HFE is a nonclassical class I molecule that associates with β2-microglobulin (β2m) and with the transferrin receptor. HFE accumulates in transferrin-containing endosomes, and its overexpression in human cell lines correlates with decreased transferrin receptor (TFR)–mediated iron uptake and decreased intracellular iron pools. A mutation that interferes with proper folding and assembly of HFE complexes results in a severe iron-overload disease hereditary hemochromatosis. We previously suggested that viruses could also interfere with iron metabolism through the production of proteins that inactivate HFE, similarly to classical class I proteins. In particular, we demonstrated in a transient expression system that human cytomegalovirus (HCMV) US2 targeted HFE for proteasomal degradation. Here we demonstrate that the stable expression of HCMV US2 in HEK 293 cells constitutively expressing HFE leads to loss of HFE expression both intracellularly and on the cell surface, and the significant reduction of classical class I expression. Both HFE and classical class I molecules are targeted to degradation via a similar pathway. This HCMV US2-mediated degradation of HFE leads to increased intracellular iron pools as indicated by reduced synthesis of TfR and increased ferritin synthesis. Whether this interference with regulation of iron metabolism potentiates viral replication and/or promotes damage of HCMV-infected tissues remains to be determined. Nevertheless, the deleterious effect of US2 on the expression of HFE and classical class I major histo-compatibility complexes (MHC) provides HCMV with an efficient tool for altering cellular metabolic functions, as well as supporting the escape of virus-infected cells from cytotoxic T lymphocyte (CTL)–mediated immune responses.
Introduction
Human cytomegalovirus (HCMV) down-regulates the surface expression of major histocompatibility complex (MHC) class I molecules.1-3 Analysis of HCMV deletion mutants led to the identification of genes within the short segment of the HCMV genome, ie, US2, US3, US6, and US11 that independently mediate this effect.4-11 Two of these gene products, US2 and US11, target class I heavy chains for dislocation from the endoplasmic reticulum (ER) to the cytosol.6,7 The heavy chains are deglycosylated by N-glycanase and, subsequently, degraded by the proteasome. The general mode of action of the 2 proteins seem to be similar, but they differ in their ability to attack allelic class I heavy chain gene products.12 It has been suggested that US2 and US11 specifically down-regulate molecules that are associated with classical antigen presentation pathways because they preferentially promote the degradation of HLA-A and -B loci products relative to those encoded by HLA-C and -G loci.13-15Supporting the association of HCMV US2 with reduced antigen presentation by HCMV-infected cells and their escape from specific immune responses are data showing that US2 also down-regulates class II MHC DMα and DRα molecules.16,17 Most effects of US2 were demonstrated using an astrocytoma cell line; however, recently both US2 and US11 have been shown to down-regulate class I, but not class II, MHC molecules in human dendritic cells.18
We have previously observed that US2, but not US11, affects the stability of the nonclassical class I molecule, HFE, that regulates iron metabolism.19 It is commonly accepted that the high-affinity binding of HFE to the transferrin receptor (TfR) leads to reduced intracellular iron stores in HFE-transfected cells.20-23 A mutation in the HFE protein (Cys282Tyr), that interferes with the proper folding, assembly, and hence cell surface expression of the HFE molecule,24 is responsible for most cases of the iron overload disease, hereditary hemochromatosis (HH).25 Not surprisingly,β2-microglobulin (β2m)-knockout (KO) mice,26,27 as well as HFE KO mice,28 suffer from iron overload similar to that seen in patients with HH.
Using a panel of human HFE monoclonal antibodies (mAbs) and cell lines infected with recombinant vaccinia viruses (rVVs) expressing HFE, we previously showed that the assembly and stability of HFE complexes do not depend on a functional transporter of peptides and that HFE/β2m complexes, as well as TfR-associated HFE/β2m complexes, are stable and mature in human cell lines overexpressing HFE.19 The introduction of HCMV US2 expression by rVV coinfection prevented the expression of HFE complexes in both human and mouse cell lines. This demonstrated that the effect of HCMV US2 was not cell line specific. In these US2-infected cells the expression of free HFE heavy chains, as well as conformed HFE/β2m and TfR-associated HFE/β2m complexes, was reconstituted by the addition of proteasome inhibitors.
In this report we demonstrate that HCMV US2 down-regulates the expression of both class I MHC and HFE-transfected HEK 293 cells. Furthermore, we show that in HFE overexpressing HEK 293 cells, TfR expression is up-regulated concomitant with down-regulation in the expression of the iron storage protein, ferritin. The introduction of HCMV US2 expression reverses these effects. Similarly, HCMV US2 expression in HEK 293 cells results in increased ferritin levels. Thus, a single viral protein acts to both subvert the immune response and to interfere with basic cellular metabolic functions. Both effects may potentiate viral replication, as well as promote damage of particular tissues that are chronically infected with HCMV.
Materials and methods
Cell cultures
HEK 293 cells and stable transfectants were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 2 mM glutamine and 10% fetal calf serum (FCS). Media and supplements were purchased from Biological Industries (Beit Haemek, Israel).
Stable transfection
HEK 293 cells were transfected with human HFE cDNA (a kind gift from Dr J. Feder, Progenitor; CA24) by calcium phosphate-DNA coprecipitation and selected in G418-containing media as described previously.29 HEK 293 cells and one of the HFE-transfected clones were further cotransfected with HCMV US2/pcDNA3.1 Zeo (Invitrogen, The Netherlands). Individual clones were selected and expanded.
Antibodies
The following antibodies were used in the course of experiments: antihuman HFE 2F5 that recognizes HFE/β2m heterodimers and HFE cytoplasmic tail (CT) that recognizes free HFE heavy chains (described previously19); V1-10 (antihuman TfR; a kind gift of Dr Z. Eshhar, The Weizmann Institute of Science, Rehovot, Israel); H68.4 (anti-TfR directed against a conserved region in the N-terminal domain of the protein; Zymed Laboratories, CA); rabbit antihuman ferritin (DAKO, Denmark); W6/32 (anti-HLA-A, -B, -C30); rabbit anti-US2 and rabbit antihuman class I heavy chain antibodies were a kind gift from Dr H. Ploegh (Harvard Medical School, Boston, MA); DO1 (antihuman p53, a kind gift of Dr M. Oren, The Weizmann Institute of Science).
Fluorescence-activated cell sorter (FACS) analysis
Cells were harvested, incubated at 4°C with the first antibody for 60 minutes, washed, and then incubated for 45 minutes with the second antibody (fluorescein isothiocyanate [FITC]-conjugated goat antimouse immunoglobulin G [IgG]; Jackson ImmunoResearch Laboratories, PA). The cells were washed with phophate-buffered saline (PBS), and fluorescence intensity was measured with a Becton Dickinson Cell Sorter (Becton Dickinson, CA).
Metabolic labeling, immunoprecipitation, and Western blot analysis
Standard procedures were used.19 For metabolic labeling and immunoprecipitation, cells were split 1 day before metabolic labeling. Briefly, cells were starved for 120 minutes in methionine + cysteine-free medium, labeled in the same medium containing 3.7 MBq/mL (100 μCi/mL)35[S]-methionine (PerkinElmer Life Sciences, MA), and chased as indicated. The protease inhibitors MG132 (30 μM; Calbiochem-Novabiochem, Germany) was added to the cells 80 minutes prior to radioactive pulsing, as well as during the pulse and the chase. The cells were lysed with a buffer containing 0.5% NP40 (Sigma Chemical, MO), 50 mM Tris (tris[hydroxymethyl]aminomethane), pH 7.5, and 150 mM NaCl and immunoprecipitated with the indicated antibodies and protein A or protein G agarose (Boehringer Mannheim GmbH, Germany). All the antibodies used in immunoprecipitation were titrated by sequential immunoprecipitations and consequently were added in access so that more than 90% of the labeled antigen was immunoprecipitated in the first round of immunoprecipitation. The total protein concentration was determined with Bradford reagent (Sigma Chemical), and equivalent protein amounts were immunoprecipitated. The immunoprecipitates were fractionated on sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) as indicated, and the dried gels were exposed to X-OMAT AR X-ray films (Eastman Kodak Company, NY). For Western blot analysis, cell lysates were fractionated, and the proteins were transferred to nitrocellulose Hybond-C membrane (Amersham Pharmacia Biotech, United Kingdom). The membrane was incubated with 5% dry milk/PBS, probed with the specific antibody, and detected with the appropriate horseradish peroxidase–conjugated secondary antibodies, and enhanced chemiluminescence substrates (Amersham Pharmacia Biotech, United Kingdom).
RNA analysis and RT-PCR
Cytoplasmic RNA was prepared as previously described.29 Reverse transcription (RT) of RNA was carried out for 1 hour at 42°C, in 20 μL RT buffer containing 1 μg total RNA, 0.5 μg Oligo dT primer (Promega, WI), nucleotide mixture (1 mM), and 15 units AMV reverse transcriptase (Promega), and 24 units RNasin ribonuclease inhibitor (Promega). Polymerase chain reaction (PCR) was carried out in the appropriate buffer with 1 μL cDNA, 20 to 25 pmol of the relevant primers, 0.8 mM nucleotide mixture, and 1.5 units Taq DNA polymerase (Biological Industries). The following primers were used: HFE sense, 5′-TGGCAAGGGTAAACAGATCC-3′; α-sense, 5′-CTCAGGCACTCCCTCTCAACC-3′; HCMV US2 sense, 5′-CGGAATTCCGCGCATGAACAATCTCTGGAAAGC-3′; α-sense, 5′-GCTCTAGAGCTCAGCACACGAAAAACCG-3′; actin sense, 5′-GTTTGAGACCTTCAACACCCC-3′; α-sense, 5′-GTGGCCATCTCTTGCTCGAAGTC-3′. After appropriate number of PCR cycles (MiniCycler; MJ Research, MA), 10 μL reaction mix was fractionated on agarose gel.
Results
TfR synthesis is induced and ferritin synthesis is reduced in HFE-transfected HEK 293 cells
HEK 293 cells were transfected with HFE, and selected clones were further analyzed for cell surface expression of HFE and HLA class I complexes. The expression of HFE/β2m heterodimers was analyzed with anti-HFE mAb, 2F5, that recognizes an epitope on the HFE/β2m heterodimer that is masked following association with TfR.19 The expression of HLA was analyzed with mAb W6/32 that recognizes a public epitope on HLA-A, -B, and -C complexes. Figure1A demonstrates that 293/HFE cells express high levels of HFE/β2m complexes; however, the expression of HLA complexes is reduced relative to the parental HEK 293 cells. A possible explanation for these observations is that HFE and HLA class I heavy chains compete for binding with the available β2m. Indeed, comparison of the relative amounts of conformed HLA complexes with free HLA heavy chains (Figure 1B) in 293/HFE versus HEK 293 cells by a quantitative immunoprecipitation revealed that the relative amount of conformed HLA-A, -B, and -C complexes is reduced, whereas that of free class I heavy chains is increased in 293/HFE cells, supporting the hypothesis that there is competition for β2m.
Cell surface and intracellular expression of HFE and class I MHC molecules in HFE-transfected HEK 293 cells.
HEK 293 and HFE-transfected HEK 293 cells were analyzed by FACS for cell surface expression of HFE and HLA (A) or metabolically labeled with 35[S]methionine for 1 hour and lysed, and cell lysates were immunoprecipitated (B) with Abs directed against hHFE (2F5) HLA (W6/32), and HLA heavy chains (αHC). Abs were used in excess as indicated in “Materials and methods.” Bold-lined histograms indicate staining with the first Ab; regular-lined histograms, background staining.
Cell surface and intracellular expression of HFE and class I MHC molecules in HFE-transfected HEK 293 cells.
HEK 293 and HFE-transfected HEK 293 cells were analyzed by FACS for cell surface expression of HFE and HLA (A) or metabolically labeled with 35[S]methionine for 1 hour and lysed, and cell lysates were immunoprecipitated (B) with Abs directed against hHFE (2F5) HLA (W6/32), and HLA heavy chains (αHC). Abs were used in excess as indicated in “Materials and methods.” Bold-lined histograms indicate staining with the first Ab; regular-lined histograms, background staining.
Next, we analyzed whether the overexpression of HFE affects intracellular iron homeostasis by determining the levels of the iron-transporter receptor, TfR, and the iron-storage protein, ferritin, in 5 individual HFE-expressing clones. The levels of newly synthesized ferritin were reduced, whereas TfR levels were increased in all clones (Figure 2). These data suggest that HEK 293 cells overexpressing functional HFE complexes have reduced intracellular iron pools, leading to induced translation of TfR and reduced translation of ferritin. Similar data were obtained with HFE-transfected HeLa cells22,31-33 but not in TfR-deficient Chinese hamster ovary (CHO) cells that have been reconstituted for the expression of human HFE, human TfR, and human β2m.34
HFE-transfected HEK 293 cells demonstrate induced synthesis of TfR and reduced synthesis of ferritin.
HEK 293 and 293/HFE cells (clones 1-5) were metabolically labeled with35[S]methionine for 1 hour, and the cell lysates were immunoprecipitated with Abs directed against HFE/β2m, 2F5, and ferritin. Cell lysates were fractionated and tested by Western blot analysis for the expression of TfR with H68.4 Ab.
HFE-transfected HEK 293 cells demonstrate induced synthesis of TfR and reduced synthesis of ferritin.
HEK 293 and 293/HFE cells (clones 1-5) were metabolically labeled with35[S]methionine for 1 hour, and the cell lysates were immunoprecipitated with Abs directed against HFE/β2m, 2F5, and ferritin. Cell lysates were fractionated and tested by Western blot analysis for the expression of TfR with H68.4 Ab.
HCMV US2 down-regulates the expression of HFE and class I molecules via a similar mechanism
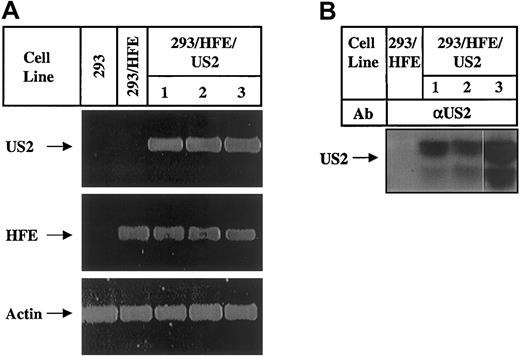
One of the HFE-transfected HEK 293 clones was further transfected with HCMV US2 cDNA. Three individual clones that expressed theUS2 gene as determined by RT-PCR (Figure3A) and immunoprecipitation (Figure 3B) were chosen for further studies. Cell surface expression of HFE and HLA class I complexes was analyzed and revealed a complete or a partial reduction in HFE expression but no significant changes in the level of HLA expression (Figure 4A). Most likely, the absence of change in cell surface HLA class I expression results from the assembly of residual HLA class I heavy chains (Figure 1B) with β2m made available by HFE degradation. Otherwise, US2 is effective in reduction of class I as demonstrated by the 3-fold reduction in cell surface HLA class I complexes expressed by US2-transfected HEK 293 cells (Figure 4B). The decrease in cell surface HLA class I is correlated with reduced level of newly synthesized free HLA class I heavy chains (Figure 4C, top) and total HLA molecules (Figure 4C, bottom) in these cells. Next, the intracellular levels of HFE and HLA class I molecules were analyzed in 293/HFE/US2 cells. Figure5A demonstrates that the levels of newly synthesized free HFE chains, HFE/β2m, TfR-associated HFE/β2m complexes are dramatically reduced. The clonal variation in US2-mediated reduction of intracellular HFE (Figure 5) was consistent with that observed for cell surface HFE reduction (Figure 4A). Figure5B shows that the level of newly synthesized free HLA class I heavy chains was greatly reduced, whereas the level of conformed class I complexes was unchanged. The latter is in agreement with results shown in Figure 4A. The detection of comparable levels of conformed class I complexes in immunoprecipitates from cell lysates and on the cell surface agrees with the idea that US2-mediated HFE degradation in 293/HFE/US2 cells promotes class I complex formation by increasing the supply of β2m. Whereas US2 mediated reduction in the levels of total HFE and total HLA, it did not affect the level of a nonrelevant protein, p53, in US2-transfected 293/HFE cells (Figure 5C), demonstrating that US2 is not functioning nonspecifically. The overall data presented in Figures 4 and 5 suggest that both HFE and HLA class I molecules are down-regulated in US2-expressing HEK 293 cells. The data demonstrate that, despite the similar effect of US2 on HFE and class I molecules, the outcome, in this system, is not identical and that US2 appears to have a more dramatic effect on free HFE chains and conformed HFE complexes than on HLA molecules.
Characterization of HEK 293 cells transfected with HFE and US2.
HEK 293, 293/HFE, and US2-transfected 293/HFE clones (1-3) were analyzed for expression of US2, HFE, and actin mRNA by RT PCR (A) and US2 protein by metabolic labeling and immunoprecipitation with rabbit anti-US2 (αUS2) Abs (B).
Characterization of HEK 293 cells transfected with HFE and US2.
HEK 293, 293/HFE, and US2-transfected 293/HFE clones (1-3) were analyzed for expression of US2, HFE, and actin mRNA by RT PCR (A) and US2 protein by metabolic labeling and immunoprecipitation with rabbit anti-US2 (αUS2) Abs (B).
US2 expression leads to decreased cell surface levels of HFE and HLA class I molecules in 293/HFE/US2 and 293/US2 cells, respectively.
The 293/HFE/US2 (clones 1-3) cells were analyzed by FACS (A) with anti-HFE mAb 2F5 and anti-HLA mAb W6/32. 293/US2 (clones 1, 2) cells were analyzed by FACS (B) with anti-HLA mAb W6/32, metabolically labeled with 35[S]methionine for 1 hour, followed by immunoprecipitation with Abs directed against free HLA class I HC (C, top), and analyzed for expression of total HLA class I (C, bottom) by Western blot hybridization of fractionated cell lysates with Abs directed against free HLA class I HC. Bold-lined histograms indicate staining with first Ab; regular-lined histograms, background staining.
US2 expression leads to decreased cell surface levels of HFE and HLA class I molecules in 293/HFE/US2 and 293/US2 cells, respectively.
The 293/HFE/US2 (clones 1-3) cells were analyzed by FACS (A) with anti-HFE mAb 2F5 and anti-HLA mAb W6/32. 293/US2 (clones 1, 2) cells were analyzed by FACS (B) with anti-HLA mAb W6/32, metabolically labeled with 35[S]methionine for 1 hour, followed by immunoprecipitation with Abs directed against free HLA class I HC (C, top), and analyzed for expression of total HLA class I (C, bottom) by Western blot hybridization of fractionated cell lysates with Abs directed against free HLA class I HC. Bold-lined histograms indicate staining with first Ab; regular-lined histograms, background staining.
US2 expression leads to decreased synthesis and total level of HFE and HLA class I molecules.
293/HFE/US2 (clones 1-3) cells were metabolically labeled with35[S]methionine for 1 hour, followed by sequential immunoprecipitation with anti-HFE Abs [αhHFE(2F5)] (2F5 followed by anti-HFE CT [αHFE (CT)]) and anti-TfR (αTfR) (A) or anti-HLA (αHLA) Abs (B). HFE/β2m complexes were detected by mAbs 2F5, free HFE heavy chains were detected with anti-HFE CT, and TfR-associated HFE complexes were detected with anti-TfR (V1-10) mAbs. HLA complexes were detected with W6/32, and free class I heavy chains were detected with anti-HLA HC. Cell lysates from HEK 293, 293/HFE, and 293/HFE/US2 were fractionated on SDS-PAGE (C) and analyzed by Western blot hybridization for the expression of total HFE (by anti-HFE CT), total HLA class I (by anti-HLA HC), and p53 (DO1).
US2 expression leads to decreased synthesis and total level of HFE and HLA class I molecules.
293/HFE/US2 (clones 1-3) cells were metabolically labeled with35[S]methionine for 1 hour, followed by sequential immunoprecipitation with anti-HFE Abs [αhHFE(2F5)] (2F5 followed by anti-HFE CT [αHFE (CT)]) and anti-TfR (αTfR) (A) or anti-HLA (αHLA) Abs (B). HFE/β2m complexes were detected by mAbs 2F5, free HFE heavy chains were detected with anti-HFE CT, and TfR-associated HFE complexes were detected with anti-TfR (V1-10) mAbs. HLA complexes were detected with W6/32, and free class I heavy chains were detected with anti-HLA HC. Cell lysates from HEK 293, 293/HFE, and 293/HFE/US2 were fractionated on SDS-PAGE (C) and analyzed by Western blot hybridization for the expression of total HFE (by anti-HFE CT), total HLA class I (by anti-HLA HC), and p53 (DO1).
HCMV US2 targets HFE to proteasome-dependent degradation
We have previously demonstrated that US2 HCMV targets HFE to proteasome-mediated degradation in HeLa cells coinfected with rVV-HFE and rVV-US2. However, although previous publications demonstrated the accumulation of a deglycosylated class I heavy chain in some US2-transfected cell lines that have been treated with proteasome inhibitors,35 we could not detect a smaller HFE fragment in infected cells that have been treated with MG132 or lactacystin. A better analysis of the mechanism of US2-mediated HFE degradation could be performed in 293/HFE/US2 cells. Figure6 demonstrates that the expression of free HFE and HFE/β2m complexes was reconstituted following treatment of cells with the proteasome inhibitor MG132. The same results were obtained following treatment with lactacystin (data not shown). Similar to our previous results,19 it appears that the conformed complexes are not retranslocated to the cytosol in MG132-treated cells as indicated by the strong signal observed in the 2F5 immunoprecipitate. However, a smaller band that corresponds in size to N-glycanase–treated HFE was observed in the free HFE immunoprecipitate (Figure 6B), whereby the mechanism of US2-mediated degradation of HFE is similar to that of classical class I heavy chains. The heavy chains are translocated to the cytosol, deglycosylated, and, subsequently, degraded. Therefore, in the presence of a proteasome inhibitor, although a deglycosylated form of HFE is stabilized, most of the molecules are glycosylated and about half of the molecules remain conformed as demonstrated previously.19
US2 targets HFE for proteasome-mediated degradation.
293/HFE and 293/HFE/US2 cells were untreated or treated with the proteasome inhibitor MG132 followed by metabolic labeling with35[S]methionine for 30 minutes, chase for 50 minutes, and immunoprecipitation. Cell lysates were treated with N-glycanase (0.2 units) as indicated, and immunoprecipitated with mAb 2F5 (anti-HFE/β2m, A) followed by anti-HFE CT (anti-HFE heavy chains, B). Immunoprecipitates were separated on 8% SDS-PAGE. Short (A,B) and long exposure of the boxed area (C) are presented. * indicates a nonspecific band.
US2 targets HFE for proteasome-mediated degradation.
293/HFE and 293/HFE/US2 cells were untreated or treated with the proteasome inhibitor MG132 followed by metabolic labeling with35[S]methionine for 30 minutes, chase for 50 minutes, and immunoprecipitation. Cell lysates were treated with N-glycanase (0.2 units) as indicated, and immunoprecipitated with mAb 2F5 (anti-HFE/β2m, A) followed by anti-HFE CT (anti-HFE heavy chains, B). Immunoprecipitates were separated on 8% SDS-PAGE. Short (A,B) and long exposure of the boxed area (C) are presented. * indicates a nonspecific band.
Expression of US2 HCMV reconstitutes TfR and ferritin levels
Because HFE overexpression results in an iron-starved phenotype (Figure 2), we expected that the degradation of HFE because of US2 expression would lead to reconstitution of intracellular iron homeostasis. Figure 7A shows that indeed, in the 3 US2-expressing clones, the synthesis of TfR is reduced and the synthesis of ferritin is increased as compared with the 293/HFE cells. Quantitative analysis of the total ferritin and TfR levels in US2-expressing cells by sequential immunoprecipitations with anti-TfR and antiferritin antibodies revealed that the effect on ferritin was more dramatic than the effect on TfR expression (data not shown). Thus, the US2-mediated degradation of HFE leads to a complete reconstitution of intracellular iron homeostasis. We observed that in the US2 transfectants ferritin levels were higher than in HEK 293 cells, suggesting that US2 was affecting more than the transfected HFE. To investigate this further we analyzed the level of TfR (Figure 7B, top), and ferritin (Figure 7B, bottom) in US2-transfected HEK 293 cells. The results demonstrate that US2-expressing cells have increased levels of ferritin in comparison to the parental cells but not consistent changes in TfR. These changes in ferritin are unlikely to be due to a direct effect of US2 on ferritin because these 2 proteins exist in different cellular compartments; however, they may be due to US2-mediated degradation of endogenous HFE. Thus, it appears that ferritin regulation is more sensitive than TfR regulation to changes in the level of intracellular iron pools, and that US2 expression results in altered intracellular iron homeostasis in the HEK 293 cell line despite the low expression level of endogenous HFE.
Regulation of TfR and ferritin synthesis in US2-transfected HEK 293 and 293/HFE cells.
HEK 293, 293/HFE, and 293/HFE/US2 (clones 1-3) cells (A) and 293/US2 (clones 1,2) cells (B) were metabolically labeled with35[S]methionine for 1 hour and immunoprecipitated with Abs directed against human TfR (V1-10) and ferritin. Immunoprecipitates were separated by 13% SDS-PAGE.
Regulation of TfR and ferritin synthesis in US2-transfected HEK 293 and 293/HFE cells.
HEK 293, 293/HFE, and 293/HFE/US2 (clones 1-3) cells (A) and 293/US2 (clones 1,2) cells (B) were metabolically labeled with35[S]methionine for 1 hour and immunoprecipitated with Abs directed against human TfR (V1-10) and ferritin. Immunoprecipitates were separated by 13% SDS-PAGE.
Discussion
HCMV is a ubiquitous herpesvirus that infects a variety of tissues and cell types, in particular epithelial and endothelial cells, but also fibroblasts, smooth muscle cells, and macrophages.36,37 HCMV infection causes a mild disease that can be associated with hepatitis in immunocompetent hosts,38 39 but it is a life-threatening disease in immunosuppressed patients, including transplantation and AIDS patients.
It is commonly accepted that viruses have evolved a variety of means of interfering with immune responses. Specifically, HCMV encodes several proteins that interfere with class I MHC–mediated antigen presentation, leading to evasion from cellular immune responses.35,40 Because HCMV infection is often associated with diseases of the gastrointestinal tract,41 the site of expression of HFE, and because HFE is closely related to classical class I MHC molecules that are targets for some HCMV encoded molecules, we postulated that these viral-encoded proteins may have a broader scope of action.
We have previously demonstrated19 that coinfection of rVV-expressing HFE and US2 prevents the expression of free HFE, HFE/β2m complexes, and TfR-associated HFE complexes in both human and mouse cell lines. In this study we demonstrate that HCMV US2 down-regulates the expression of HFE in HFE-transfected HEK 293 cells (Figures 4A and 5A,C). The expression of free HFE chains, conformed HFE/β2m, and TfR-associated HFE/β2m complexes was reconstituted in the presence of proteasome inhibitors (Figure 6). These data are consistent with the model of US2-mediated retranslocation of class I proteins for proteasomal degradation.
US2-mediated regulation of HFE has wide-reaching implications for iron metabolism. Expression of HFE in transfected cells results in decreased levels of ferritin and increased levels of TfR (Figure 2), changes that are consistent with an iron-starved phenotype. This situation is not surprising because HFE is thought to down-regulate intracellular iron.22,31-33 In US2-transfected cells HFE levels are reduced. Consequently, we observed that the expression of ferritin and TfR are reversed, such as ferritin synthesis is enhanced and TfR synthesis is inhibited (Figure 7A). US2 induced ferritin not only in cells overexpressing HFE but also in parental HEK 293 cells. In parallel to reduction of HFE, US2 induced reduction of HLA class I proteins (Figures 4B-C and 5B-C) as previously reported.40Thus, one viral protein may be involved in the subversion of immune responses and regulation of factors involved in iron hemostasis. The 2 effects might contribute to viral maintenance, to survival of virus-infected cells, and also to the damage of chronically infected tissues because of iron accumulation that results from the reduction in HFE expression and induced iron uptake.
The effect of US2 on HFE and on HLA heavy chains is similar but not identical. Although US2 almost completely abolishes intracellular and cell surface expression of HFE heavy chains and HFE complexes, it affects mainly free HLA heavy chains and does not significantly change the overall expression of conformed class I complexes. There are several possible explanations for these observations: US2 has higher affinity for HFE than for HLA molecules. The high concentration of HFE in the transfected cells may preferentially facilitate HFE association with US2 and its consequential degradation. In this case free HFE chains are no longer available for assembly with β2m, and most of the US2 is associated with HFE and is not available for association with residual HLA heavy chains. Alternatively, the HLA observed may consist of HLA-C–encoded molecules that are not affected by US2 expression.14 A less likely possibility is that US2 binds to both free and conformed HFE complexes, leading to translocation of both to the cytosol and subsequent degradation, whereas conformed HLA complexes are not translocated or are less efficiently translocated to the cytosol. The question of whether folded molecules can be translocated to the cytosol through the Sec61 complexes was recently discussed42; however, there is no proof yet that such a mechanism occurs in mammalian cells.
Current data show that HCMV US2 targets HLA-A and HLA-B locus products, but not HLA-C and HLA-G locus products for proteasome-dependent degradation despite the high homology between these genes at the predicted US2-binding site.13,43 However, in specific systems, US2 targets class II DR and DM complexes for degradation, despite their very low homology to HLA-A and HLA-B locus products.16 These observations raised the possibility that US2 was specifically selected for its ability to block class I and class II presentation pathways. Hence, MHC molecules that are expressed in specific tissues and might have specific functions such as HLA-G (expressed in the trophoblasts and protects from natural killer [NK]–mediated activity44) or HLA-C (known to have reduced surface stability45) may be somehow resistant to this effect. However, the data presented in this article show that US2 has a more dynamic effect. It targets to degradation a nonclassical class I molecule that is not homologous to class I at the predicted US2 binding site43 and does not play any direct role in the classical pathway of antigen presentation. How a viral protein can associate with completely different sequence elements remains an enigma. However, Chevalier et al17demonstrated recently that the US2 residues involved in binding to MHC class I differ subtly from those involved in binding to class II proteins. The US2 binding site on HFE was not characterized yet, but it might be HFE specific. Thus, the virus uses particular sequence elements in the same protein for altering functions that might promote its survival and replication. On one hand, it interferes with classical antiviral immune responses (mediated by class I and II complexes), and, on the other hand, it induces cellular iron uptake that might support its growth.
The fact that HCMV infection is often associated with tissue injury in the same tissues that HFE is highly expressed is particularly interesting in view of observations that iron supports viral replication and chronic infections. Hepatic iron concentration has consistently been observed as being directly correlated with the response to interferon therapy in the treatment of chronic hepatitis C virus infection.46 Moreover, treatment of patients with hepatitis C and iron overload with iron chelators improved their response to interferon therapy.47 In vitro studies demonstrated that iron enhances hepatitis C virus replication in cultured human hepatocytes.48 Replication of human immunodeficiency virus type 1 (HIV-1) can be influenced by iron as demonstrated by the fact that iron chelators inhibit virus replication.49,50 Iron chelators also inhibit CMV infection and CMV-induced pathogenic changes.51 Zoll et al52 showed recently that the Mengovirus leader protein interferes with antiviral immune responses through inhibition of iron/ferritin-mediated activation of nuclear factor κB (NFκB), a different pathway that correlates virus replication and spread with intracellular iron levels. Thus, increasing the cellular iron pool by down-regulation of HFE expression may be beneficial for viral persistence. The direct effect of HCMV infection on cellular iron metabolism is currently being analyzed.
We thank Dr C. Enns (Oregon Health Sciences University, Oregon) for the fruitful discussions; Drs Z. Eshhar, M. Oren (The Weizmann Institute of Science, Rehovot, Israel), and H. Ploegh (Harvard Medical School, Boston, MA) for the antibodies; and Dr N. Smorodinsky and M. Yaacubovicz for their collaboration in the generation of the antihuman HFE mAbs.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-07-2158.
Supported by the United States-Israel Binational Science Foundation (BSF), the EC Contract No. QLG1-CT-1999-00665, the Israel Ministry of Health, and by the Mozelsio Fund for Pediatric Cancer Research. S.V.-B.A. is a recipient of an ICRETT fellowship (International Union Against Cancer UICC, Switzerland).
Correspondence:Rachel Ehrlich, Department of Cell Research and Immunology, Tel Aviv University, Ramat Aviv 69978, Israel; e-mail: rachele@post.tau.ac.il.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

![Fig. 1. Cell surface and intracellular expression of HFE and class I MHC molecules in HFE-transfected HEK 293 cells. / HEK 293 and HFE-transfected HEK 293 cells were analyzed by FACS for cell surface expression of HFE and HLA (A) or metabolically labeled with 35[S]methionine for 1 hour and lysed, and cell lysates were immunoprecipitated (B) with Abs directed against hHFE (2F5) HLA (W6/32), and HLA heavy chains (αHC). Abs were used in excess as indicated in “Materials and methods.” Bold-lined histograms indicate staining with the first Ab; regular-lined histograms, background staining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045001.jpeg?Expires=1765897610&Signature=caRVH8TKgDgQYSAF8dJooxn5RaCTBz735ykrx0IZTI1jUpsm07RAS2zM-Chih1YIul35zcR8Cda0WXxzRV6dBnHJSifDI1UmNdg2f4GA7B-HB0eW6IdaGmf7W~0PUihtdQKEiXdyz-CPSuNkS5S2RCYhaogyZE7g~B3yTBuk2mfatJ1ij1cbCVcvQhcoGH1v6TnckYpd0x5PkPvzJpPAjWe37moifhgqhbrKUEV8xWQabL8oY8KkdEZ1mCgj1nU2pw~BTgkYt~ripKltH2H8dzqQlQjesZQG-3PUawaCFuXtqKbzt13I~EPkxw9aGOIf-ZCAf5bI7Tsb7GCI1WiHwg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 2. HFE-transfected HEK 293 cells demonstrate induced synthesis of TfR and reduced synthesis of ferritin. / HEK 293 and 293/HFE cells (clones 1-5) were metabolically labeled with35[S]methionine for 1 hour, and the cell lysates were immunoprecipitated with Abs directed against HFE/β2m, 2F5, and ferritin. Cell lysates were fractionated and tested by Western blot analysis for the expression of TfR with H68.4 Ab.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045002.jpeg?Expires=1765897610&Signature=Czh72uColgB2gT8Elmd~jmiJ2b~7ZK6LnlZcifuyx64C61fbg2QSs1RctJUNEPpdNjuawELnxzK3rHPYnRdQMTZQdMIFA4BQ727HaV6oHNrjPsLcb7WIDT5z4-sty34ANnHmgWsW9XednEMKpfccseX9ZRttgPcmlwUyss78PivyghQnL26jWqNZJIIWlPM~tYhx9jAcq38Ac~ojyidZQMgpvd0lZsrgt0qD13u1xfnLi1OnlnrAomJBGT1RDBFrLOCKYGp~f1VXCbvMRe0UAHqJcy8YnVsys64j2alIrG~AGBrlDxsc6EHd~Fqb7ynH5KYhtndqNQTaW82xxUvyRA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 4. US2 expression leads to decreased cell surface levels of HFE and HLA class I molecules in 293/HFE/US2 and 293/US2 cells, respectively. / The 293/HFE/US2 (clones 1-3) cells were analyzed by FACS (A) with anti-HFE mAb 2F5 and anti-HLA mAb W6/32. 293/US2 (clones 1, 2) cells were analyzed by FACS (B) with anti-HLA mAb W6/32, metabolically labeled with 35[S]methionine for 1 hour, followed by immunoprecipitation with Abs directed against free HLA class I HC (C, top), and analyzed for expression of total HLA class I (C, bottom) by Western blot hybridization of fractionated cell lysates with Abs directed against free HLA class I HC. Bold-lined histograms indicate staining with first Ab; regular-lined histograms, background staining.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045004.jpeg?Expires=1765897610&Signature=mXij1C5ldje8GQNRQqcqCO3~YO5kCyE6NqW8zNgzNyYNx6Oewod5h9ClE6Nfiki0FyYermThyw39NSlTbKEGSoKXgvYR4XfHOJnS-yV7MLOEvrHezhtjB7iZpvJ1i18-0IaKCHc8ORVv5QDL11il2eDGyrmvgrpigWo4UB-Idg-BF2X4CFK6Uns71hbJKm~cFNQfVLxeA852Z29hC7y4NWzMoAaSra7omAhhVjqJxHY6q-UKHHeGdmQ9f4rjUb5hKrC5Pxuvgsf3B4PYGTMvlqEAEBgmq6i2MZSxR0JD29uSkOG0SCntmq2QoAqRNWsfeBeQ2cZaro47r4gZYt~JdA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 5. US2 expression leads to decreased synthesis and total level of HFE and HLA class I molecules. / 293/HFE/US2 (clones 1-3) cells were metabolically labeled with35[S]methionine for 1 hour, followed by sequential immunoprecipitation with anti-HFE Abs [αhHFE(2F5)] (2F5 followed by anti-HFE CT [αHFE (CT)]) and anti-TfR (αTfR) (A) or anti-HLA (αHLA) Abs (B). HFE/β2m complexes were detected by mAbs 2F5, free HFE heavy chains were detected with anti-HFE CT, and TfR-associated HFE complexes were detected with anti-TfR (V1-10) mAbs. HLA complexes were detected with W6/32, and free class I heavy chains were detected with anti-HLA HC. Cell lysates from HEK 293, 293/HFE, and 293/HFE/US2 were fractionated on SDS-PAGE (C) and analyzed by Western blot hybridization for the expression of total HFE (by anti-HFE CT), total HLA class I (by anti-HLA HC), and p53 (DO1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045005.jpeg?Expires=1765897610&Signature=ktRYFwkQ7tnGEc94xNpobGJVhJXYDeTt-fghRj7HJCd5s1PS-4JaFLaeV6vjgxisJfo1n7a0UaGrLMVzXpYMXIWcmDoGpuE3SkUyGp7jEeTzyCatMfzoXz5fZ3EAL7~vnPdry9dtR21d5teAqDnTIXh30ga21icX6OowlNHTFnUAAsakylrHyst34e-JyQqFtEIvyEo4gwJi3Ick0rLA4ohLzUwJsCQEJGjtISRmeTpajlse2zYzlT3fk~mjUuwM0qzMoJ-p4NyZoMChrkGReQG6xdsZtDq94iaOauH3hbOS8AUnvYCSWG~eRDSm8hpEufaQNZFNwJ2~eKz-yp76Dg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 6. US2 targets HFE for proteasome-mediated degradation. / 293/HFE and 293/HFE/US2 cells were untreated or treated with the proteasome inhibitor MG132 followed by metabolic labeling with35[S]methionine for 30 minutes, chase for 50 minutes, and immunoprecipitation. Cell lysates were treated with N-glycanase (0.2 units) as indicated, and immunoprecipitated with mAb 2F5 (anti-HFE/β2m, A) followed by anti-HFE CT (anti-HFE heavy chains, B). Immunoprecipitates were separated on 8% SDS-PAGE. Short (A,B) and long exposure of the boxed area (C) are presented. * indicates a nonspecific band.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045006.jpeg?Expires=1765897610&Signature=TnBoTRTVO7U7m~bX4RgyrWs5ANfTw3Zk~ZeHyyrRFaRMHeeI6frIfH~siVdPVuWs0Z-twRRWLoNVjwFheyfU7vun12T1N5pVnDn-I0e0OUro2kMDIsy9wlS4JeJHZpjtVhPvEzKa63t2Ua3zr~18BFjewTKekNeYgJK~YsLjqVc4uZfRmgT4jCBHfVJldWQ5IvsxJPa0Dp~wAd3NoYvaNGaItHhOjpb6ruC3boX-0t3jMQQ-ne-OaGu6PN~evo-hN0HXc~PI4kqexB4CtuvxLVaWaUxWyNv~2ADl1~d5NOeA~cWH4~P8sE~ivWn6OhtZJ6Z4MQnXMZTyg5EfNF2jGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 7. Regulation of TfR and ferritin synthesis in US2-transfected HEK 293 and 293/HFE cells. / HEK 293, 293/HFE, and 293/HFE/US2 (clones 1-3) cells (A) and 293/US2 (clones 1,2) cells (B) were metabolically labeled with35[S]methionine for 1 hour and immunoprecipitated with Abs directed against human TfR (V1-10) and ferritin. Immunoprecipitates were separated by 13% SDS-PAGE.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-07-2158/3/m_h80734045007.jpeg?Expires=1765897610&Signature=w2lUsJcRKEcVreDv7bJV07MDpBXaYNBiYC90JK~l-7wBm2OESOypXK1gfvOujrI-C81Eg3gwbX2HoBnCp~83AV9kbvnTTuqcjW8sE9w01E-ey~JPWYF8GUuJmkq5T3kZrtnUtsvF8ln~xbVbGTDokmdjohvTnlexyRfBAkw-Fs74qqGVihgtcWgNUQGNAB9haAO5l3HQOj0e3CwkaCKhkdfaT8T0Q7BysouTg4j75pnGLkNIeTHo3atf2-~nn01ZtU4Hx9xfISB8mtlYjdUET9~1h4SHWf0MraNnYpJu1TmeCJl6bT3XrxI1dlSC0OAOzQeC2SLxI2ULkiJdbrzgnA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal