed in Nepveu61). Both acetylation of CDP/cut via p300/CBP and phosphorylation of CDP/cut are posttranslational modifications that have been postulated to regulate CDP/cut function.56 62 We hypothesize that CDP/cut uses a different one (or more) of its 4 DNA-binding elements to bind to the CDP/cut motifs in different myeloid promoters during neutrophil maturation. Differential modification, involving either phosphorylation and/or acetylation, of CDP/cut-DNA complexes in the promoters of these genes likely results in the observed differential repression exerted by CDP/cut during neutrophil development. A defect in the ability to specifically modify the CDP/cut-DNA complex in the LF promoter is thus likely to be responsible, in part, for the observed persistent binding of CDP/cut to the LF promoter in ATRA-induced NB4 cells. Since LF is expressed later in myeloid maturation than any of the other 3 genes examined in this study, we predict that CDP/cut binding to the site in the LF promoter would be modulated by a different mechanism of posttranslational modification that would likely be shared with other secondary granule protein genes.
In summary, we have demonstrated specific roles for both C/EBPα and C/EBPε in modulating the expression of LF gene expression in the maturing neutrophil. We have, in addition, demonstrated that the defect associated with lack of LF expression in an APL cell model does not involve defective C/EBP binding. That defect may be explained, in part, by the persistent binding of the repressor CDP/cut to the LF promoter.
The authors would like to thank Dr Steve Ackerman and members of his laboratory for valuable technical advice and Dr Arch Perkins for critically reading the manuscript. We also thank members of the Myeloid Stem Cell group at Yale University School of Medicine for helpful insights and discussions.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/ blood-2002-09-2767.
Supported by National Institutes of Health awards RO1-DK53471 and PO1-HL63357 (N.B.) and by the Anna G. and Argall L. Hull Cancer Research Award (A.K.-G.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Nancy Berliner, Department of Internal Medicine, Section of Hematology, WWW-428, Yale University School of Medicine, 333 Cedar St, New Haven, CT, 06510; e-mail:nancy.berliner@yale.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal