Anaplastic large cell lymphoma (ALCL) with t(2;5)(p23;q35) and Hodgkin disease (HD) share many cellular features, including expression of CD30. We compared gene expression profiles of 4 ALCL (Karpas 299, SU-DHL-1, DEL, SR-786) and 3 HD cell lines and found thatBCL3, which encodes a nuclear protein belonging to the IκB family of inhibitors of nuclear factor–κB (NF-κB) transcriptional factors, was expressed at higher levels in ALCL than HD. Northern and Western blotting analyses confirmed the high-level expression of BCL3 in ALCL at both mRNA and protein levels. We established a real-time reverse transcriptase–mediated polymerase chain reaction assay to measure the BCL3 mRNA level and found a predominant level of BCL3 expression in t(2;5)+ ALCL; the levels of cell lines and clinical materials were comparable to or higher than that of a B-cell chronic lymphocytic leukemia carrying t(14;19)(q32;q13). Southern blotting and fluorescence in situ hybridization disclosed that the BCL3gene copies were amplified in SU-DHL-1, whereas Karpas 299 carried 4 BCL3 gene loci. The BCL3 gene contains 2 cytosine-guanine dinucleotide (CpG) islands, and the intragenic 3′ CpG was entirely demethylated in SU-DHL-1 and DEL. In contrast to HD, in which NF-κB was constitutively activated, ALCL cells consistently showed (p50)2 homodimer binding activity on electrophoretic mobility shift assay. It is suggested that the high-level nuclear Bcl-3 sequestrates the (p50)2 homodimer to the nucleus, which may account for the contradictory effect of CD30 stimulation on ALCL and HD. We propose that BCL3 is overexpressed by genetic and epigenetic modifications, potentially contributing to the development of t(2;5)+ ALCL.
Introduction
Anaplastic large cell lymphoma (ALCL) was initially defined on the basis of the anaplastic appearance of tumor cells, characteristic intrasinusoidal spread, and consistent expression of the cytokine receptor CD30.1 However, heterogeneity in the cytology as well as in the clinical features has been described.2,3 Anaplastic large cell lymphoma kinase–positive (ALK+) ALCL was subsequently identified as a distinctive disease entity within the heterogeneous group of ALCL.4,5 Most cases with ALK+ ALCL carry t(2;5)(p23;q35), which leads to the generation of nucleophosmin (NPM)/ALK fusion protein.2,3,6 Introduction of theNPM/ALK fusion gene in mouse cell lines leads to a transformed phenotype, and several downstream signaling pathways through which this particular fusion protein exerts its oncogenic action have been identified.7-9
Hodgkin disease (HD) is another lymphoid neoplasm, in which neoplastic Reed-Sternberg (RS) cells express the CD30 molecule on their cell surface.10 Studies of HD-derived cell lines and micromanipulated RS cells have revealed recurrent molecular abnormalities of HD.11 An initial study demonstrated constitutive nuclear factor–κB (NF-κB) DNA binding activity in the nucleus,12 and later studies showed that a proportion of HD cell lines and primary RS cells carried mutations or deletions within the IKBA gene,13-16leading to their being functionally null for IκBα activity. Although the genetic lesions of the IKBAgene are not common to all RS cells, as yet unidentified defects in the IκB family are likely responsible for the constitutive NF-κB activity, thereby contributing to the pathogenesis of HD.
Although both ALCL and HD express CD30 at high levels, stimulation of this molecule exerts contradictory effects on these 2 cell types.17-20 ALCL cells upon stimulation with a CD30 agonistic antibody show apoptotic cell death along with the impairment of activation of prosurvival NF-κB.20 In contrast, HD cells are resistant to CD30-induced apoptosis20; constitutive activation of NF-κB is likely to be responsible for the protective effect. In this study, we found that BCL3, which encodes a member of the IκB protein family of inhibitors of NF-κB,21-29 was expressed at higher levels in ALCL than HD. The BCL3 gene copy was amplified in 1 ALCL cell line, and the CpG island that exists within the gene30was demethylated in 2. BCL3 was initially identified as a candidate proto-oncogene located adjacent to the breakpoint of t(14;19)(q32;q13) in a proportion of B-cell chronic lymphocytic leukemia (B-CLL)30-33; however, a role of the Bcl-3 protein in T-cell survival by positive regulation of NF-κB has been suggested.34 A possible linkage between theBCL3 expression levels and differential NF-κB activities in ALCL and HD is discussed.
Materials and methods
Cells
The ALCL cell lines were Karpas 299, SU-DHL-1, DEL, and SR-786, which were provided by Drs Y. Matsuo (Fujisaki Cell Center, Okayama, Japan) and H. Drexler (DSMZ [Deutsche Sammlung von Mikroorganismen und Zellkulturen], Braunschweig, Germany).6,35 The HD cell lines were KM-H2 (which was established and characterized in our laboratory),36 L428, and HDLM-2; the latter 2 were provided by Dr Matsuo.35 Other hematologic tumor cell lines were FL-18 and FL-618, follicular lymphoma; Ramos, Burkitt lymphoma; HBL-2 and KIS-1, diffuse large B-cell lymphoma; RPMI 8226, plasma cell myeloma; Kasumi-1, acute myeloid leukemia; K562, chronic myeloid leukemia; Jurkat, acute T-cell leukemia; and HUT 102, adult T-cell leukemia. The cell lines used in this study were cultured in RPMI 1640 medium supplemented with 10% or 20% fetal calf serum under the standard culture conditions. Clinical materials of hematologic tumors were selected from a collection in our laboratory, including t(2;5)+ ALCL (case nos. 646, 1029, and 1078)37and ALCL that lacked the translocation. Molecular analysis of a B-CLL carrying t(14;19)(q32;q13) was described in detail previously.32 The cells for extraction of RNA were stored as frozen viable cell suspensions with 10% dimethyl sulfoxide in liquid nitrogen.
Complementary DNA array hybridization
Total cellular RNA was extracted from the cell lines using an RNeasy kit (Qiagen, Tokyo, Japan). After incubation with 5 units of RNase-free DNase, the RNA was reverse transcribed in the presence of [32P]deoxyadenosine triphosphate ([32P]dATP) using the Atlas Pure Total RNA Labeling System (Clontech, Palo Alto, CA). Hybridization of the cDNA with Atlas Human 1.2 Array (Clontech) was performed according to the manufacturer's instructions. The membranes were exposed to a BAS2000 imaging analyzer (Fuji Photo Film, Tokyo, Japan), and the hybridization signals were quantified by ArrayGauge software version 1.2 (Fuji Photo Film). Data from each array were normalized by the median value to eliminate the variability due to the sample labeling or to the exposure duration. Hierarchic clustering analysis of the array data was performed using GeneSpring software version 4.1 (Silicon Genetics, Redwood City, CA).
Southern and Northern blot analyses and long-distance polymerase chain reaction
Southern and Northern blotting were carried out as described previously.32 The cDNA probe for BCL3 was clone cLK2 containing a 1.8-kb cDNA fragment.31 Probe A representing the 5′ cytosine-guanine dinucleotide (CpG) island ofBCL3 was a 1.4-kb PstI fragment containing exon 1 and part of intron 1, whereas probe B for the 3′ CpG was a 0.92-kb PstI/BamHI containing exon 7 and neighboring introns.30 The BCL6 probe represented the major translocation cluster.38Long-distance polymerase chain reaction (LD-PCR) amplifying a DNA fragment encompassing the NPM/ALK fusion point was as described previously.37 The PCR was carried out using an automated thermal cycler (GeneAmp PCR System 2400, Applied Biosystems, Foster City, CA).
Real-time reverse transcriptase–mediated polymerase chain reaction
Real-time PCR analysis based on the TaqMan methodology was performed using an ABI Prism 7700 Sequence Detection System (Applied Biosystems). The sequences of oligonucleotide primers and a fluorogenic probe for BCL3 mRNA were 5′-TCGACGCAGTGGACATTAAGAG-3′ (forward); 5′-ACATTTGCGCGTTCACGTT-3′ (reverse); and 5′-TCATCCACGCCGTGGAAAACAACAG-3′ (probe). Complementary DNA was synthesized from 1 μg of total cellular RNA using random primers. The DNA standard template containing the BCL3 cDNA sequence was generated by cloning into pGEM-T EASY vector (Promega, Madison, WI). An aliquot of cDNA or standard template DNA (1 μL) was added to the PCR reaction mixture containing 1 × TaqMan Universal PCR Master Mix (Applied Biosystems), 400 nM of each primer, and 100 nM of probe in a total reaction volume of 50 μL. After an initial incubation for 2 minutes at 50°C and 10 minutes at 95°C to activate theTaq polymerase, 50 cycles of 15 seconds at 95°C and 1 minute at 60°C were carried out. The CT (threshold cycle) parameter was defined as the fractional cycle number at which the fluorescence generated by cleavage of the probe passed a preset threshold. The standard curve, in which CT decreased in linear proportion to the log of the template copy number, was established by serially diluted pGEM-T-BCL3 plasmid DNA. The CT values of test materials were plotted on standard curve, and the corresponding copy number was calculated by Sequence Detector version 1.6 software (Applied Biosystems). The amount ofBCL3 cDNA copy number of the test materials was divided by the endogenous reference, 18S ribosomal RNA (rRNA) (TaqMan Ribosomal RNA Control Reagents; Applied Biosystems), and the BCL3/18S rRNA ratio was further normalized with that of adult T-cell leukemia HUT 102 cells. All assays were performed in triplicate.
Western blot analysis
Cells were lysed in 1 × loading buffer containing protease inhibitor cocktail.39 The lysate was loaded onto 10% sodium dodecyl sulfate–polyacrylamide gels and electrotransferred onto Immobilon polyvinylidene difluoride (PVDF) transfer membranes (Millipore, Bedford, MA). The membranes were blocked in phosphate-buffered saline–Tween (PBS-T) buffer containing dried milk and then incubated with the following antibodies purchased from Santa Cruz Biotechnology (Santa Cruz, CA): polyclonal rabbit anti–Bcl-3 antiserum raised against a peptide at the carboxy-terminus of human Bcl-3 (sc-185); monoclonal immunoglobulin G1 (IgG1) mouse anti-p65 (sc-8008); monoclonal IgG1 mouse anti-p50 (sc-8414); monoclonal IgG2a mouse anti-p52 (sc-7386); and polyclonal goat anti-ALK antiserum (sc-6344). After extensive washing in PBS-T buffer, the blots were incubated for 1 hour with horseradish peroxidase–conjugated secondary antiserum followed by enhanced chemiluminescence reaction (Amersham Pharmacia Biotech, Piscataway, NJ).
Electrophoretic mobility shift assay for NF-κB activity
Cells were lysed in ice-cold buffer C (50 mM HEPES-KOH [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid–potassium hydroxide] [pH 7.8], 420 mM KCl, 0.1 mM EDTA [ethylenediamine tetraacetic acid] [pH8.0], 5 mM MgCl2, 20% glycerol, 1 mM dithiothreitol) supplemented with protease inhibitors.38 The whole-cell lysate was passed through a 25-gauge needle several times and centrifuged, and the supernatant was subjected to electrophoretic mobility shift assay (EMSA). The sequence of the oligonucleotide probe corresponding to a κB site of the immunoglobulin κ light chain gene was 5′-AGTTGAGGGGACTTTCCCAGGC-3′. Binding reactions were performed in a total volume of 10 μL containing 5 μg extract, 2 μL of 5 × binding buffer (Promega), and 1 μL of 32P–end-labeled probes.38Incubations were carried out for 30 minutes at 25°C, and the resulting complexes were resolved on 5% nondenaturing polyacrylamide gels in 0.5 × TBE. For competition experiments, incubations were performed with a 100-fold excess of unlabeled probe. For antibody supershift assays, the lysates were preincubated for 10 minutes at 25°C with 4 μg antibody before the addition of the radiolabeled gel shift probe. Antibodies used for the supershift assay were from Santa Cruz Biotechnology: monoclonal IgG1 mouse anti-p65 (sc-8008X); polyclonal goat anti-p50 (sc-1190X); and monoclonal IgG2a mouse anti-p52 (sc-7386X).
Fluorescence in situ chromosomal hybridization
Metaphase spreads were prepared as described previously.39 A bacterial artificial chromosome (BAC) clone containing the BCL3 locus (clone no. 84C16 of the RPCI-11 Human BAC Library; Invitrogen, Carlsbad, CA) was labeled with digoxigenin-11–deoxyuridine triphosphate (digoxigenin-11-dUTP) (Roche Diagnostics, Mannheim, Germany) by nick translation. Hybridization and washing were performed according to the manufacturer's instructions, and hybridization signals were detected by a fluorescein isothiocyanate (FITC)–conjugated antidigoxigenin antibody (Roche Diagnostics). The chromosomes and nuclei were counterstained with propidium iodide. Fluorescence in situ chromosomal hybridization (FISH) results were analyzed with a fluorescence microscope (Olympus, Tokyo, Japan).39 A charge-coupled device camera (CoolSNAP/OL; Olympus) attached to the fluorescence microscope and LuminaVision software (Mitani, Fukui, Japan) were used to capture and process images.
Bisulfite DNA sequencing
Bisulfite conversion of DNA was performed using a CpGenome DNA Modification Kit (Intergen, Purchase, NY). Bisulfite-modified DNA (50 ng) was subjected to PCR amplification of the CpG island ofBCL3 using a reverse primer 5′-CAAAATCAACCAAACCATC-3′ and a forward primer 5′-GAGTAGAGTTTGGAGAAAT-3′. The PCR products were ligated into pGEM-T EASY vector, and transformation and extraction of DNA were performed by established methods. Nucleotide sequencing to determine the methylation status of the insert was performed with a BigDye Terminator Cycle Sequencing Kit (Applied Biosystems), and the sequencing reactions were resolved on an automated sequencer (ABI Prism 310 Genetic Analyzer, Applied Biosystems).
Indirect immunofluorescence
Cells were pelleted onto poly-L-lysine–coated coverslips (Nacalai Tesque, Kyoto, Japan) by a cytocentrifuge (600 rpm, 3 minutes). The cells were washed with PBS and fixed for 15 minutes with 3.0% paraformaldehyde in PBS at room temperature. The fixed cells were treated with 0.1% Triton X-100 for 4 minutes and cold methanol for 5 minutes. The cells were then permeabilized with 0.05% PBT solution (0.05% Tween 20 in PBS containing 0.1% fetal calf serum [FCS]) for 5 minutes and blocked with 2 Blocking (DAKO, Carpinteria, CA). After 3 washes with 0.05% PBT, the specimens were incubated with primary antibodies (polyclonal rabbit anti–Bcl-3; polyclonal rabbit anti-p65 (sc-372), Santa Cruz Biotechnology; polyclonal rabbit anti-p50 (no. 06-414), Upstate Biotechnology, Lake Placid, NY), which were diluted 1:100 in 0.05% PBT for 1 hour at room temperature. They were washed 3 times with 0.05% PBT and incubated with a secondary antibody conjugated to Alexa 488 (Molecular Probes, Eugene, OR) for 20 minutes at room temperature. After rinsing in PBS, the DNA was stained with DAPI (4,6 diamidino-2-phenylindole) (Vysis, Downers Grove, IL). Preparations were examined and photographed using a camera-equipped fluorescence microscope (Olympus).
Results
Hierarchic clustering that differentiates ALCL and HD
Table 1 summarizes the characteristics of ALCL and HD cell lines used in this study. Three of the 4 ALCL cell lines carried cytogenetically identified t(2;5)(p23;q35), whereas the translocation of DEL was t(5;6)(q35;p21). Genomic DNA prepared from the 4 ALCL lines was subjected to LD-PCR amplifying a DNA fragment encompassing theNPM/ALK fusion point. The results showed that the 4 cell lines were positive for amplification and the sizes of the PCR products were unique to each cell line. Western blot analysis confirmed restrictive expression of ALK protein in ALCL cells. Deletion and mutation within the IKBA gene of KM-H2 and L428 were detected by appropriate reverse transcriptase (RT)–PCR and sequencing analysis. The genotype was determined by receptor gene-rearrangement studies.
Characteristics of ALCL and HD cell lines
| Cell lines . | Genetic lesions . | Genotype . | LD-PCR ofNPM/ALK . | ALK protein expression . |
|---|---|---|---|---|
| ALCL cell lines | ||||
| Karpas 299 | t(2;5)(p23;q35) | T cell | +(3.1 kb) | + |
| SU-DHL-1 | t(2;5)(p23;q35) | T cell | +(2.1 kb) | + |
| DEL | t(5;6)(q35;p21) | B cell* | +(1.4 kb) | + |
| SR-786 | t(2;5)(p23;q35) | T cell | +(1.4 kb) | + |
| HD cell lines | ||||
| KM-H2 | Deletion in theIKBA gene | B cell | – | – |
| L428 | Point mutation in the IKBA gene | B cell | – | – |
| HDLM-2 | Not available | T cell | – | – |
| Cell lines . | Genetic lesions . | Genotype . | LD-PCR ofNPM/ALK . | ALK protein expression . |
|---|---|---|---|---|
| ALCL cell lines | ||||
| Karpas 299 | t(2;5)(p23;q35) | T cell | +(3.1 kb) | + |
| SU-DHL-1 | t(2;5)(p23;q35) | T cell | +(2.1 kb) | + |
| DEL | t(5;6)(q35;p21) | B cell* | +(1.4 kb) | + |
| SR-786 | t(2;5)(p23;q35) | T cell | +(1.4 kb) | + |
| HD cell lines | ||||
| KM-H2 | Deletion in theIKBA gene | B cell | – | – |
| L428 | Point mutation in the IKBA gene | B cell | – | – |
| HDLM-2 | Not available | T cell | – | – |
Immunoglobulin heavy chain gene was rearranged.
+ indicates positive; –, negative.
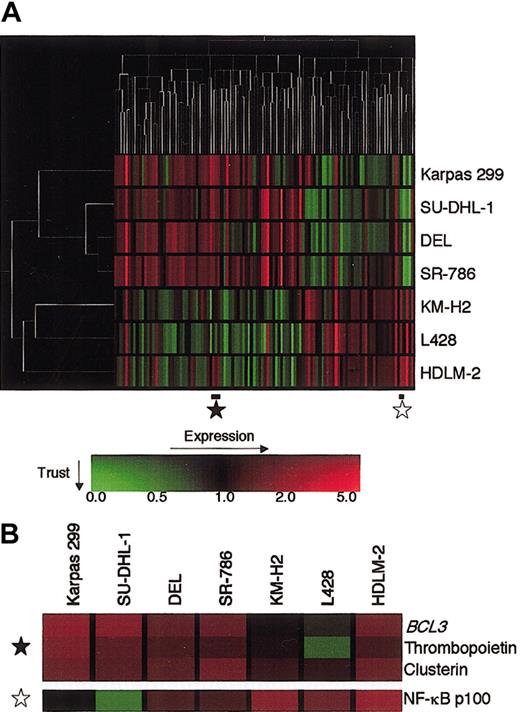
We profiled 4 ALCL and 3 HD cell lines using Atlas Human 1.2 Array containing 1176 cDNA fragments of known human genes, including several housekeeping genes. After global normalization, we selected a total of 110 genes that were differentially expressed between the 2 disease groups at a P value of less than 0.1 by the Student t test. Hierarchic clustering analysis based upon the levels of these genes was applied to both axes using the GeneSpring software. As shown in Figure 1A, the analysis effectively separated ALCL and HD and highlighted the gene expression pattern characterizing the 2 groups. The ALCL branch diverged to Karpas 299 and to the remaining 3 cell lines, generating a tight cluster, whereas the T-cell type HDLM-2 was separated from the other 2 B-cell type HD cell lines. Examination of the gene axis revealed that 69 genes were expressed to a higher degree in ALCL than HD, whereas 41 genes were expressed at a higher level in HD than ALCL. The ALCL-associated genes included BCL3, which lay close to clusterin and whose preferential expression in ALCL was described previously40 (Figure 1B). In contrast, the gene for NF-κB p100, the precursor of p52, was included in the HD-associated gene group (Figure 1B).
Hierarchic clustering analysis of gene expression of 4 ALCL and 3 HD cell lines.
(A) Expression data of 110 genes were selected to optimally separate ALCL and HD. Analysis was performed with GeneSpring software. The unrooted tree of each axis, where the length of the branches represents a similarity in the distance of expression profiles, shows the relationship of the samples (left) and the genes (top). The gene expression values and trust are color-coded as indicated by the scale on the bottom: red indicates the expression level above a median level across all samples, green indicates below the median, and the brightness represents the trust of each value. (B) Enlargement of proportions of the expression profile indicated by stars in panel A. In this matrix, each column represents a sample and each row a gene.
Hierarchic clustering analysis of gene expression of 4 ALCL and 3 HD cell lines.
(A) Expression data of 110 genes were selected to optimally separate ALCL and HD. Analysis was performed with GeneSpring software. The unrooted tree of each axis, where the length of the branches represents a similarity in the distance of expression profiles, shows the relationship of the samples (left) and the genes (top). The gene expression values and trust are color-coded as indicated by the scale on the bottom: red indicates the expression level above a median level across all samples, green indicates below the median, and the brightness represents the trust of each value. (B) Enlargement of proportions of the expression profile indicated by stars in panel A. In this matrix, each column represents a sample and each row a gene.
High-level expression of BCL3 in ALCL cells
To confirm the higher levels of BCL3 expression in ALCL than HD, we performed Northern blotting using the BCL3 cDNA clone as a probe and Western blotting using an anti–Bcl-3 polyclonal antibody. As shown in Figure 2, the levels of BCL3 expression at both the mRNA and protein levels were in good accordance with those of Atlas array analysis. The level of T-cell type HDLM-2 was higher than the other 2 B-cell type HD cell lines. A comparison with other types of cell lines revealed a predominant level of BCL3 expression in ALCL.
Expression of BCL3 mRNA and Bcl-3 protein in ALCL, HD, and other hematologic tumor cell lines.
(A) Northern blotting analysis of RNA extracted from ALCL, HD, and other hematologic tumor cell lines. Total RNA samples (10 μg) were electrophoresed on a MOPS (3-[N-Morpholino]propanesulphonic acid)–formaldehyde gel, and the patterns of the 28S and 18S ribosomal RNA bands were observed to verify the comparable amounts of RNA in each lane (bottom). (B) Western blotting analysis of ALCL, HD, and other hematologic tumor cell lines for Bcl-3 protein expression. Cell lysates were prepared from 1 × 106 cells, and an aliquot was loaded. Bcl-3 migrates around 58 kDa in sodium dodecyl sulfate (SDS)–polyacrylamide gels.
Expression of BCL3 mRNA and Bcl-3 protein in ALCL, HD, and other hematologic tumor cell lines.
(A) Northern blotting analysis of RNA extracted from ALCL, HD, and other hematologic tumor cell lines. Total RNA samples (10 μg) were electrophoresed on a MOPS (3-[N-Morpholino]propanesulphonic acid)–formaldehyde gel, and the patterns of the 28S and 18S ribosomal RNA bands were observed to verify the comparable amounts of RNA in each lane (bottom). (B) Western blotting analysis of ALCL, HD, and other hematologic tumor cell lines for Bcl-3 protein expression. Cell lysates were prepared from 1 × 106 cells, and an aliquot was loaded. Bcl-3 migrates around 58 kDa in sodium dodecyl sulfate (SDS)–polyacrylamide gels.
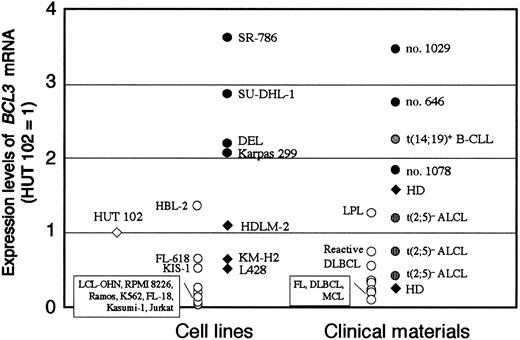
We next developed real-time RT-PCR to quantify the BCL3 mRNA levels. The forward primer was designed for exons 4 through 5 and the reverse primer for exon 6 to prevent amplification of genomic DNA. The PCR product was sequenced to verify that the expected fragment was amplified. The calculated BCL3 mRNA copy number was normalized by the level of 18S rRNA of each sample, and the ratio ofBCL3/18S rRNA of HUT 102 cell line was arbitrarily defined as 1. Figure 3 summarizes theBCL3 mRNA levels in a variety of hematologic tumor cell lines and clinical materials. The levels of the 4 ALCL cell lines and 3 clinical materials with t(2;5)+ ALCL were comparable to or higher than that of a B-CLL carrying t(14;19)(q32;q13), in which highBCL3 mRNA expression resulting from the translocation was previously determined.32 Among t(2;5)+ ALCL cells, SR-786 showed the highest level of BCL3 mRNA. Other types of lymphoid and myeloid disease expressed BCL3 mRNA at low levels. These data clearly confirmed that t(2;5)+ ALCL cells express a predominant level of BCL3 among hematologic tumors.
Real-time quantitative RT-PCR to measure theBCL3 mRNA levels in hematologic tumor cells.
The BCL3/18S rRNA ratio of each test material was normalized to that of HUT 102 (= 1). The t(2;5)+ ALCL, t(2;5)− ALCL, and HD are highlighted with appropriate symbols. B-CLL indicates B-cell chronic lymphocytic leukemia; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; MCL, mantle cell lymphoma; and LPL, lymphoplasmacytoid lymphoma. Molecular analyses of t(14;19)+ B-CLL and t(2;5)+ ALCL (cases no. 646, 1029, 1078) were described previously.32 37
Real-time quantitative RT-PCR to measure theBCL3 mRNA levels in hematologic tumor cells.
The BCL3/18S rRNA ratio of each test material was normalized to that of HUT 102 (= 1). The t(2;5)+ ALCL, t(2;5)− ALCL, and HD are highlighted with appropriate symbols. B-CLL indicates B-cell chronic lymphocytic leukemia; FL, follicular lymphoma; DLBCL, diffuse large B-cell lymphoma; MCL, mantle cell lymphoma; and LPL, lymphoplasmacytoid lymphoma. Molecular analyses of t(14;19)+ B-CLL and t(2;5)+ ALCL (cases no. 646, 1029, 1078) were described previously.32 37
BCL3 gene copy number in ALCL cells
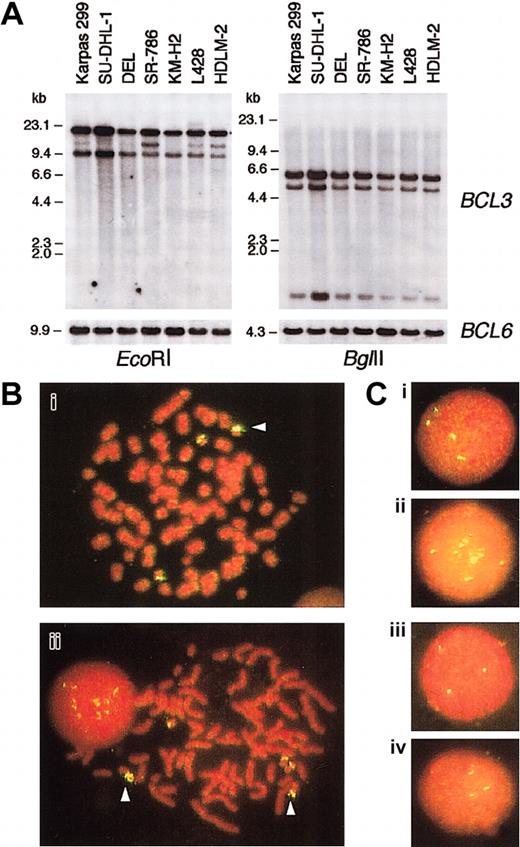
To investigate the molecular mechanism underlying the highBCL3 expression in ALCL, we first performed Southern blotting analysis of DNA extracted from ALCL cell lines using theBCL3 cDNA probe (Figure 4A). Although no rearrangement was detected within the region covered by the restriction enzymes used, hybridizing bands of SU-DHL-1 cells were intensified compared with those of other cell lines.
BCL3 gene copies in ALCL cell lines.
(A) Southern blot analysis of DNA extracted from ALCL and HD cell lines. The membranes were sequentially hybridized with BCL3cDNA and BCL6 genomic DNA probes.HindIII-digested λ phage DNA was a molecular weight marker. (B) Chromosomal FISH analysis of Karpas 299 (i) and SU-DHL-1 (ii). Metaphase preparations were hybridized with a BAC clone containing the BCL3 locus, which was labeled with digoxigenin/antidigoxigenin-FITC. The chromosomes and nuclei were counterstained with propidium iodide (PI). The arrowheads indicate duplicated (i) and amplified (ii) BCL3 gene copies. (C) Interphase FISH analysis of Karpas 299 (i), SU-DHL-1 (ii), DEL (iii), and SR-786 (iv). Original magnification Bi-ii, × 1000; Ci-iv, × 1000.
BCL3 gene copies in ALCL cell lines.
(A) Southern blot analysis of DNA extracted from ALCL and HD cell lines. The membranes were sequentially hybridized with BCL3cDNA and BCL6 genomic DNA probes.HindIII-digested λ phage DNA was a molecular weight marker. (B) Chromosomal FISH analysis of Karpas 299 (i) and SU-DHL-1 (ii). Metaphase preparations were hybridized with a BAC clone containing the BCL3 locus, which was labeled with digoxigenin/antidigoxigenin-FITC. The chromosomes and nuclei were counterstained with propidium iodide (PI). The arrowheads indicate duplicated (i) and amplified (ii) BCL3 gene copies. (C) Interphase FISH analysis of Karpas 299 (i), SU-DHL-1 (ii), DEL (iii), and SR-786 (iv). Original magnification Bi-ii, × 1000; Ci-iv, × 1000.
We next performed FISH analysis of metaphase spreads of ALCL cells using a fluorescence-labeled BAC clone containing the BCL3locus. The chromosome numbers of the 4 ALCL cell lines were in the near-triploid range. As shown in Figure 4B, SU-DHL-1 cells carried 3 normal-appearing chromosome 19's and 2 marker chromosomes containing multiple BCL3 gene copies. The amplification ofBCL3 gene copies was also observed in interphase nuclei (Figure 4C). Karpas 299 cells showed 4 hybridizing signals on interphase nuclei (Figure 4C), and the BCL3 gene was duplicated on one chromosome 19 (Figure 4B). In contrast, interphase cells of DEL and SR-786 carried 3 fluorescence signals (Figure 4C); on metaphase cell analysis, the latter had 3 copies of normal chromosome 19, whereas 2 of the 3 chromosome 19's of the former were affected by undetermined structural aberrations (data not shown). Thus, a gene dosage effect was potentially responsible for the high-levelBCL3 expression in SU-DHL-1 and Karpas 299.
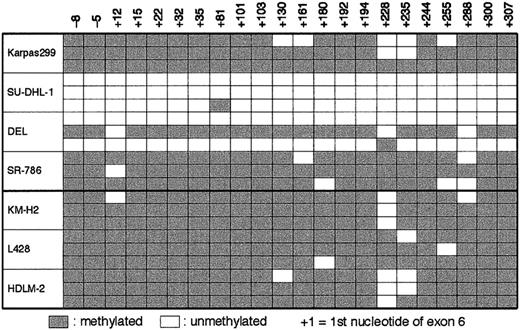
The 3′ CpG island of BCL3 was demethylated in SU-DHL-1 and DEL cells
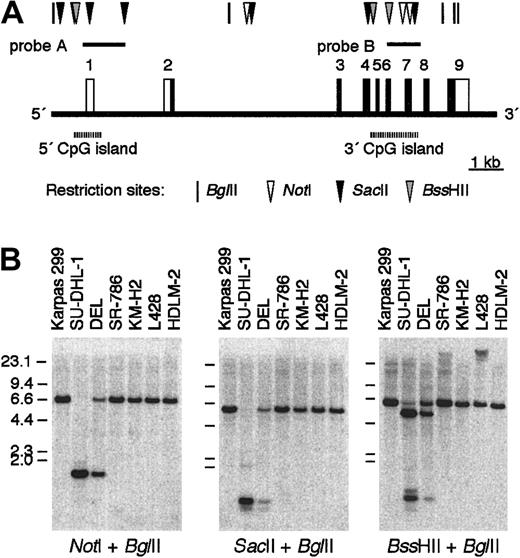
A characteristic structural feature of BCL3 is that it contains 2 independent CpG islands30 (Figure5A). The 5′ CpG island containing exon 1 was unmethylated in all the tissues tested,30 and this previous observation was confirmed in DNA from a variety of hematologic tumor cell lines (data not shown). In contrast, the methylation status of the 3′ CpG island, which encompasses exons 4 through 7 covering approximately 1.6 kb of DNA, varied among cell types and tissues.30 We therefore studied whether ALCL and HD cells exhibit different methylation patterns of the 3′ CpG and whether the degree of methylation is related to the level of expression ofBCL3. Because 6 methylation-sensitive restriction enzyme sites—that is, 2 NotI, 2 SacII, and 2BssHII sites—were identified within the 3′ CpG, we performed a double digest of DNA with BglII and either one of the 3 enzymes and hybridized the DNA with probe B representing the 3′ CpG. The results showed no digestion of DNA from the hematologic tumor cell lines tested, with the exceptions of 2 ALCL cell lines, SU-DHL-1 and DEL (Figure 5B; the data of the other cell lines are not shown). Because the restriction sites were distributed within a 1.2-kb region, the 3′ CpG of these 2 cell lines was determined to be entirely unmethylated.
Methylation status of the 3′ CpG island ofBCL3.
(A) A map of BCL3 focusing upon the 5′ and 3′ CpG islands according to the draft sequence registered in the database (accession no. NT011109). Restriction sites for BglII, NotI,SacII, and BssHII are indicated. (B) Southern blotting analysis of DNA from ALCL and HD cell lines digested byBglII and either one of the 3 methylation-sensitive restriction enzymes. The blots were hybridized with probe B representing the 3′ CpG island.
Methylation status of the 3′ CpG island ofBCL3.
(A) A map of BCL3 focusing upon the 5′ and 3′ CpG islands according to the draft sequence registered in the database (accession no. NT011109). Restriction sites for BglII, NotI,SacII, and BssHII are indicated. (B) Southern blotting analysis of DNA from ALCL and HD cell lines digested byBglII and either one of the 3 methylation-sensitive restriction enzymes. The blots were hybridized with probe B representing the 3′ CpG island.
We next prepared bisulfite-treated DNA of the ALCL and HD cell lines, in which unmethylated cytosine was selectively converted to uracil. A 360-bp fragment within the 3′ CpG island, which contained 22 CpG sites, was amplified by PCR using appropriately designed primers. Sequencing analysis revealed that 3 of 3 and 2 of 3 clones of SU-DHL-1 and DEL, respectively, showed conversion from C to T at almost all the CpG sites, whereas the sequenced areas of Karpas 299, SR-786, and the 3 HD cell lines were more than 90% methylated (Figure6). We applied the bisulfite DNA sequencing to 3 clinical materials of t(2;5)+ ALCL. The frequencies of demethylation calculated from 10 independent clones for each sample were 0.9% (no. 646), 8.6% (no. 1029), and 7.3% (no. 1078).
Sequencing analysis of bisulfite-treated DNA from ALCL and HD cell lines for the 3′ CpG island of BCL3.
Numbers indicating the nucleotide positions refer to the 5′ boundary ofBCL3 exon 6. The unmethylated CpG sites are indicated by open cells, whereas the methylated CpG sites are indicated by gray cells.
Sequencing analysis of bisulfite-treated DNA from ALCL and HD cell lines for the 3′ CpG island of BCL3.
Numbers indicating the nucleotide positions refer to the 5′ boundary ofBCL3 exon 6. The unmethylated CpG sites are indicated by open cells, whereas the methylated CpG sites are indicated by gray cells.
Comparison of NF-κB activities between ALCL and HD cells
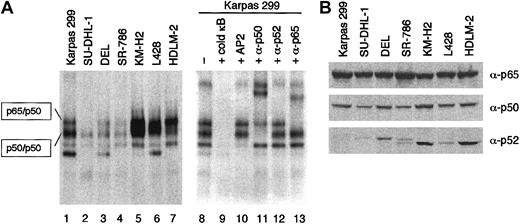
Because BCL3 encodes a unique nuclear protein belonging to the IκB family of inhibitors of NF-κB transcriptional factors,21-29 high-level expression of BCL3 is likely to affect the NF-κB activity of ALCL cells. Whole-cell lysates of ALCL and HD cell lines were incubated with the32P-labeled oligonucleotide probe containing a κB binding site and analyzed by EMSA (Figure 7A). High levels of NF-κB binding activity in the HD cell lines were clearly demonstrated as reported previously.12 On the other hand, the ALCL cells exhibited considerable levels of (p50)2 homodimer activity determined by supershift analysis. However, the shifted bands representing the p65/p50 heterodimer were much less intense than those of HD; SU-DHL-1 cells lacked a measurable level of p65/p50 activity. The activities of both (p50)2 and p65/p50 of Karpas 299 cells were higher than those in the remaining 3 ALCL cells, potentially accounting for the divergence of ALCL cell lines on array analysis.
Comparison of NF-κB activity between ALCL and HD.
(A) NF-κB DNA binding activity of ALCL (lanes 1 to 4) and HD (lanes 5 to 7) cells was analyzed by EMSA. The whole-cell lysates were incubated with 32P-labeled oligonucleotide probe containing a κB site and resolved on a 5% nondenaturing polyacrylamide gel. Supershift assay of Karpas 299 cells using antibodies against p65, p50, and p52 determined the components of NF-κB binding complexes (lanes 11 to 13). Lane 9, addition of an excess amount of unlabeled probe; lane 10, addition of the unlabeled AP2 probe (5′-GATCGAACTGACCGCCCGCGGCCCGT-3′) to eliminate nonspecific binding. (B) Western blotting analysis of ALCL and HD cell lines for the expression of p65, p50, and p52. Cell lysates were prepared from 1 × 106 cells, and an aliquot was loaded.
Comparison of NF-κB activity between ALCL and HD.
(A) NF-κB DNA binding activity of ALCL (lanes 1 to 4) and HD (lanes 5 to 7) cells was analyzed by EMSA. The whole-cell lysates were incubated with 32P-labeled oligonucleotide probe containing a κB site and resolved on a 5% nondenaturing polyacrylamide gel. Supershift assay of Karpas 299 cells using antibodies against p65, p50, and p52 determined the components of NF-κB binding complexes (lanes 11 to 13). Lane 9, addition of an excess amount of unlabeled probe; lane 10, addition of the unlabeled AP2 probe (5′-GATCGAACTGACCGCCCGCGGCCCGT-3′) to eliminate nonspecific binding. (B) Western blotting analysis of ALCL and HD cell lines for the expression of p65, p50, and p52. Cell lysates were prepared from 1 × 106 cells, and an aliquot was loaded.
To clarify the difference in NF-κB activity between ALCL and HD, we analyzed the levels of p65, p50, and p52 by Western blotting analysis. As shown in 7B, there was no significant difference in the expression levels of p65 and p50. The levels of p52 in ALCL were quite low in accordance with the result of Atlas array analysis (Figure 1B).
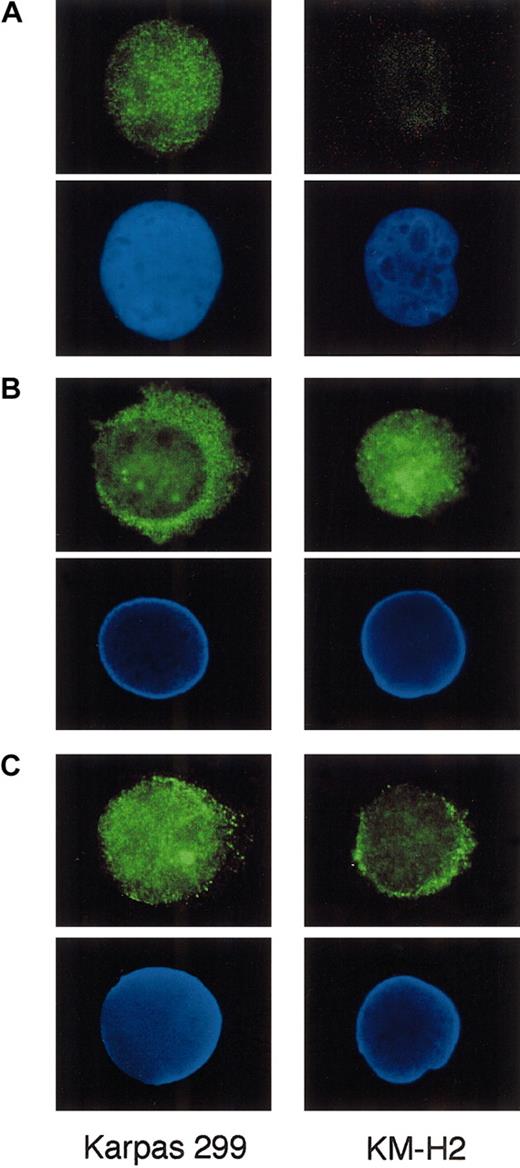
We finally stained Karpas 299 and KM-H2 cells for Bcl-3, p65, and p50 by indirect immunofluorescence to study whether these proteins exhibit differential subcellular localization between ALCL and HD. Cytocentrifuged preparations of the cells were incubated with primary antibodies against these proteins and labeled with the Alexa 488–conjugated secondary antibody. As shown in Figure8A, Karpas 299 cells displayed bright nuclear staining against Bcl-3, confirming the nuclear localization of this particular protein belonging to the IκB family. In contrast, p65 was predominantly localized in the cytoplasm of Karpas 299, whereas KM-H2 exhibited prominent nuclear staining for p65 (Figure 8B).
Indirect immunofluorescence of Karpas 299 and KM-H2 cells against Bcl-3, p65, and p50.
Cytocentrifuged cells were incubated with anti–Bcl-3 (A), p65 (B), and p50 (C) antibodies and labeled with the Alexa 488–conjugated secondary antibody. The positions of nuclei, as revealed by DAPI staining in the lower panels. Original magnification × 1000.
Indirect immunofluorescence of Karpas 299 and KM-H2 cells against Bcl-3, p65, and p50.
Cytocentrifuged cells were incubated with anti–Bcl-3 (A), p65 (B), and p50 (C) antibodies and labeled with the Alexa 488–conjugated secondary antibody. The positions of nuclei, as revealed by DAPI staining in the lower panels. Original magnification × 1000.
Immunofluorescence against p50 was detected in both the nucleus and cytoplasm of these 2 cell types; the nuclei of Karpas 299 were more intensely labeled than those of KM-H2 (Figure 8C). These results suggest that high-level nuclear Bcl-3 in ALCL cells may significantly affect the localization of NF-κB components.
Discussion
The cDNA array technology has identified global gene expression differences among subtypes of hematologic tumors and provided new diagnostic and prognostic markers.40-44 However, the studies have not been able to predict whether these markers are implicated in the oncogenic process of particular tumors or whether the expression of each gene reflects only the characteristics of normal counterpart cells from which the tumor developed. Here, we showed the gene expression profiles differentiating t(2;5)+ ALCL and HD, both of which share many clinical and pathologic features. The array analysis and additional studies at both the mRNA and protein levels clearly demonstrated predominant expression levels ofBCL3 in t(2;5)+ ALCL. We next found that theBCL3 gene of ALCL cells was altered by gene amplification and/or demethylation of the intragenic CpG island, potentially accounting for the high-level expression of the gene. These 2 alterations, however, were not common to all t(2;5)+ ALCL cells tested; SR-786, showing the highest level of BCL3mRNA, lacked amplification or demethylation. Nevertheless, it is most likely that high-level expression of BCL3 plays an oncogenetic role in the development of t(2;5)+ALCL.
The Bcl-3 protein contains 7 ankyrin repeats and shares structural features with the members of the IκB protein family.21-29 In contrast to other IκB proteins, Bcl-3 localizes within the nucleus and is predominantly associated with p50 or p52 homodimers.21-29 Although p50 and p52 have no defined transactivating domains, the association with Bcl-3 can lead to the conversion of these homodimers to positive regulators of gene expression.24,25 On the other hand, other studies suggested that Bcl-3 promotes removal of the suppressive p50 homodimer from DNA, thereby allowing active NF-κB heterodimers to bind to the κB sites.21 23 Thus, it remains to be determined whether Bcl-3 ultimately functions as a coactivator or antirepressor or both.
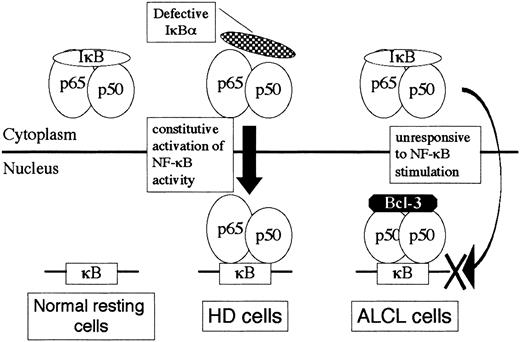
Evidences for the involvement of NF-κB activity in HD cells are numerous.11-16 The proposed mechanisms underlying the constitutive activation of NF-κB in HD include molecular defects in the components of the IκB kinase cascade, mutation or loss of IκB at the DNA level, and modification of NF-κB rendering it insensitive to IκB inhibition.11 Very recently, amplification of theREL gene copies in HD was reported.45 In contrast, little is known about the role of NF-κB in ALCL. Because ALCL cells invariably express CD30, which is a member of the tumor necrosis factor receptor superfamily,10 the CD30/NF-κB signaling pathway is likely to function in ALCL cells; indeed, ALCL cells express the NF-κB subunits at comparable levels to those in HD.20 In the present study, a comparison between ALCL and HD revealed the following characteristics of ALCL: (1) (p50)2 homodimer activity was consistently detected, (2) p52 expression levels were low, (3) p65/p50 heterodimer activity was low or below the level of detection on EMSA, and (4) nuclear Bcl-3 expression was markedly elevated. Based upon these observations, we propose a model for the interaction of NF-κB/IκB/Bcl-3 molecules in ALCL cells (Figure9). In contrast to HD cells, in which NF-κB is constitutively activated through defects of IκB at some levels, high-level Bcl-3 sequestrates (p50)2 homodimer to the nucleus in ALCL. Upon stimulation of NF-κB, the active p65/p50 heterodimer translocated to the nucleus may not be capable of binding to the κB sites that have been occupied by the (p50)2/Bcl-3 ternary complex. This model would agree well with the observation that CD30 activation of ALCL cells leads to apoptosis along with impairment of activation of prosurvival transcription factor NF-κB.20 We further propose that the differential gene expression profiles of ALCL and HD may be in part a reflection of the distinctive NF-κB activity between the 2.
A model for NF-κB/IκB/Bcl-3 interaction in HD and ALCL cells.
A hypothetical model for differential NF-κB binding activity between HD and ALCL focusing upon the inhibitory molecules IκBα and Bcl-3.
A model for NF-κB/IκB/Bcl-3 interaction in HD and ALCL cells.
A hypothetical model for differential NF-κB binding activity between HD and ALCL focusing upon the inhibitory molecules IκBα and Bcl-3.
BCL3 was originally identified as a putative oncogene by cloning the breakpoint of t(14;19)(q32;q13), which is a recurrent chromosomal translocation in B-CLL.31-33 As a result of translocation, BCL3 on 19q13 is juxtaposed to the α constant regions of immunoglobulin heavy chain gene (IgH) on 14q32, leading to markedly increased expression of BCL3mRNA.32 Transgenic mice bearing an IgHenhancer–driven BCL3 show splenomegaly and accumulation of B cells in the lymph nodes, bone marrow, and peritoneal cavity, although the mice ultimately do not develop B-cell lymphoid tumors.46 On the other hand, BCL3 was recently shown to enhance T-cell survival.34 Acquired immune response to foreign antigens is a process of activation, differentiation, and proliferation of effector cells, followed by apoptosis of antigen-specific T cells. The molecular mechanisms implicated in apoptosis of activated T cells include “cytokine withdrawal” regulated by the Bcl-2–related protein, Bim, and induction of Fas ligand expression leading to death of neighboring Fas+ T cells.47 In contrast, activated T cells can survive in the presence of materials associated with infections, called adjuvants, maintaining the numbers of responding T cells.34 A cDNA array analysis comparing gene expression in activated T cells that had or had not been exposed to bacterial products disclosed that BCL3 was activated in response to the exposure to adjuvants.34 The role of BCL3and NF-κB activity in the T-cell survival was confirmed byBCL3 transduction experiments with retroviruses.34 Although it remains to be determined howBCL3 transcription is induced by adjuvants, these studies on immunologically activated T cells as well as our present study on ALCL, which is included in T-cell neoplasms, strongly suggest thatBCL3 is involved in the proliferation and survival of not only B cells but also T cells.
The BCL3 gene contains 2 CpG islands.30 The 5′ CpG, covering the promoter and the transcription initiation site, was unmethylated in all the tissues and hematologic tumor cell lines tested. Sequencing analysis of the promoter of BCL3 of ALCL disclosed a germ-line configuration in this region (data not shown), indicating that genetic alteration of the promoter at the nucleotide level does not account for the high-level expression of BCL3in ALCL. In contrast, the 3′ CpG was unmethylated in sperm DNA and methylated to varying degrees in several other tissues,30whereas essentially methylated in DNA from peripheral blood leukocytes30 and most hematopoietic tumor cells. The function of 3′ CpG is currently unknown. A study of the apolipoprotein E gene showed that transcription of the gene was not related to methylation status of the 3′ CpG.48 On the other hand, methylation of the intronic CpG of mouse insulinlike growth factor type 2 receptor gene (Igf2r) acted as an imprinting signal and was positively correlated to the transcriptional activity ofIgf2r.49 We showed here that the 2 particular ALCL cells expressing a high level of BCL3 mRNA carried almost entirely unmethylated 3′ CpG islands. Thus, it is possible that transcription of BCL3 is negatively controlled, at least in part, by methylation of the 3′ CpG.
The previous studies and our present work suggest that t(2;5)+ ALCL cells are characterized by the presence of NPM/ALK fusion protein predominantly in the cytoplasm, expression of the CD30 molecules on the cell surface, and high-level expression of Bcl-3 in the nucleus. Because there is no link between the signaling cascades activated by NPM/ALK and those by CD30 stimulation, t(2;5) leading to the generation of NPM/ALK may occur at a stage of T-cell differentiation that expresses CD30, and therefore CD30 expression itself may not be involved in the development of ALCL9; indeed, ALCL as well as HD cells carry the CD30 gene with a germ-line configuration.50 In contrast, the BCL3 gene of ALCL cells showed genetic and epigenetic modifications, both of which are likely associated with oncogenesis. We suggest that the high-level expression of BCL3 is an important process in the development of ALCL and contributes to the cellular features of this particular lymphoid tumor.
The authors thank Mitsubishi Kagaku Bio-clinical Laboratories (Tokyo, Japan) for its technical assistance with the FISH analysis.
Prepublished online as Blood First Edition Paper, November 27, 2002; DOI 10.1182/blood-2002-08-2464.
Supported by grants-in-aid from the Ministry of Education, Culture, Sports, Science and Technology (no. 13470204) of Japan.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Hitoshi Ohno, Department of Hematology and Oncology, Graduate School of Medicine, Kyoto University, 54-Shogoin-Kawaramachi, Sakyo-ku, Kyoto 606-8507, Japan; e-mail:hohno@kuhp.kyoto-u.ac.jp.

![Fig. 2. Expression of BCL3 mRNA and Bcl-3 protein in ALCL, HD, and other hematologic tumor cell lines. / (A) Northern blotting analysis of RNA extracted from ALCL, HD, and other hematologic tumor cell lines. Total RNA samples (10 μg) were electrophoresed on a MOPS (3-[N-Morpholino]propanesulphonic acid)–formaldehyde gel, and the patterns of the 28S and 18S ribosomal RNA bands were observed to verify the comparable amounts of RNA in each lane (bottom). (B) Western blotting analysis of ALCL, HD, and other hematologic tumor cell lines for Bcl-3 protein expression. Cell lysates were prepared from 1 × 106 cells, and an aliquot was loaded. Bcl-3 migrates around 58 kDa in sodium dodecyl sulfate (SDS)–polyacrylamide gels.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/7/10.1182_blood-2002-08-2464/3/m_h80734087002.jpeg?Expires=1767870842&Signature=tPWFUSpthlsWH13xmBywpFQ3q4br4LQYG-LKMt9OdgVtVcjjO6qwROFK37CbuulUKF8mLRv2YeSDcoHC3vF8zmdEGtRv2QqO7ln0dAyK4Fk3cFOSlwzlSh-aL6tv2SpzerFW5jjXOvOXWXaXyU66maOfLGiTijRPPR4qg5-ys-8Eq-rpRxZXsKNPBe9ZysM~JFidnqUZOA5djKw-4F84EVqJ0Idj5-3Bhz4oYcqrL0VxxUNNwTX4gYur~TZhrgLSmzEIOlheyC3jMCIt6hPWzNoGStAqwC5~Kas3fLcEPitXaGkOMx4THcgfIbdgxdzoTN~u0-7WCy9ihYYuqY7ePg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal