Through its Src homology 3 (SH3) and SH2 domains, the Src kinase Lyn interacts with a small number of phosphoproteins, such as Shc, Cbl, and Vav, which regulate cell cycle and the cytoskeleton. Using Lyn's Unique, SH3, and SH2 domains as bait in a yeast 2–hybrid screen, we isolated a novel gene product with features of a scaffolding protein. We named it Felic because it contains a domain homologous to the tyrosine kinase Fes and the cytoskeletal proteinezrin and forms a Lyn interaction with the GTPase Cdc42 (Felic). Felic was expressed in both hematopoietic and nonhematopoietic tissues. Because it represents an alternative splice product related to the Cdc42-interacting protein 4, CIP4, we also refer to Felic as CIP4b. Felic contains an SH3 recognition site RXPXXP and multiple tyrosine residues. In insulin or serum-stimulated HEK293 cells, Felic became tyrosine phosphorylated. Like CIP4, Felic associated with Cdc42 in its activated form only. Unlike CIP4, Felic does not possess a C-terminal SH3 domain. Coprecipitation studies show that Felic bound to Lyn or activated forms of Cdc42. Overexpression of Felic or CIP4 inhibited NIH 3T3 cell invasiveness in a Matrigel assay. Because Lyn and Cdc42 are involved in phagocytosis, we examined the distribution of Felic in RAW macrophages during particle ingestion. Felic was recruited more efficiently than CIP4 to the phagocytic cups. Altogether, these data suggest that CIP4/Felic constitute a novel family of cytoskeletal scaffolding proteins, integrating Src and Cdc42 pathways. The absence of an SH3 domain in Felic provides a structural basis for functional differences.
Introduction
Extracellular signals lead to the activation of intracelluar kinases and subsequent formation of signaling networks and cascades, which regulate cell growth, differentiation, movement or adhesion, and survival.1 Signaling networks rely on proteins serving as scaffolds for molecular recruitment of adaptors, enzymes, and substrates.2 A wide range of ligands activates Src-related protein tyrosine kinases.3 In turn, Src kinases associate with other proteins via Src homology 2 (SH2)–phosphotyrosine or SH3-proline interactions. Signaling cascades involving Shc-Grb2-Ras-Erk, Cbl-PI 3-kinase, Fak-Paxillin-Tensin, or Vav-Rac GEF are associated with Src activation.4-13
The Src-related kinase Lyn is found predominantly in B lymphocytes and myeloid cells. Lyn−/− mice display disturbed B-cell lymphopoiesis and autoimmunity, and defective mast cell degranulation.14,15 In these mice or in the DT40 Lyn−/− cells, there is absent or reduced phosphorylation of Shc, Cbl, Vav, or HS-1 following stimulation.7,14-17Studies of Lyn in B-cell behavior show a complex pattern of Lyn in supporting both growth stimulatory and inhibitory responses.18,19 However, genetic or pharmacologic targeting of Lyn results in loss of lymphoid or myeloid leukemic cell growth.20 Using the DT40 cell lines, we determined that granulocyte colony-stimulating factor (G-CSF) receptor signaling requires the presence of Lyn for G1/S phase progression.21
Because the involvement of Lyn in growth signaling pathways is complex and depends on the expression of its binding partners, we sought to identify relevant binding proteins in a yeast 2–hybrid screen of a B-cell cDNA library. A unique cDNA clone was identified and verified as a bona fide interactor of Lyn. This cDNA encodes the tyrosine kinase Fes and the cytoskeletal protein ezrin and forms a Lyn interaction with the GTPaseCdc42 (Felic), a scaffolding protein that possesses a Fes-related coiled-coil domain, homology to ezrin/moesin/radixin (ERM), and can mediate the interaction of Lyn with the activated form of the Cdc42 GTPase. FcγR-mediated phagocytosis is dependent on the local reorganization of the actin cytoskeleton, which ostensibly depends in part on both Cdc42 and Src-family kinases.22-24Consistent with this, Felic became tyrosine phosphorylated and was found to accumulate at sites of phagocytosis of immunoglobulin G (IgG)–opsonized particles in a mouse macrophage cell line. Felic is an alternatively spliced product of a gene that encodes Cdc42-interacting protein-4 (CIP4).25 CIP4 contains an SH3 domain and has been found to associate with microtubules and Wiskott-Aldrich syndrome protein (WASP).26 27Unlike Felic, however, CIP4 did not accumulate in the phagocytic cup. Altogether, these data suggest that Felic/CIP4 comprises a novel family of cytoskeletal scaffolding proteins, which links Lyn and Cdc42 pathways. Felic differs from the SH3-containing CIP4 isoform, which may account for only Felic localizing to the early phagosome.
Materials and methods
Yeast 2–hybrid screening and cloning of human cDNA
The Unique, SH3, and SH2 domains of human Lyn cDNA, corresponding to amino acids 1 to 226, were amplified using primers engineered to contain EcoRI and BamHI restriction sites. This amplified fragment was ligated into pGBT9 (Clontech, Palo Alto, CA) as reported and characterized elsewhere.8 A cDNA library prepared from Daudi B cells and cloned into the pGAD vector (Clontech) was screened. Following transformation of the YRG-2 yeast with pGBT9/LynU32 and the pGAD B cell library, positive clones were identified by β-galactosidase assay as described.8These putative Lyn interactors were restreaked onto fresh glucose and then galactose plates. Following confirmation that the positive clones were not transactivating, DNA was prepared from the positive yeast colonies and used to transform JEB181 leu-negative bacteria. DNA sequencing was performed using dye terminator chemistry automated sequencing in the Division of Immunogenetics at Rangos Research Building or the University of Pittsburgh Sequencing Facility. Using mRNA harvested by Trizol reagent (Life Technologies, Rockville, MD) and Dynabeads (Dynal Biotech, Lake Success, NY) from Daudi B cells (provided by Dr Theresa Whiteside, University of Pittsburgh Cancer Institute), 5′ rapid amplification of cloned ends (RACE) was performed with the Marathon system (Clontech). Clones were isolated and sequenced. The GenBank database was searched using the BLAST algorithm. Full-length cDNAs for CIP4, Felic, Lyn, V12Cdc42, L61Cdc42, Cdc42, and 17NCdc42 were subcloned into the pCMVFLAG vector (IBI, Eastman Kodak, New Haven, CT) or pcDNA3.1 vector (Invitrogen, Carlsbad, CA). Constructs of hemagglutin (HA)–tagged CIP4 and Felic were made. The cDNAs for Cdc42 contained the HA epitope tag and were provided by Dr Margaret Chou (University of Pennsylvania).
Cell culture and expression
Daudi, HEK293, HL60, U937, TF-1, and NIH 3T3 cells were grown in medium containing RPMI and 10% fetal calf serum (FCS) and supplemented with 2 mM l-glutamine. TF-1 cells were grown in medium containing recombinant human granulocyte macrophage (GM)–CSF. HEK293 cells were grown to near confluence. RAW264.7 macrophages were maintained in α–modified Eagle medium (α-MEM) medium supplemented with 10% FCS. Using the calcium phosphate system or Lipofectin-Plus (Gibco, Rockville, MD), HEK293 cells were transfected with the following constructs: pCMVFLAG-Felic, pCMVFLAG-CIP4, pCMV-FLAG, pcDNA3.1-Lyn, pCDNA3.1-V12Cdc42HA, pcDNA3.1-N17Cdc42HA, pcDNA3.1-L61Cdc42HA, and pcDNA3-Cdc42HA. Transfection of RAW macrophages with the pcDNA3.1-FelicHA and pcDNA3.1-CIP4HA constructs was accomplished with FuGENE 6 (Roche Diagnostics, Indianapolis, IN), according to the manufacturer's instructions. Cells were used in phagocytic assays within 24 hours after transfection.
Northern blot analysis
Total RNA (20 μg) prepared by Trizol reagent was loaded onto a 1.2% agarose-formaldehyde gel, and then transferred onto Nytran nitrocellulose filter (Schleicher and Schuell, Keene, NH). In addition, human Multiple Tissue Northern Blot II was purchased from Clontech and used for Northern blotting. The hybridization probe was a randomly primed, 32P-labeled 159–base pair (bp) polymerase chain reaction (PCR) fragment of Felic (nucleotides 1328-1487 from translation start).
Transfection and immunoprecipitation studies
Cells were harvested 48 hours after transfection and lysed in 1% nonidet P-40 (NP40) solution containing 137 mM NaCl, 20 mM Tris, pH 7.5, 1 mM EDTA (ethylenediaminetetraacetic acid), 10% glycerol, and 10 μg/mL aprotinin, 10 μg/mL leupeptin, 2 mM phenylmethylsulfonyl fluoride (PMSF), and 2 mM NaVO3. An equivalent amount of protein lysates, as determined by Bradford assay (Bio-Rad, Hercules, CA), was subjected to immunoprecipitation with antibodies directed against Lyn, HA, or FLAG epitopes (Santa Cruz Biotechnology, Santa Cruz, CA). Immune complexes were captured with either protein A– or protein G–Sepharose (Santa Cruz Biotechnology) and washed 5 times with phosphate-buffered saline (PBS)/1% NP40. After being resuspended in 2 × Laemmli buffer and boiled for 5 minutes, the immunoprecipitated proteins were loaded onto 5% to 15% sodium dodecyl sulfate (SDS) polyacrylamide gels. After being electrophoresed, proteins were electro-transferred onto polyvinylidene fluoride (PVDF) membrane (ImmoblinP, Medford, MA). After blocking with 5% bovine serum albumin (BSA) in PBS with 0.1% Tween20, membranes were incubated with anti-HA monoclonal antibody, anti-FLAG monoclonal antibody, anti-Syk polyclonal antibody, or anti-Lyn polyclonal antibody. After washing, blots were incubated with secondary antibody, goat anti–rabbit Ig coupled to horseradish peroxidase, or goat anti–mouse Ig coupled to horseradish peroxidase (Bio-Rad). Visualization was done with enhanced chemiluminescence (Amersham, Arlington Heights, IL).
Microscopic analysis of phagocytosis
For phagocytic assays, sheep red blood cells (SRBCs) and 0.8-μm latex beads were opsonized as described.28Zymosan particles were opsonized with human serum for 1 hour at 37°C. IgG-opsonized particles were then added to transfected RAW macrophages at a 10:1 ratio and incubated in cold medium for 10 minutes to allow binding. Following, the cells were incubated with warm medium for 7 to 10 minutes to allow formation of phagocytic cups, and fixed immediately with ice-cold 4% paraformaldehyde for 20 minutes and then with room temperature fixative for one hour. For phagocytosis of serum-opsonized zymosan, RAW macrophages were activated with 100 nM phorbol myristate acetate (PMA) for 20 minutes prior to phagocytosis. For immunofluorescence, cells were permeabilized with 0.1% TritonX-100 for 10 minutes, blocked with 5% donkey serum, and followed with mouse anti-HA antibody. Donkey antimouse antibody conjugated with Cy3 was from Jackson ImmunoResearch Laboratories (West Grove, PA) and used at 1:1000. Confocal microscopy and image processing was performed as described.28
Matrigel invasiveness assay
A 12-well BioCoat Matrigel Invasion Chamber was purchased from Becton Dickinson (San Diego, CA). Following manufacturer's directions, it was allowed to thaw at room temperature. The wells were rehydrated with Dulbecco modified Eagle medium (DMEM). Suspensions of NIH 3T3 cells transfected with either empty vector, HA-CIP4, or HA-Felic were added to the wells at 2.5 × 104 cells/mL. After 24 hours of incubation at 37°C, the cells from the upper surface of the membrane were removed. The filters were fixed and stained with 0.5% Crystal Violet (Sigma). Cover slips were prepared for cell counting and photography. Data were analyzed for statistical significance, using Microsoft Excel software (Bellingham, WA) and the Student pairedt test for independent means.
Results
Isolation and cloning of Felic
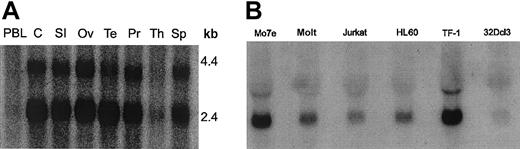
To identify new binding partners for Lyn, we screened a Daudi B cell library, since Lyn plays a major role in B-cell signal transduction.28 From the B-cell library screen of more than 100 000 transformants, we isolated 17 clones that interacted with the Lyn bait, which comprised the Unique-SH3-SH2 domains of Lyn. Positives were picked and restreaked onto fresh glucose and then galactose plates. DNA was prepared from positive yeast colonies and used to transform JEB181 bacteria (which were leu-negative and thus could be rescued by the library plasmid, which encodes for β-isopropylmalate dehydrogenase that is involved in leucine biosynthesis). Of the 17 clones, 6 appeared to be true interactors upon rescreening. Automated DNA sequencing was then performed, and 4 of the 6 gave open reading frames of more than 30 amino acids. We concentrated on sequencing the strongest positive, clone 10-1, which consisted of 435 nucleotides. With the exception of a 29-nucleotide insertion AGATCTGGGCCCACCCCCACCC CCATCAC, GenBank searching showed identical sequence to a partial cDNA, thyroid hormone receptor interactor 10, and at that time, no homology to any known proteins. Using a different source of Daudi B cells, we performed reverse transcriptase (RT)–PCR and 5′ RACE PCR (Marathon cDNA amplification kit). The complete cDNA consisted of 1782 nucleotides, which encoded a 593–amino acid protein of approximately 68 kDa. Using a probe consisting of clone 10-1, we examined hematopoietic and nonhematopoietic tissues. Northern blot analysis showed transcripts of 2.3 to 2.4 kilobase (kb) and 3.3 kb in both types of tissues (Figure 1).
Northern blot analysis of Felic.
Hematopoietic and nonhematopoietic tissues were analyzed by Northern blotting with a randomly labeled probe of Felic. (A) Total RNA of multiple normal human tissues was evaluated: PBL indicates peripheral blood lymphocytes; C, Colon; SI, small intestine; Ov, ovary; Te, testis; Pr, prostate; Th, thymus; and Sp, spleen. Two transcripts are present, except for T cells (thymus and peripheral blood lymphocytes). (B) Total RNA was harvested from myeloid cell lines Mo7e, HL60, TF-1, and 32Dcl3 and lymphoid cell lines Molt and Jurkat, which were analyzed. Comparable amounts of RNA were loaded, as verified by ethidium bromide staining (data not shown).
Northern blot analysis of Felic.
Hematopoietic and nonhematopoietic tissues were analyzed by Northern blotting with a randomly labeled probe of Felic. (A) Total RNA of multiple normal human tissues was evaluated: PBL indicates peripheral blood lymphocytes; C, Colon; SI, small intestine; Ov, ovary; Te, testis; Pr, prostate; Th, thymus; and Sp, spleen. Two transcripts are present, except for T cells (thymus and peripheral blood lymphocytes). (B) Total RNA was harvested from myeloid cell lines Mo7e, HL60, TF-1, and 32Dcl3 and lymphoid cell lines Molt and Jurkat, which were analyzed. Comparable amounts of RNA were loaded, as verified by ethidium bromide staining (data not shown).
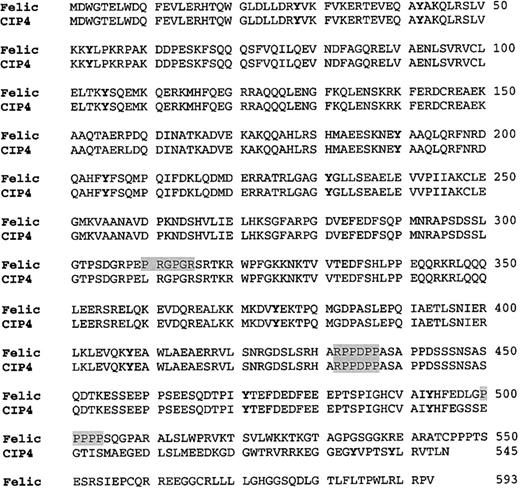
Amino acid analysis of Felic and its relationship to CIP4
When the complete cDNA was isolated and sequenced, GenBank searching showed that Felic was highly homologous to CIP4. The difference between these 2 gene products was due to the 29- nucleotide insertion occurring after codon 495 of CIP4. The cDNA was named Felic, because it contains regions that are highly homologous to the amino-terminus of the nonreceptor protein tyrosine kinaseFes/Fer, to the ezrin/radixin/moesin cytoskeletal protein family and it demonstrates a Lyninteraction with activated forms of Cdc42 (Figure 2). Of note, at positions 432 to 437, RPPDPP constitutes a favored SH3 binding site (ie, PXXP) for Src SH3 domains.29,30 A second RXPXXP motif is found only in Felic at positions 310 to 315. A minimal SH3 binding site, the PXXP motif, is found again only in Felic (positions 500-505); however, Lyn does coprecipitate with CIP4 (data not shown). In addition, a proximal Arg residue favors a salt bridge with the corresponding Asp residue found in the SH3 domain of Lyn. Multiple RPXXP or RXPXXP motifs are found in the proline-rich region of Cbl, which we and others have isolated from a cDNA library with Lyn as bait in the yeast 2–hybrid screen.8 16
Comparison of the amino acid sequence of Felic and CIP4.
Primary amino acid sequence of Felic is aligned with CIP4. The proline-rich motif, which provides a putative SH3 docking site, is shaded. Potential tyrosine phosphorylation sites are featured by boldface type. The Fes/CIP4 domain, also known as the RAEYL motif or the S pombe Cdc15 N-terminal domain, occurs at positions 1 to 56. The region with 30% homology to ezrin/radixin/moesin motif is found at positions 329 to 410. The Cdc42-binding region occurs at positions 338 to 467. CIP4 contains an SH3 domain at its C-terminus at positions 489 to 545.
Comparison of the amino acid sequence of Felic and CIP4.
Primary amino acid sequence of Felic is aligned with CIP4. The proline-rich motif, which provides a putative SH3 docking site, is shaded. Potential tyrosine phosphorylation sites are featured by boldface type. The Fes/CIP4 domain, also known as the RAEYL motif or the S pombe Cdc15 N-terminal domain, occurs at positions 1 to 56. The region with 30% homology to ezrin/radixin/moesin motif is found at positions 329 to 410. The Cdc42-binding region occurs at positions 338 to 467. CIP4 contains an SH3 domain at its C-terminus at positions 489 to 545.
RT-PCR screening of Daudi B cell mRNA also yielded CIP4 clones. The cDNAs differ in that Felic contains a 29 nucleotide insertion, which occurs after codon 495. As a result of the insertional frameshift, Felic does not have a C-terminal SH3 domain, and a stop codon occurs at codon 594. Genomic cloning of the human Felic/CIP4 locus revealed that the 29 nucleotide sequence is found at the 5′ end of exon 13. The isolation of both Felic and CIP4 clones from Daudi B cells and the identification of the additional nucleotides at an exonic 5′ end suggested that these proteins represented alternatively spliced products of the same gene. Southern blot analysis of human genomic DNA, digested with 4 different restriction enzymes, showed a single band.31
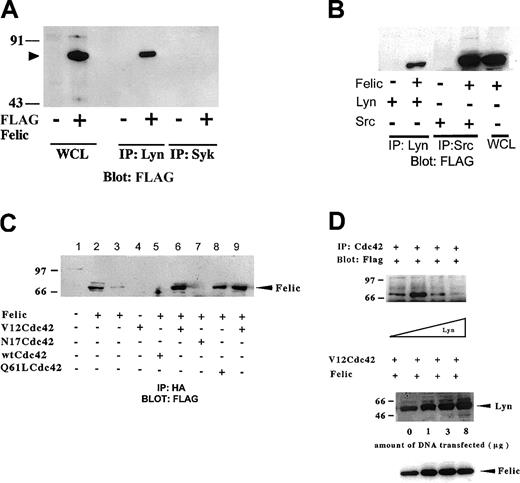
In vitro association of Felic with Lyn and V12Cdc42
To confirm the yeast 2–hybrid interaction between Lyn and Felic, we constructed eukaryotic expression vectors with FLAG-tagged Felic or CIP4 and cotransfected these with full-length Lyn expression vector or wild-type, inactive, or active forms of HA-tagged Cdc42 in HEK293 cells. Coprecipitation studies showed that Lyn associated with Felic (Figure 3A). Because Felic is expressed in epithelial tissues, such as lung, ovary, and intestine where Src, not Lyn, is found,32-34 we verified that Src could also associate with Felic (Figure 3B). We also confirmed that Felic associated with the active forms of Cdc42 (V12Cdc42 and 61LCdc42) and not the GDP-bound or inactive form of Cdc42 (N17Cdc42) (Figure 3C). Increasing levels of Lyn inhibited the association of Felic with V12Cdc42, suggesting an allosteric effect (Figure 3D).
In vitro association of Felic, Lyn, and Cdc42.
(A) HEK293 cells were transfected with cDNA for pCMVFLAG-Felic and pCDNA3.1-Lyn. 24 h after transfection, cells were harvested, and proteins immunoprecipitated (IP) with polyclonal antibody against Lyn were resolved on 10% SDS-PAGE. After being transferred onto PVDF, the membrane was blotted with monoclonal antibody (mAb) against FLAG. Detection was performed using enhanced chemiluminscence. Blotting of whole cell lysates (WCLs) of transfected cells show position of Felic (arrowhead). To confirm specific binding between Lyn and Felic, an isotypic control, rabbit anti-Syk polyclonal antibody, was used and failed to coprecipitate FLAG-tagged Felic from Felic-overexpressing cells. (B) HEK293 cells were transfected with cDNA for Felic and Src. In these experiments, proteins immunoprecipitated with polyclonal antibody against c-Src were resolved by SDS-PAGE, followed by transfer to PVDF and blotting with anti-FLAG mAb. (C) HEK293 cells were transfected with cDNA for Felic and wild-type and mutant forms of Cdc42. V12Cdc42 and Q61L Cdc42 are constitutively activated forms, whereas N17Cdc42 is the inactive form. The cDNAs for Cdc42 forms encoded the HA tag. At 24 hours after transfection, cells were harvested and proteins immmunoprecipitated with mAb against HA were resolved by 10% SDS-PAGE, transferred onto PVDF filter, and blotted with mAb against FLAG. Lane 2 represents WCLs of Felic-transfected cells. Lane 9 represents a duplicate of experimental conditions shown in lane 6. Lanes 6, 8, and 9 show association of Felic with activated Cdc42. Lanes 5 and 8 show the absence of association of Felic with inactive or wild-type forms of Cdc42. (D) HEK293 cells were transfected with cDNA for pCMVFLAG-Felic and pCDNA3.1-V12Cdc42HA and varying amounts of pcDNA3.1-Lyn (0-8 μg). Plasmid pcDNA3 was added to keep the total cDNA transfected equivalent at 25 μg between transfection conditions. Proteins were immunoprecipitated with mAb against HA, and blotting was performed with FLAG mAb. Equivalent amount of Felic was expressed as demonstrated by blotting.
In vitro association of Felic, Lyn, and Cdc42.
(A) HEK293 cells were transfected with cDNA for pCMVFLAG-Felic and pCDNA3.1-Lyn. 24 h after transfection, cells were harvested, and proteins immunoprecipitated (IP) with polyclonal antibody against Lyn were resolved on 10% SDS-PAGE. After being transferred onto PVDF, the membrane was blotted with monoclonal antibody (mAb) against FLAG. Detection was performed using enhanced chemiluminscence. Blotting of whole cell lysates (WCLs) of transfected cells show position of Felic (arrowhead). To confirm specific binding between Lyn and Felic, an isotypic control, rabbit anti-Syk polyclonal antibody, was used and failed to coprecipitate FLAG-tagged Felic from Felic-overexpressing cells. (B) HEK293 cells were transfected with cDNA for Felic and Src. In these experiments, proteins immunoprecipitated with polyclonal antibody against c-Src were resolved by SDS-PAGE, followed by transfer to PVDF and blotting with anti-FLAG mAb. (C) HEK293 cells were transfected with cDNA for Felic and wild-type and mutant forms of Cdc42. V12Cdc42 and Q61L Cdc42 are constitutively activated forms, whereas N17Cdc42 is the inactive form. The cDNAs for Cdc42 forms encoded the HA tag. At 24 hours after transfection, cells were harvested and proteins immmunoprecipitated with mAb against HA were resolved by 10% SDS-PAGE, transferred onto PVDF filter, and blotted with mAb against FLAG. Lane 2 represents WCLs of Felic-transfected cells. Lane 9 represents a duplicate of experimental conditions shown in lane 6. Lanes 6, 8, and 9 show association of Felic with activated Cdc42. Lanes 5 and 8 show the absence of association of Felic with inactive or wild-type forms of Cdc42. (D) HEK293 cells were transfected with cDNA for pCMVFLAG-Felic and pCDNA3.1-V12Cdc42HA and varying amounts of pcDNA3.1-Lyn (0-8 μg). Plasmid pcDNA3 was added to keep the total cDNA transfected equivalent at 25 μg between transfection conditions. Proteins were immunoprecipitated with mAb against HA, and blotting was performed with FLAG mAb. Equivalent amount of Felic was expressed as demonstrated by blotting.
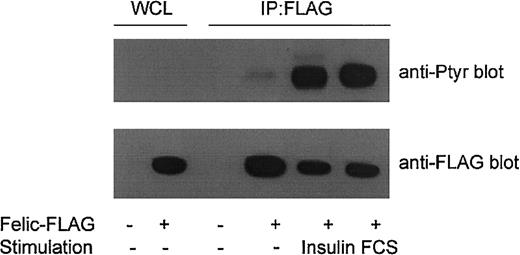
Tyrosine phosphorylation of Felic
Felic contains 11 potential tyrosine phosphorylation sites. Of these, Y472 is the most likely phosphoacceptor site because the extended sequence (TPIYTEFDEDFEEE) is rich in acidic residues. To determine whether Felic is tyrosine phosphorylated, HEK293 cells were transfected with the FLAG-tagged Felic construct. After 30 hours, transfected cells were serum starved overnight, and then stimulated with 10% FCS or 100 ng/mL insulin for 15 minutes. An antiphosphotyrosine blot demonstrated stimulus-induced tyrosine phosphorylation of immunoprecipitated Felic (Figure4).
Tyrosine phosphorylation of Felic.
HEK293 cells were transfected for 30 hours with pCMVFLAG-Felic (Felic-FLAG +) or empty vector (−). Cells were then grown overnight in RPMI depleted of serum and 2% bovine serum albumin, then stimulated with 10% FCS or 100 ng/mL insulin for 15 minutes at 37°C. Lysates were prepared and immunoprecipitated (IP) with FLAG-antibody, and blotted with antiphosphotyrosine mAb 4G10 (Upstate Biotechnology, Lake Placid, NY). Lower panel shows comparable levels of immunoprecipitated Felic. WCLs show expression of Felic.
Tyrosine phosphorylation of Felic.
HEK293 cells were transfected for 30 hours with pCMVFLAG-Felic (Felic-FLAG +) or empty vector (−). Cells were then grown overnight in RPMI depleted of serum and 2% bovine serum albumin, then stimulated with 10% FCS or 100 ng/mL insulin for 15 minutes at 37°C. Lysates were prepared and immunoprecipitated (IP) with FLAG-antibody, and blotted with antiphosphotyrosine mAb 4G10 (Upstate Biotechnology, Lake Placid, NY). Lower panel shows comparable levels of immunoprecipitated Felic. WCLs show expression of Felic.
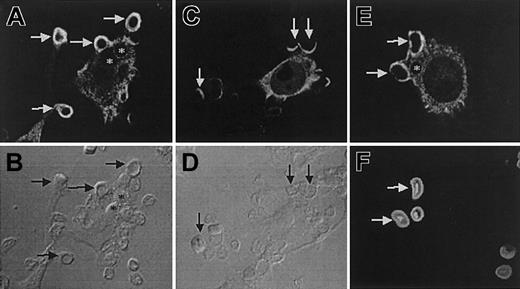
Felic localizes to the phagocytic cup
The role of Felic in regulating cytoskeletal properties is strongly suggested by the presence of the Cdc42 binding protein domain, its expression in both hematopoietic and epithelial tissues, and its ability to undergo serum-induced tyrosine phosphorylation. Felic/CIP4 also contain a region between residues 329 and 410, which displays 30% homology to the C-terminal region of ezrin/radixin/moesin (ERM), a family of proteins, that links membrane receptors to the actin cytoskeleton. FcγR-mediated phagocytosis is an actin-dependent process that appears to be regulated by Cdc42 and Src-family kinases.22-24 Therefore, we examined the subcellular localization of Felic and CIP4 in transfected RAW264.7 cells undergoing phagocytosis of IgG-opsonized SRBCs. Using immunofluorescence, Felic and CIP4 are mostly distributed to the cytoplasm in unstimulated RAW macrophages, although it was also possible to find Felic and CIP4 in the cell cortex. Importantly, Felic is effectively recruited to sites of phagocytosis during particle binding and pseudopod extension (Figure 5A-B). This association, like that of actin, is temporary since no detectable Felic was observed on formed phagosomes (Figure 5A, asterisks). Compared with Felic, CIP4 weakly accumulated in phagocytic cups (Figure 5C), suggesting that while Felic and CIP4 differ only at the extreme C-terminus, this distinction may lead to a functional difference between the 2 splice variants. Independent of protein expression, similar results were also obtained with green fluorescent protein (GFP)–tagged proteins of Felic and CIP4 (data not shown). Felic was also recruited to phagocytic cups containing IgG-opsonized 0.8-μm latex beads that mimic the size of bacteria, indicating that the recruitment of Felic likely occurs during bacterial internalization (not illustrated). Finally, the involvement of Felic in phagocytosis is not limited to Fcγ receptor–mediated phagocytosis, since phagocytic cups containing serum-opsonized zymosan, which engage complement receptors, were enriched in Felic (Figure 5E).
Differential distribution of Felic and CIP4 during phagocytosis.
RAW264.7 macrophages were transfected with HA-Felic (A-B,E-F) or HA-CIP4 (C-D) and allowed to bind and initiate phagocytosis of IgG-opsonized SRBCs (A-D) or serum-opsonized zymosan (E-F). The cellular distribution of HA-Felic and HA-CIP4 was often cytosolic. HA-Felic was strongly recruited to phagocytic cups during phagocytosis of IgG-opsonized SRBCs (A-B, arrows) and serum-opsonized zymosan (E-F, arrows) and quickly dissociated from sealed phagosomes (A-B,E-F, asterisks). In contrast to Felic, CIP4 was weakly recruited to the sites of phagocytosis (C-D, arrows). Panels B, D, and F show the corresponding differential interference contrast images of panels A, C, and E, respectively. Original magnifications × 1000.
Differential distribution of Felic and CIP4 during phagocytosis.
RAW264.7 macrophages were transfected with HA-Felic (A-B,E-F) or HA-CIP4 (C-D) and allowed to bind and initiate phagocytosis of IgG-opsonized SRBCs (A-D) or serum-opsonized zymosan (E-F). The cellular distribution of HA-Felic and HA-CIP4 was often cytosolic. HA-Felic was strongly recruited to phagocytic cups during phagocytosis of IgG-opsonized SRBCs (A-B, arrows) and serum-opsonized zymosan (E-F, arrows) and quickly dissociated from sealed phagosomes (A-B,E-F, asterisks). In contrast to Felic, CIP4 was weakly recruited to the sites of phagocytosis (C-D, arrows). Panels B, D, and F show the corresponding differential interference contrast images of panels A, C, and E, respectively. Original magnifications × 1000.
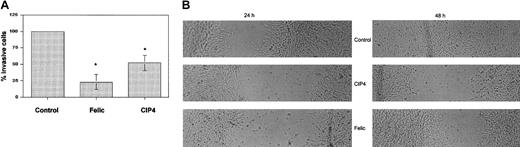
Effect of Felic/CIP4 on cell invasiveness
To determine whether Felic or CIP4 affects migration, we analyzed this behavior using the Matrigel-coated filter assay. NIH 3T3 cells were transfected with either empty vector as a control, HA-CIP4, or HA-Felic. Comparable levels of ectopic protein expression were seen. Following 24 hours of incubation in Matrigel, filters were fixed and analyzed. Compared with control cells, both Felic and CIP4 inhibited NIH 3T3 invasiveness, with a statistically significant greater effect for Felic (Figure 6A). Inhibition of cell migration at 48 hours was also seen in confluent NIH 3T3 cells transfected with CIP4 or Felic, which had been disturbed by pipette streaking (Figure 6B).
Effect of Felic/CIP4 on cell migration.
(A) NIH3T3 cells expressing either Felic or CIP4 were compared with control cells in a Matrigel filter invasiveness assay. Data presented are the average ± standard error of the percentage of migrating cells. Results were statistically significant (*P < 0.05) using Student paired t test. (B) NIH3T3 cells transfected with empty vector (Control), Felic, or CIP4 were allowed to grow to confluence, then streaked horizontally with a sterile pipette tip. Microscopy with digital photography was performed at 24 and 48 hours after streaking. Original magnifications, × 100.
Effect of Felic/CIP4 on cell migration.
(A) NIH3T3 cells expressing either Felic or CIP4 were compared with control cells in a Matrigel filter invasiveness assay. Data presented are the average ± standard error of the percentage of migrating cells. Results were statistically significant (*P < 0.05) using Student paired t test. (B) NIH3T3 cells transfected with empty vector (Control), Felic, or CIP4 were allowed to grow to confluence, then streaked horizontally with a sterile pipette tip. Microscopy with digital photography was performed at 24 and 48 hours after streaking. Original magnifications, × 100.
Discussion
Here we have reported the isolation, cloning, and characterization of Felic, a novel scaffolding protein that integrates the Src and Cdc42 pathways. Felic differs from its related CIP4 isoform as a result of an out-of-frame 29-nucleotide insertion at codon 496. CIP4 was first identified as a binding partner for the activated form of Cdc42 and then for WASP.25,27 We isolated the 3′ end of Felic in a yeast 2–hybrid screen using the noncatalytic portion of Lyn. Both Felic and CIP4 contain a region, corresponding to amino acids 338 to 417, that binds the activated form of Cdc42. This domain does not resemble the classic GBD or CRIB region. Furthermore, Felic and CIP4 also contain an N-terminal region, which has been found to associate with microtubules.27 A proline-rich motif, found in both Felic and CIP4 (amino acids 432-437), provides a mechanism for their interaction with Lyn.
Of note, CIP4 contains a C-terminal SH3 domain, which is not found in Felic. Felic contains a unique sequence of 96 amino acids at its C-terminus. To determine any functional differences between Felic and CIP4, we studied their subcellular location in phagocytosing macrophages. Felic, but not CIP4, was recruited along with Lyn to the phagocytic cup, an early stage in phagosome formation. Because Felic was first isolated from a Daudi B cell library, Felic is probably involved uniquely in other cytoskeletal functions. Overexpression of either Felic or CIP4 inhibited the migration of 3T3 cells in Matrigel or after physical separation. We therefore propose that Felic and CIP4 constitute a family of scaffolding proteins with different roles in cytoskeletal organization. Because the SH3 domain of CIP4 binds WASP, we hypothesize that CIP4 can form a complex with Lyn, Cdc42-GTP, and WASP. Microinjection of CIP4 with its SH3 domain deleted was associated with the macrophage's inability to form podosomes, suggesting a critical role for CIP4/WASP interaction.26 In turn, Lyn can phosphorylate WASP, an event enhanced by activated Cdc42.35 While tyrosine phosphorylation of WASP has not been linked to a change in its function, binding of activated Cdc42 permits WASP to unfold, which promotes binding of Arp2/3 and subsequent actin nucleation.36,37 Since Lyn and other Src kinases phosphorylate Vav and Vav2, which results in their activation as GTP exchange factors for Rac and Cdc42,11,38 it is possible that Lyn/CIP4/WASP/Cdc42 constitute a signaling network critical for actin polymerization. The role of Felic in modifying this network or other components of a Lyn/Felic/Cdc42 network remains to be elucidated. An important clue to differences between CIP4 and Felic (CIP4b) is the presence or absence of the C-terminal SH3 domain. Interestingly, 2 additional alternative splice forms have been isolated.31One, CIP4c, also lacks the SH3 domain, while the other, CIP4h, contains it. SH3 domains are found in a wide variety of signaling molecules. Constitutive association, albeit weak, between SH3-containing proteins and proteins containing a polyproline helix loop facilitates formation of multicomponent signaling complexes. One outcome of multicomponent signaling complexes is to direct intracellular location. Besides WASp, CIP4 can bind proline-rich c-Cbl (K. Sullivan and S.J.C., unpublished observations, May 2002). Therefore, the absence of an SH3 domain in Felic may account for its ability to traffic to the early phagosome.
We thank Drs Margaret Chou and Richard Cerione for providing us with the cDNAs for Cdc42 and Dr Ram Menon for providing bovine insulin. We thank Kevin Sullivan for additional preliminary data.
R.J.B. is a recipient of a studentship from the Canadian Institutes of Health Research. A.G. is a Cure for Lymphoma Fellow. S.G. is an International Scholar of the Howard Hughes Medical Institute, a recipient of a Canadian Institute of Health Research (CIHR) Distinguished Scientist Award, and the current holder of the Pitblado Chair in Cell Biology.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-03-0851.
Supported by grants from the American Cancer Society, American Heart Association Grant-in-Aid, National Institutes of Health (NIH) FIRST Award, and NIH Independent Scientist Award to S.J.C.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Seth Corey, Section Pediatric Leukemia/Lymphoma, Division of Pediatrics, MD Anderson Cancer Center, 1515 Holcombe Blvd, Houston, TX 77030; e-mail:sjcorey@mdanderson.org.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal