Protein C is a member of the vitamin K– dependent protein family. Proteins in this family have similar γ-carboxyglutamic acid (Gla)–rich domains, but their affinities for negatively charged phospholipid membranes vary more than 1000-fold. We have shown that it is possible to enhance anticoagulant activity and membrane affinity of protein C by selective mutagenesis of the Gla domain. In this study, 3 new mutants, Q10G11N12 (QGN), S23E32D33Y44 (SEDY), and Q10G11N12S23E32D33Y44 (QGNSEDY), were created. In plasma-based coagulation assays, the activated form of QGNSEDY (QGNSEDY-APC) demonstrated approximately 20-fold higher anticoagulant activity than wild-type activated protein C (WT APC), while QGN-APC and SEDY-APC did not. Both normal activated factor V (FVa) and FVa Leiden (Arg506Gln) were degraded much more efficiently by QGNSEDY-APC than by WT APC in the presence as well as in the absence of protein S. Binding of protein C variants to negatively charged phospholipid membranes was investigated using light scattering and the BIAcore technique. QGNSEDY demonstrated 3- to 7-fold enhanced binding as compared with WT protein C, suggesting the membrane affinity to be influenced by several residues located at different parts of the Gla domain. The anticoagulant activity as well as phospholipid binding ability was only enhanced when multiple regions of the Gla domain were modified. The results provide insights into the molecular mechanisms that are involved in determining the binding affinity of the interaction between Gla domains and phospholipid membranes. The unique properties of QGNSEDY-APC suggest this APC variant possibly to have greater therapeutic potential than WT APC.
Introduction
Negatively charged phospholipid membranes play an important role in blood coagulation, providing a suitable surface for the assembly of enzyme-cofactor complexes, which efficiently convert circulating zymogens to active enzymes. Phosphatidylserine (PS) is a key component in the negatively charged membrane that supports the clotting reactions.1-3 In most cells, PS is normally mainly present in the inner leaflet of the cell membrane, but certain cellular processes such as platelet activation result in the exposure of PS on the cell surface. The vitamin K–dependent coagulation proteins in plasma bind to the surface of negatively charged phospholipid with a common mechanism involving the γ-carboxyglutamic acid (Gla)–rich domains of these proteins.4 Vitamin K is required in the posttranslational carboxylation of glutamic acid residues to Gla.5 The Gla domains comprise approximately 45 amino acid residues, and they are structurally very similar in the different vitamin K–dependent proteins.6 Despite the extensive structural similarity between different Gla domains, they bind negatively charged phospholipid with very different affinity, with dissociation constants (Kd's) ranging more than 1000-fold.6 This variation in phospholipid binding ability is related to amino acid sequence differences at relatively few positions. This has made it possible to modulate the phospholipid binding abilities of this group of proteins by site-directed mutagenesis.7-9
The nonvitamin K–dependent cofactors of blood coagulation also interact with the phospholipid membranes, certain proteins being membrane spanning, whereas others, such as factor V (FV) and factor VIII (FVIII), contain binding sites for negatively charged phospholipid membranes.2,10 FV circulates as a procofactor to activated FV (FVa), which functions as a cofactor to activated FX (FXa) in the activation of prothrombin to thrombin. Activated FVIII (FVIIIa) serves a similar function in the FIXa-mediated activation of FX.2 3
The protein C anticoagulant system, which includes the vitamin K–dependent proteins protein C and protein S, regulates the activities of FVa and FVIIIa by limited proteolysis (reviewed by Dahlbäck et al11 and Esmon et al12). Protein C is activated on the endothelial cell surface by the thrombin-thrombomodulin complex, and the serine protease activated protein C (APC) down-regulates coagulation by cleaving a limited number of peptide bonds in each of FVa and FVIIIa. Protein S serves as a cofactor to APC by increasing the affinity of APC for the membrane13 and also by changing the distance of the active site of APC relative to the membrane.14,15 In FVa degradation, APC-mediated cleavages at Arg506 and Arg306 result in inhibition of FVa activity, the Arg506 cleavage resulting in partial inhibition of FVa activity,16,17 whereas the Arg306 cleavage completely inactivates FVa.16-18 Protein S is an important cofactor for the Arg306 cleavage, whereas the Arg506 is less affected by protein S.19 In the degradation of FVIIIa, the effect of APC is stimulated by a synergistic APC cofactor activity between protein S and the intact form of FV.20
A common mutation in FV, which replaces Arg506 with a Gln (FV Leiden),21 results in a hypercoagulable condition known as APC resistance.22,23 This is the most common inherited risk factor for venous thrombosis. Heterozygous deficiencies of protein C or protein S are other genetic risk factors for venous thrombosis.23 The association between defects of the protein C system and increased risk of thrombosis demonstrates the biologic importance of the protein C system. Protein C is a candidate for treatment of certain thrombotic states.24,25 Moreover, APC has recently been shown to be efficient in the treatment of sepsis, a life-threatening condition associated with hypercoagulation and generalized inflammatory reactions.26 APC not only has anticoagulant properties but also anti-inflammatory effects, which are poorly understood.24 Whether the positive therapeutic effects depend on both the anticoagulant and anti-inflammatory properties of APC is not known.
Protein C interacts with negatively charged phospholipid membranes with relatively low affinity.6 We have previously reported that site-directed mutagenesis of positions 10 to 12, 32, and 33 in protein C result in protein C variants with enhanced membrane-binding ability and anticoagulant activity.7 8 We now report on novel Gla domain mutations resulting in the creation of APC variants with very high anticoagulant activity. The results provide insights into the link between sequence variations of the Gla domain and the membrane-binding properties and the importance of membrane binding for expression of APC anticoagulant activity.
Materials and methods
Materials
Human FXa and human prothrombin were from Kordia (Leiden, Netherlands). Human protein S was purified from human plasma.27 Human plasma protein C was purified as described28 with an additional monoclonal antibody step.29 α-Thrombin was from Haematologics (Essex Junction, VT). Human FV was purified from human plasma as described.30 31 Activated partial thromboplastin time (APTT) reagent Platelin LS and thromboplastin (tissue factor) reagent Simplastin Excel were from Biomerieux (Durham, NC). Chromogenic substrates S-2238 and S-2366 were kindly provided by Chromogenix (Milan, Italy).l-α-phosphatidylserine (PS) from brain andl-α-phosphatidylcholine (PC) from egg were from Avanti Polar Lipids (Alabaster, AL). Octyl d-glucoside was from Sigma-Aldrich (St Louis, MO). The AHV5146 monoclonal antifactor V antibody, which reacts with the heavy chain of FVa, was from Haematologics. The Vectastain Elite ABC kit was obtained from Vector Laboratories (Burlingame, CA).
Phospholipid vesicles used in the FVa-degradation assay and for the light-scattering measurements were prepared by sonication. The phospholipids (10:90, or 20:80 wt/wt, PS/PC) were dissolved in CHCl3/CHOH (9:1, vol/vol) in glass tubes and dried under a mild flow of nitrogen. The phospholipids were resuspended in 2 mL 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), 150 mM NaCl, pH 7.7 (HN buffer), and vigorously vortexed for 1 minute. The phospholipid suspension was then sonicated for 10 minutes at 37°C using XL-2020 SONICATOR, an ultrasonic liquid processor from MISONIX (NY). The phospholipid vesicles used for light-scattering experiments (20:80, wt/wt, PS/PC) were isolated by gel filtration chromatography on Sepharose 4B, as described.32The phospholipid concentration was determined with the Phospholipid B kit (Wako Chemicals).
Recombinant proteins
Mutagenesis.
Mutagenesis was performed using polymerase chain reaction (PCR), essentially as previously described.8 To construct the protein C variant D23S/Q32E/N33D/H44Y (SEDY), the cDNA for the protein C variant Q32E/N33D (ED)8 was first used as template to introduce the D23S (Asp23Ser) mutation. The codon for amino acid at position 23 was changed to AGC using PCR, the sense and antisense mutant PCR primers containing 15 matching nucleotides on each side of the mutant codon. The resulting cDNA for the SED variant was further mutated at position 44, replacing the codon for His with a codon for Tyr (TAC) by PCR-based mutagenesis, as described above. The 360–base pair (bp) cDNA fragment containing the SEDY mutations was isolated after HindIII-SalI digestion and ligated together with the SalI-XbaI fragment representing the remaining part of the protein C cDNA intoHindIII-XbaI–cleaved pRc/CMV vector. The H10Q/S11G/S12N (QGN) protein C variant was created with similar methodology using mutant PCR primers introducing codons for Gln (CAA), Gly (GGC), and Asn (AAC) at positions 10 to 12. After the PCR, the cDNA was digested with BsrBI and the 200 bp 5′ fragment isolated and ligated with the BsrBI-XbaI fragment representing the remaining protein cDNA intoHindIII-XbaI–digested pRc/CMV vector. To construct the H10Q/S11G/S12N/D23S/Q32E/N33D/H44Y (QGNSEDY) variant, theHindIII-BsrBI fragment from QGN was ligated with the BsrBI-XbaI fragment from SEDY into theHindIII-XbaI–digested pRc/CMV vector. All cDNA fragments containing mutations were sequenced before transfection.
Stable expression, purification, and characterization of recombinant proteins.
The cDNAs corresponding to wild-type (WT) protein C and protein C variants QGN, SEDY, and QGNSEDY, all inserted into the eukaryotic expression vector pRc/CMV, were transfected into HEK 293 cells (CRL-1573 ATCC), high-expressing colonies selected, and recombinant protein C purified, as previously described.33The purity and integrity of the isolated protein C was evaluated by 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The GNED protein C variant used as control has been characterized previously.8 Protein C concentrations were quantified by measurement of absorbance at 280 nm using an extinction coefficient of 14.5 (280 nm, 1%, 1 cm). The purified proteins were subjected to amino acid analysis after acid hydrolysis and Gla analysis after basic hydrolysis using methods previously detailed.34
Activation of protein C and catalytic activity against small substrates.
The recombinant protein C variants were activated by thrombin, and the thrombin was removed by chromatography, essentially as previously described.8 APC concentrations were estimated by measuring absorbance at 280 nm. Amidolytic activities were determined using chromogenic substrate S-2366.
Functional activity analysis
Coagulation assays.
The anticoagulant effects of APC variants were measured in 2 plasma-based assay systems, one using an APTT reaction (intrinsic pathway), the other a tissue factor (thromboplastin; extrinsic pathway)–initiated clotting reaction. An Amelung KC10 apparatus was used. In the APTT assay, 50 μL normal human pooled plasma and 50 μL APTT LS reagent were mixed and incubated at 37°C for 200 seconds. Coagulation was initiated by the addition of 50 μL CaCl2 (25 mM). The APC was included in the CaCl2 reagent, and the concentrations given in the figures refer to the concentration in the CaCl2 mixture. The extrinsic pathway system used Simplastin Excel diluted 1:50 in Tris (tris(hydroxymethyl)aminomethane)–buffered saline (TBS) buffer containing 25 mM CaCl2. The Simplastin reagent contains tissue factor and negatively charged phospholipids. In the assay, 50 μL plasma was incubated with 50 μL APC at 37°C for 200 seconds before the addition of 100 μL of the diluted Simplastin Excel reagent that initiated the clotting reaction. Clotting times were reported as the average and range of 2 determinations.
Factor Va degradation assay.
Plasma-purified FVa as well as FV from diluted plasma were used as substrates for APC in the degradation assay, and the FVa activity was measured in a prothrombinase assay. In the prothrombinase-based FVa assay, a prothrombinase (PTase) mixture containing 0.5 μM prothrombin and 50 μM phospholipid vesicles (10:90, wt/wt, PS/PC) was prepared in 25 mM HEPES, 150 mM NaCl, and 2 mM CaCl2, pH 7.7, containing 0.5 mg/mL ovalbumin (HNO buffer). Purified FV (0.8 nM) was activated by thrombin (final concentration, 0.5 U/mL) at 37°C for 10 minutes. To measure the FVa activity, FXa (final concentration, 5 nM) and the FVa samples were added to the PTase mix, and after 2 minutes the prothrombin activation was stopped by 40-fold dilution in ice-cold EDTA (ethylenediaminetetraacetic acid) buffer. The EDTA buffer contained 50 mM Tris, 100 mM NaCl, 20 mM EDTA, and 1% polyethylene glycol (PEG) 6000, pH 7.9. The amount of thrombin formed was measured kinetically with a chromogenic substrate, S2238. Normal plasma diluted between 1:50 and 1:1600 was used as standard in the assay. In the degradation of FVa, FVa (final concentration, 0.8 nM) was incubated with increasing concentrations of APC in the presence of 25 μM phospholipid vesicles (10:90, wt/wt, PS/PC) in 25 mM HEPES, 150 mM NaCl, and 5 mM CaCl2, pH 7.7, containing 5 mg/mL bovine serum albumin (BSA; HNBSACa buffer). At defined intervals, aliquots were drawn and diluted 1:5 in ice-cold HNBSACa buffer and kept on ice until tested in the PTase-based FVa assay. To test if similar results were obtained with FVa in diluted plasma, normal or APC-resistant plasma (from an individual with homozygosity for FV Leiden) was used. The plasmas were diluted 25-fold with HNBSACa buffer and incubated with 0.5 U/mL thrombin for 10 minutes at 37°C, which was found to yield full FV activation. APC and phospholipid vesicles were added as described in the purified system and the FVa inhibition followed as described above. To determine the effect of protein S on FVa degradation, purified human protein S was included in the FVa-degradation assay at a final concentration of 100 nM.
Western blotting
The APC-mediated proteolysis of FVa was performed as described previously.35 In brief, thrombin-activated plasma-purified FVa (0.8 nM) was incubated for 25 minutes at 37°C with 0.04 nM WT APC or QGNSEDY-APC in the presence of 25 μM phospholipid (10:90, PS/PC). The samples were analyzed by Western blotting using the AHV5146 monoclonal antibody and the Vectastain Elite ABC kit, as described.35
Protein binding to phospholipid membranes
Light scattering.
Protein-membrane interaction was measured with light scattering at 90 degrees to the incident light using a Fluoromax-3 (Jobin Yvon) spectrofluorometer.32 In brief, the intensity of the light scatter of phospholipid alone (I1) and after addition of protein (I2) was measured. The ratio of the molecular weight of the protein-vesicle complex (M2) to that of vesicles alone (M1) was estimated from the following relationship: I2/I1 = (M2/M1)2(∂∂n/∂∂c2/∂n/∂c1)2, where ∂n/∂c is the refractive index of the respective species. The final phospholipid concentration was 5 μg/mL, and the buffer was TBS containing 5 mM CaCl2 and 1 mg/mL BSA. At the end of the experiment, the calcium dependence and reversibility of the binding was tested by the addition of EDTA to 10 mM final concentration. The binding data were evaluated to calculated Kdusing equation 1:
C is the concentration of free protein, and fmax is the maximum binding signal, which was estimated at high concentrations of protein C or bovine prothrombin. The assumption is made that the free protein equals the added protein. The data were alternatively analyzed as previously described32 with essentially identical results.
Surface plasma resonance.
The interaction between the protein C variants and immobilized phospholipid was also investigated using a BIAcore 2000 biosensor instrument (Biacore, Uppsala, Sweden) as described.36 The phospholipid vesicles used for these experiments were prepared by extrusion. The phospholipid vesicles were then captured on the surface of L1 sensor chip.37 Extruded liposomes with different compositions were used in the different flow cells. In cell 1, 50 μL of 0.05 mM PC vesicles (100%) was injected at a flow rate of 5 μL/min. In cell 2, 50 μL of 0.125 mM PS/PC (20%:80%) was injected at a flow rate of 2 μL/min. The buffer used was 10 mM HEPES, 150 mM NaCl, pH 7.4. The degree of coverage of the surface was measured by increase in resonance units (RUs). The slightly different conditions for PC and PS/PC vesicles were chosen because they resulted in similar levels of RUs (around 7000 RUs). The capture process permits the liposome to retain the lipid bilayer structure.38 After liposome binding, a 15 μL injection of 8 mg/mL BSA resulted in an increased signal of only 100 RUs, indicating that the dextran is no longer accessible but covered by lipid. The protein C variants in 10 mM HEPES, 150 mM NaCl, 5 mM CaCl2, pH 7.4, containing 1 mg/mL BSA were injected at different concentrations (flow rate of 30 μL/min; association phase), and the dissociation was then followed after change to buffer alone. Membranes were regenerated by injection of 5 mM EDTA (pH 8.0).
Results
Expression and purification of recombinant protein C variants
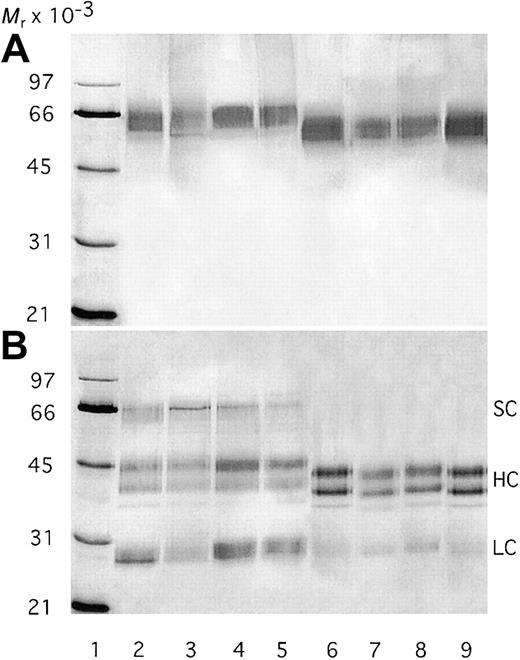
The expression levels of the different protein C variants were similar: 3 to 5 mg/L. The proteins were collected under serum-free conditions and purified with overall recoveries of 35% to 40%. The Gla content was determined after basic hydrolysis and found to be 6.3, 6.8, 7, 6.7, and 10.0 for plasma protein C, WT protein C, QGN-, SEDY-, and QGNSED–protein C, respectively. On SDS-PAGE, mutant and WT protein C migrated to similar positions consistent with a relative molecular mass (Mr) of approximately 60 000 (Figure 1). After reduction, the α, β, and γ isoforms of the heavy chain as well as the light chain are seen. In addition, all variants contained a certain amount of the single-chain form. After activation by thrombin, the heavy-chain isoforms shifted to slightly lower-molecular-weight positions, consistent with full activation. In addition, the single-chain bands disappeared.
SDS-PAGE analysis of recombinant protein C variants.
The purified protein C variants were subjected to 10% SDS-PAGE before and after activation by thrombin under both nonreducing (A) and reducing conditions (B). The proteins were visualized by silver staining. One microgram of protein was applied to each lane. Lanes 2 to 5 represent nonactivated protein C, whereas activated proteins were applied to lanes 6 to 9. Lanes 2 and 6 contain WT protein C; lanes 3 and 7, the QGN variant; lanes 4 and 8, the SEDY variant; and lanes 5 and 9, the QGNSEDY variant. Lane 1 contains the molecular weight markers. SC indicates single chain; HC, heavy chain; LC, light chain.
SDS-PAGE analysis of recombinant protein C variants.
The purified protein C variants were subjected to 10% SDS-PAGE before and after activation by thrombin under both nonreducing (A) and reducing conditions (B). The proteins were visualized by silver staining. One microgram of protein was applied to each lane. Lanes 2 to 5 represent nonactivated protein C, whereas activated proteins were applied to lanes 6 to 9. Lanes 2 and 6 contain WT protein C; lanes 3 and 7, the QGN variant; lanes 4 and 8, the SEDY variant; and lanes 5 and 9, the QGNSEDY variant. Lane 1 contains the molecular weight markers. SC indicates single chain; HC, heavy chain; LC, light chain.
Phospholipid binding of protein C variants
Two approaches were used to quantify the protein-membrane interaction, one being surface plasmon resonance, the other light scattering. In the surface plasmon resonance–based assay, phospholipid membranes of defined composition were immobilized on an L1 sensor chip. The method allowed simultaneous investigation of several phospholipid compositions. Two conditions were used, one being pure PC and the other a mixture of 20% PS and 80% PC (20:80, wt/wt). Protein C was injected, and the binding to the immobilized membrane was followed over time. The response obtained with pure PC-containing membranes was subtracted from that obtained with the PS/PC membranes. To compare the various protein C variants, they were all injected at a concentration of 0.25 μM and the binding followed over time (Figure2A). Under these conditions, WT protein C yielded a weak binding signal in contrast to the mutant QGNSEDY, which resulted in a rapid increase in binding signal. To put the protein C variants into the perspective of membrane-binding ability of previous protein C variants prepared in our laboratory, the GNED variant was tested under the same conditions. This protein C variant was previously shown with light-scattering techniques to have around 10-fold higher affinity for PS/PC membranes than WT protein C.8 The GNED variant bound better than WT protein C but less efficiently than the QGNSEDY variant. The QGN variant yielded a pattern similar to that of WT protein C, whereas SEDY bound slightly better than WT protein C but less efficiently than GNED- and QGNSEDY–protein C.
Binding of protein variants to immobilized phospholipid evaluated using BIAcore.
The various protein C variants were infused at a flow rate of 30 μL/min over 2 different phopholipid membranes, one being 100% PC, the other being PS/PC, 20:80, wt/wt. The signals (resonance units [RU]) obtained from PC membranes were subtracted from the PS/PC-derived signals to obtain the specific binding to PS/PC membranes. (A) The 5 different protein C variants were injected at a concentration of 0.25 μM (arrows at left). After approximately 90 seconds, the protein solution was replaced with buffer (arrows at right) and the dissociation was followed. (B) WT protein C (upper panel) and QGNSEDY protein C (lower panel) were infused at the 3 indicated concentrations, and association and dissociation followed over time. (C) Increasing concentrations of WT protein C or QGNSEDY protein C were infused until a stable level was obtained, when association and dissociation is at equilibrium. The RU obtained at equilibrium was plotted against the protein C concentration, and the curves were used to calculate the Kd's. (D) Membrane binding was determined with light scatter intensity. M2 is the molecular weight of the protein-membrane complex, and M1 is that of the vesicles alone. The concentration of phospholipid was 5 μg/mL, and the protein C concentrations were varied from 0 to 1 μM, yielding protein/phospholipid (P/PL) ratios up to 16. QGNSEDY, (▴); SEDY, (▵); QGN, (■); WT protein C, (●). The means of 2 to 3 different experiments are shown with error bars representing ± SD.
Binding of protein variants to immobilized phospholipid evaluated using BIAcore.
The various protein C variants were infused at a flow rate of 30 μL/min over 2 different phopholipid membranes, one being 100% PC, the other being PS/PC, 20:80, wt/wt. The signals (resonance units [RU]) obtained from PC membranes were subtracted from the PS/PC-derived signals to obtain the specific binding to PS/PC membranes. (A) The 5 different protein C variants were injected at a concentration of 0.25 μM (arrows at left). After approximately 90 seconds, the protein solution was replaced with buffer (arrows at right) and the dissociation was followed. (B) WT protein C (upper panel) and QGNSEDY protein C (lower panel) were infused at the 3 indicated concentrations, and association and dissociation followed over time. (C) Increasing concentrations of WT protein C or QGNSEDY protein C were infused until a stable level was obtained, when association and dissociation is at equilibrium. The RU obtained at equilibrium was plotted against the protein C concentration, and the curves were used to calculate the Kd's. (D) Membrane binding was determined with light scatter intensity. M2 is the molecular weight of the protein-membrane complex, and M1 is that of the vesicles alone. The concentration of phospholipid was 5 μg/mL, and the protein C concentrations were varied from 0 to 1 μM, yielding protein/phospholipid (P/PL) ratios up to 16. QGNSEDY, (▴); SEDY, (▵); QGN, (■); WT protein C, (●). The means of 2 to 3 different experiments are shown with error bars representing ± SD.
To further characterize the binding of WT protein C and the QGNSEDY variant to the phospholipid, increasing concentrations of protein were injected over the phospholipid surface (Figure 2B). The 3 curves generated by WT protein C were obtained with protein C concentrations between 250 and 1000 nM, whereas the 3 curves obtained for QGNSEDY were generated by protein concentrations between 62.5 and 250 nM. From the shape of the binding curve and the amplitude of the signal, it is obvious that QGNSEDY interacted more efficiently with the membrane than did WT protein C. Of particular note is the observation that both the association and dissociation are faster for QGNSEDY than WT protein C. The shape of the binding curves indicated that both the association and dissociation reactions were complex and, because the mode of interaction is unknown, the rate constants could not be calculated. To circumvent this problem, another approach to determine the binding constants for the 2 proteins was used. In this experiment, increasing concentrations of protein were infused and the binding reactions were allowed to proceed until steady state was reached when the association and dissociation were equal. The observed binding responses were plotted against the protein concentrations, yielding binding curves that allowed calculation of equilibrium binding constants (Figure 2C). The data were evaluated with the BIAcore evaluation program using equation KA = Req/C(Rmax − Req). C is the concentration of protein, Rmax is the total surface binding capacity in RUs, and Req is the steady-state binding level in RUs. The Rmax andKd values were allowed to vary to obtain the best fit to the data points. In the case of QGNSEDY, the calculation yielded an Rmax of 475, which fits well with the results presented in Figure 2C. The Kd for QGNSEDY was estimated to be 0.5 μM. Rmax for the WT protein C is less certain, and the calculated Kd value of 3.5 μM was obtained with Rmax of 303. Because the exact nature of the binding sites for WT PC and QGNSEDY is unknown, we cannot assume that the Rmax should be the same for the 2 protein C variants.
The protein-membrane binding was also measured with the light-scattering technique, which gives information about the equilibrium binding constants but not the rates of binding. The experimental data are shown in Figure 2D, and the derivedKd's for WT protein C, SEDY, QGN, and QGNSEDY were 7.3 μM, 4.6 μM, 6.8 μM, and 2.2 μM, respectively. The QGNSEDY variant demonstrated higher affinity for the membrane than did WT protein C. The QGN variant behaved like WT protein C, whereas the SEDY variant yielded results intermediate of those obtained with WT protein C and the QGNSEDY variant. The GNED variant was also tested and gave similar binding as QGNSEDY. However, the binding of GNED was not fully reversed by the addition of EDTA, and a Kdwas therefore not calculated.
Anticoagulant activities of activated protein C variants in plasma
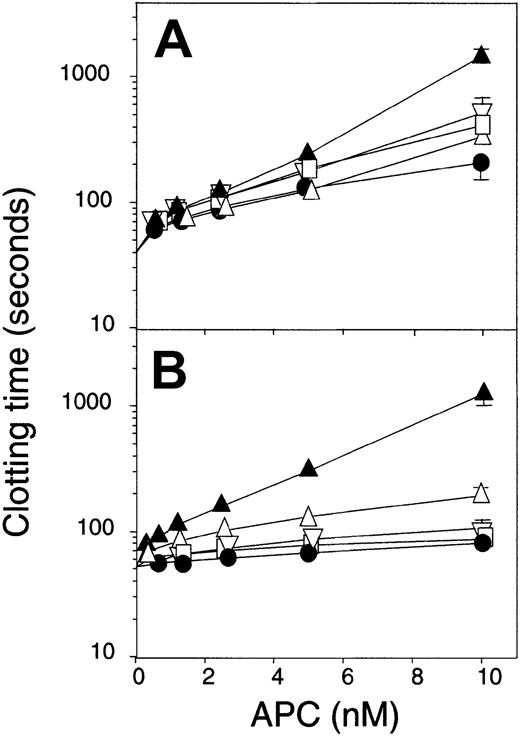
To investigate the anticoagulant potential of the various protein C variants, 2 clotting systems were tested using normal human plasma. One of the systems was based on activation of the intrinsic system (APTT), the other being dependent on initiation of coagulation by tissue factor (PT assay; Figure 3). In the APTT-based system, increasing concentrations of WT APC yielded prolongation of clotting time to approximately 200 seconds at the highest APC concentration tested. The response to the GNED variant was similar to that of WT APC, whereas both QGN and SEDY were more potent as anticoagulants than WT APC, resulting in clotting times around 400 seconds at the highest concentrations tested. The QGNSEDY-APC variant was considerably more potent than any of the other variants, with clotting times reaching more than 1000 seconds at the highest concentration tested.
Anticoagulant activity of APC variants.
Increasing concentrations of the different APC variants were added to plasma-based clotting assays using (A) APTT or (B) tissue factor–containing reagent. QGNSEDY-APC, (▴); SEDY-APC, (▵); GNED-APC, (▿); QGN-APC, (■); WT APC, (●). Note the logarithmic y-axes. Each point is the mean of 3 independent experiments performed in duplicate, and the error bars represent the standard deviation.
Anticoagulant activity of APC variants.
Increasing concentrations of the different APC variants were added to plasma-based clotting assays using (A) APTT or (B) tissue factor–containing reagent. QGNSEDY-APC, (▴); SEDY-APC, (▵); GNED-APC, (▿); QGN-APC, (■); WT APC, (●). Note the logarithmic y-axes. Each point is the mean of 3 independent experiments performed in duplicate, and the error bars represent the standard deviation.
In the PT-based assay, the results were somewhat different; the GNED-APC mutant was more active than both QGN-APC and SEDY-APC (Figure3B). These 2 APC variants were only slightly more active than WT APC. The QGNSEDY-APC variant was highly efficient as anticoagulant and yielded a clotting time of more than 1000 seconds at the highest concentration tested. Even the lowest concentration of this APC variant was more efficient in prolonging the clotting time than the highest concentration of WT APC, suggesting the QGNSEDY variant in this system to be at least 20-fold more active than WT APC.
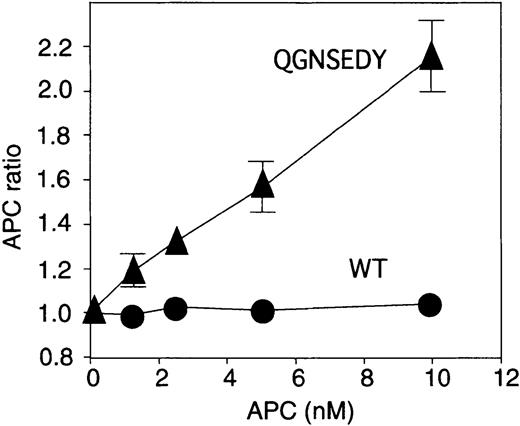
Addition of WT APC to plasma from an individual with homozygosity for FV Leiden did not prolong the clotting time, yielding an APC ratio of around 1. In contrast, the QGNSEDY-APC variant had a distinct anticoagulant effect in this plasma, resulting in an APC ratio of more than 2 at the 10 nM concentration of APC (Figure4).
Anticoagulant activity of APC variants in FV Leiden plasma.
Increasing concentrations of WT APC or QGNSEDY-APC variant were added to homozygous FV Leiden plasma-based APTT assay. The APC ratios were calculated and plotted. Each point is the mean of 3 independent experiments performed in duplicate, and the error bars represent the standard deviation.
Anticoagulant activity of APC variants in FV Leiden plasma.
Increasing concentrations of WT APC or QGNSEDY-APC variant were added to homozygous FV Leiden plasma-based APTT assay. The APC ratios were calculated and plotted. Each point is the mean of 3 independent experiments performed in duplicate, and the error bars represent the standard deviation.
Degradation of FVa by activated protein C variants
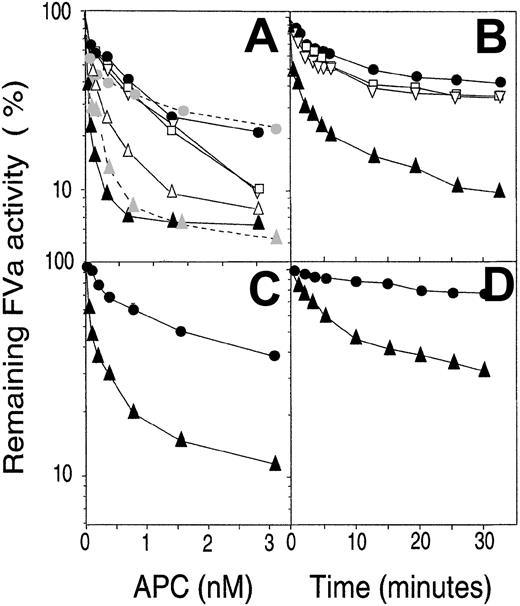
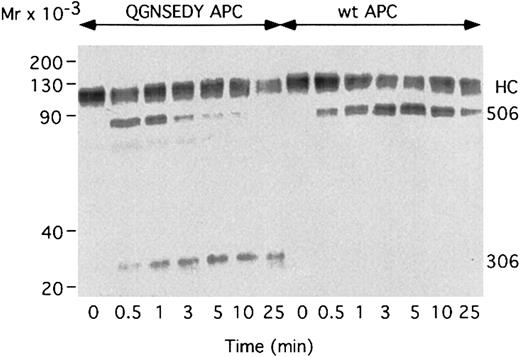
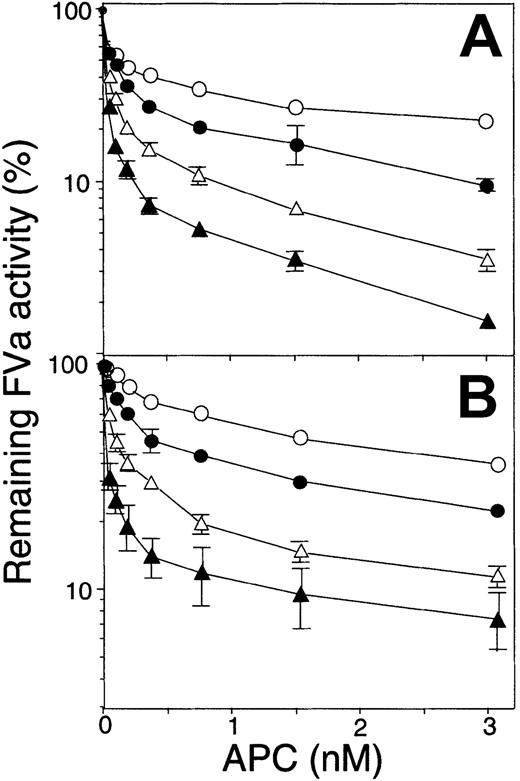
In an FVa-degradation system, either using purified FV or diluted plasma as source of FVa, increasing concentrations of WT APC resulted in partial loss of FVa activity (Figure5A). Results obtained with purified FVa and diluted plasma were similar, suggesting the latter system to adequately reflect FVa degradation. Approximately 25% FVa activity remained at the highest concentration of APC used, suggesting that the Arg506 cleavage had occurred, whereas the Arg306 site was not completely cleaved. The QGN- and SEDY-APC variants were somewhat more active than WT APC but only at the highest concentration used. In contrast, the QGNSEDY-APC variant was highly efficient in inhibiting FVa even at low concentrations. This suggests the Arg306 site to be cleaved, which resulted in complete loss of FVa activity. The GNED-APC variant demonstrated intermediate activity between WT APC and the QGNSEDY-APC variant, indicating that the Arg306 site had been cleaved. The FVa degradation was also followed over time (Figure 5B). The initial rapid loss of activity obtained in the presence of WT APC is the result of cleavage at Arg506. The FVa activity reached a plateau with 40% to 50% remaining activity after 10 to 15 minutes. The QGN- and SEDY-APC variants were only slightly more active than WT APC. In contrast, the QGNSEDY-APC was highly efficient and only around 20% FV activity remained after the first 5 minutes of incubation. The results suggest that both the Arg506 and the Arg306 sites were cleaved more rapidly by the QGNSEDY-APC variant than by WT APC. These conclusions were supported by the results of Western blotting of samples drawn from an FV degradation assay (Figure 6). In the presence of WT APC, only the Arg506 site is cleaved, which results in the generation of a 75 kDa band (corresponds to residues 1-506). In contrast, QGNSEDY-APC generates a strong 30 kDa band, which is the result of cleavages at both Arg506 and Arg306.
FVa-degrading capacity of recombinant APC variants.
To investigate the efficiency of the various APC variants in an FVa-degradation system, FVa (0.8 nM) from normal (A-B) or FV Leiden (C-D) plasma was incubated at 37°C for 10 minutes with increasing concentrations (A,C) of the various APC variants in the presence of 25 μM phospholipid vesicles (PS/PC, 10:90, wt/wt). The remaining FVa activity was measured with the PTase assay as described in “Materials and methods.” Alternatively, a time course was performed using 0.4 nM APC, and the FVa activity was followed over time using normal FVa (B) or FVa Leiden (D). Concentration courses of FVa inactivation by WT APC and QGNSEDY-APC were measured both in plasma system and plasma-purified FV system. QGNSEDY-APC, (▴); SEDY-APC, (▵); GNED-APC, (▿); QGN-APC, (■); WT APC, (●). Dotted lines with shaded symbols represent the results obtained with plasma-purified FVa. Note the logarithmic y-axes. Each point is the mean of 2 independent experiments performed in duplicate. The error bars were too small to be seen.
FVa-degrading capacity of recombinant APC variants.
To investigate the efficiency of the various APC variants in an FVa-degradation system, FVa (0.8 nM) from normal (A-B) or FV Leiden (C-D) plasma was incubated at 37°C for 10 minutes with increasing concentrations (A,C) of the various APC variants in the presence of 25 μM phospholipid vesicles (PS/PC, 10:90, wt/wt). The remaining FVa activity was measured with the PTase assay as described in “Materials and methods.” Alternatively, a time course was performed using 0.4 nM APC, and the FVa activity was followed over time using normal FVa (B) or FVa Leiden (D). Concentration courses of FVa inactivation by WT APC and QGNSEDY-APC were measured both in plasma system and plasma-purified FV system. QGNSEDY-APC, (▴); SEDY-APC, (▵); GNED-APC, (▿); QGN-APC, (■); WT APC, (●). Dotted lines with shaded symbols represent the results obtained with plasma-purified FVa. Note the logarithmic y-axes. Each point is the mean of 2 independent experiments performed in duplicate. The error bars were too small to be seen.
Western blot analysis of FVa degradation.
Plasma-derived FVa (0.8 nM) was incubated with 25 μM PS/PC, 10:90, phospholipid vesicles at 37°C and 0.04 nM WT APC or QGNSEDY-APC. At the indicated times, aliquots were drawn and subjected to Western blot analysis using the monoclonal antibody AHV5146, which reacts with an epitope located between positions 306 and 506. HC, heavy chain of Fva; 506, the 1-506 fragment; 306, the 307-506 fragment.
Western blot analysis of FVa degradation.
Plasma-derived FVa (0.8 nM) was incubated with 25 μM PS/PC, 10:90, phospholipid vesicles at 37°C and 0.04 nM WT APC or QGNSEDY-APC. At the indicated times, aliquots were drawn and subjected to Western blot analysis using the monoclonal antibody AHV5146, which reacts with an epitope located between positions 306 and 506. HC, heavy chain of Fva; 506, the 1-506 fragment; 306, the 307-506 fragment.
The conclusions drawn from the degradation of normal FVa were supported by results obtained using FVa Leiden (Figure 5C-D). In this case, FVa degradation is dependent mainly on the cleavage at the Arg306 site, because Gln replaces the Arg506. Increasing concentrations of WT APC resulted in a gradually increased loss of FVa activity, the highest APC concentration (3 nM) yielding approximately 40% remaining activity. The QGNSEDY-APC variant was considerably more efficient, and approximately 10-fold lower concentrations of QGNSEDY-APC as compared to WT APC were needed to obtain similar loss of FVa activity. A study of the time course of the FVa degradation demonstrated the slow loss of FVa activity obtained when WT APC was used. In contrast, the QGNSEDY-APC yielded a rapid inhibition of FVa activity, which was due to enhanced rate of cleavage of the Arg306 site.
Protein S was found to stimulate the rate of degradation of normal FVa as well as of FVa Leiden (Figure 7). In FVa degradation, the Arg306 cleavage is stimulated by the presence of protein S, whereas the Arg506 cleavage site is unaffected. As a consequence, the difference observed between the degradation curves obtained in the presence and absence of protein S is due to increased rate of cleavage at the Arg306 site. The QGNSEDY-APC variant was also stimulated by the presence of protein S. Similar effects were observed in the degradation of FVa Leiden, which is consistent with the protein S effect being related to stimulation of the cleavage at Arg306. It is noteworthy that in the degradation of both normal FVa and FVa Leiden, the QGNSEDY variant in the absence of protein S was more potent than WT APC in the presence of protein S.
FVa degradation of recombinant APC variants in the presence of protein S.
To investigate the influence of protein S on the efficiency of the various APC variants in an FVa-degradation system, FVa (0.8 nM) from normal (A) or FV Leiden (B) plasma was incubated at 37°C for 10 minutes with increasing concentrations of the various APC variants in the presence or absence of protein S (100 nM). The remaining FVa activity was measured with the PTase assay as described in “Materials and methods.” WT APC, (○); WT APC, with human protein S (●); QGNSEDY-APC, (▵); QGNSEDY-APC with human protein S, (▴); Note the logarithmic y-axes. Each point is the mean of 2 independent experiments performed in duplicate, and the error bars represent the standard deviation.
FVa degradation of recombinant APC variants in the presence of protein S.
To investigate the influence of protein S on the efficiency of the various APC variants in an FVa-degradation system, FVa (0.8 nM) from normal (A) or FV Leiden (B) plasma was incubated at 37°C for 10 minutes with increasing concentrations of the various APC variants in the presence or absence of protein S (100 nM). The remaining FVa activity was measured with the PTase assay as described in “Materials and methods.” WT APC, (○); WT APC, with human protein S (●); QGNSEDY-APC, (▵); QGNSEDY-APC with human protein S, (▴); Note the logarithmic y-axes. Each point is the mean of 2 independent experiments performed in duplicate, and the error bars represent the standard deviation.
Discussion
The Gla domains of different vitamin K–dependent proteins are very similar, sharing a high degree of sequence identity with certain positions being completely conserved.6 In particular the Gla residues tend to be conserved, which is consistent with their role in the calcium-dependent folding of the Gla domain. In addition, the Gla domains contain 3 hydrophobic residues at positions 4, 5, and 8 (corresponding to positions 5, 6, and 9 in prothrombin and FIX) that are hypothesized to intercalate into the hydrophobic part of the phospholipid membrane.39-41 Despite the extensive similarities between the different Gla domains, their affinities for negatively charged phospholipid membranes vary considerably over a 1000-fold range.6
There are not only differences between various proteins but also between the same protein of 2 distinct species. Thus, human protein C binds to phospholipid membranes with 10-fold higher affinity than its bovine counterpart, which is due to an amino acid residue difference at position 10. A Pro to His replacement at position 10 in recombinant bovine protein C was found to result in a 10-fold increase in membrane-binding affinity, proving that improved membrane-binding ability may be achieved by mutagenesis of certain residues in the Gla domain.7 Further support for this concept was derived from mutagenesis studies of factor VII, which converted factor VII into a high-affinity membrane-binding protein.9
Human protein C has also been the subject of similar mutagenesis studies, which resulted in several protein C variants having enhanced membrane-binding ability and, in parallel, increased anticoagulant potential.7,8 In the previously investigated human protein C variants, Gln and Asn occupying positions 32 and 33 in WT protein C were replaced with Glu and Asp, respectively, Glu at position 32 being expected to be carboxylated to Gla.8 These mutations did not in themselves affect the membrane-binding ability of the recombinant protein C variants. However, when combined with single or double mutagenesis of amino acids His10 (to Glu or Gln), Ser11 (to Gly), and Ser12 (Asn), a number of protein C variants with enhanced phospholipid- and calcium-binding ability were obtained, and several of them demonstrated increased anticoagulant activity as well.8 The present investigation is an extension of our previous work and aimed both at identification of even more potent protein C variants and the creation of protein C reagents useful for in vitro and in vivo experimentation on phospholipid binding. In addition, we wanted to test the hypothesis that multiple positions at very different positions in the Gla domain affect the membrane-binding characteristics and the anticoagulant potential of the protein.
The selection of positions for mutagenesis in this investigation was the result of a combination of sequence comparison of different Gla domains and 3-dimensional structural analysis. A recently developed model of full-length protein C was instrumental for the structural analysis.42 The 3 recombinant protein C variants, QGN, SEDY, and QGNSEDY, were chosen for several reasons. The QGN variant was only mutated at positions 10 to 12, with the purpose to investigate whether such a modification would be sufficient to increase phospholipid binding ability per se. The SEDY variant, on the other hand, was a modification of a previously investigated protein C variant denoted ED (E32D33). In this variant, S23 was introduced because several of the high-affinity phospholipid-binding Gla domains have Ser at position 23, whereas in human protein C this position is occupied with Asp. The Y44 mutation was introduced because the Gla domain of human protein C is unique in having His at position 44. Even all the protein C's from other species have Tyr at position 44. The fully mutated variant QGNSEDY was a combination of the QGN and SEDY variants, which allowed questions to be asked concerning whether multiple regions had to be modified in order to create a protein with high-affinity binding to negatively charged phospholipid.
The phospholipid binding of the purified protein C variants was investigated with light scattering as well as with surface plasmon resonance. Membrane binding of vitamin K–dependent coagulation factors have previously been determined by ellipsometry,43,44light scattering,6,45,46 and fluorescence polarization.47 Recently, the utility of BIAcore technique for evaluation of membrane-binding ability of vitamin K–dependent proteins has been demonstrated.36,38 In the BIAcore, theKd for the QGNSEDY variant was found to be around 0.5 μM, suggesting the equilibrium binding affinity to be considerably higher than that of WT protein C, which yielded aKd of 3.5 μM. The corresponding values obtained in the light-scattering experiment were 2.2 μM and 7.3 μM, respectively. The Kd values now obtained for WT protein C binding to phospholipid are in good agreement with results on record derived from light-scattering experiments.8 The BIAcore not only yielded equilibrium binding constants but demonstrated that both the rates of association and dissociation were much higher for the QGNSEDY variant than for WT protein C. The faster kinetics of membrane interaction of QGNSEDY may be of particular importance for its enhanced anticoagulant efficiency.
It was noteworthy that the QGNSEDY-APC variant was a potent anticoagulant even in a normal APTT reaction. Previously characterized protein C variants only demonstrated enhanced anticoagulant activity in systems with diluted phospholipid reagents (ie, at low concentrations of the membrane).7 8 Consistent with these observations, we now found the GNED-APC variant not to enhance anticoagulant activity in the undiluted APTT system. In contrast, the diluted tissue factor (PT) system contained low phospholipid levels and the GNED-APC was more potent as anticoagulant than WT APC, which is also consistent with results on record.
The APC variants were also evaluated in a system investigating the degradation of normal FVa and FVa Leiden (Arg506Gln). In the absence of protein S, the rate of Arg506 cleavage in normal FVa is 10-fold higher than that of Arg306 but only results in partial FVa inactivation. The slower cleavage at Arg306 results in complete inhibition of FVa activity. This cleavage is stimulated by the presence of protein S, which is believed to affect the activity in APC by dual mechanisms.19 First, protein S and APC form a complex on the phospholipid membrane, with protein S enhancing the membrane-binding ability of APC approximately 10-fold. Second, the protein S–APC interaction affects the location of the active site of APC, reducing the distance of the active site from the membrane by approximately 1.0 nm.14 15 From the degradation curves of normal FVa and FVa Leiden obtained in the absence of protein S, it is evident that the QGNSEDY-APC is highly efficient in cleaving FVa and that both the Arg506 and the Arg306 sites are cleaved at an enhanced rate by this APC variant. The main conclusion from the FVa-degradation system is that the enhanced membrane-binding ability of QGNSEDY-APC results in increased levels of active APC on the membrane, which results in enhanced efficiency of FVa degradation. The enhanced membrane affinity of QGNSEDY-APC was found to stimulate cleavage of Arg306 even in the absence of protein S, which demonstrates the importance of the membrane binding of APC for this cleavage.
APC has been demonstrated to be highly efficient in the treatment of sepsis, which is related to its anticoagulant activity as well as to a poorly defined anti-inflammatory function of APC.25 26 It is tempting to speculate that APC variants with enhanced anticoagulant activity resulting from improved membrane-binding ability may prove more efficient than WT APC in the treatment of sepsis. The low-affinity binding of APC to negatively charged phospholipid membranes may under normal healthy conditions be adequate, because protein S serves as a specific cofactor to increase the membrane binding of APC at certain locations. The situation may be different in pathologic conditions such as sepsis where higher membrane-binding ability of APC could potentially be beneficial, in particular because protein S and FV may be consumed in these conditions. The creation of the now-described QGNSEDY-APC variant will allow in vivo elucidation of the biologic consequences of enhanced membrane-binding ability of protein C and may open a path for development of APC variants with improved therapeutic potential in sepsis as well as other thromboembolic disorders.
The authors thank Dr Anna Blom and Miss Eva Norstrom for valuable advice and Mrs Astra Andersson and Mrs Ing-Marie Persson for expert technical assistance.
Prepublished online as Blood First Edition Paper, November 21, 2002; DOI 10.1182/blood-2002-06-1691.
Supported by a senior investigator award from the Swedish Foundation for Strategic Research, the Swedish Medical Research Council (grant no. 07143), the Albert Påhlsson Trust, and research funds from the University Hospital, Malmö.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Björn Dahlbäck, Department of Laboratory Medicine, Division of Clinical Chemistry, Lund University, The Wallenberg Laboratory, University Hospital, Malmö, SE-205 02 Malmö, Sweden; e-mail:bjorn.dahlback@klkemi.mas.lu.se.

![Fig. 2. Binding of protein variants to immobilized phospholipid evaluated using BIAcore. / The various protein C variants were infused at a flow rate of 30 μL/min over 2 different phopholipid membranes, one being 100% PC, the other being PS/PC, 20:80, wt/wt. The signals (resonance units [RU]) obtained from PC membranes were subtracted from the PS/PC-derived signals to obtain the specific binding to PS/PC membranes. (A) The 5 different protein C variants were injected at a concentration of 0.25 μM (arrows at left). After approximately 90 seconds, the protein solution was replaced with buffer (arrows at right) and the dissociation was followed. (B) WT protein C (upper panel) and QGNSEDY protein C (lower panel) were infused at the 3 indicated concentrations, and association and dissociation followed over time. (C) Increasing concentrations of WT protein C or QGNSEDY protein C were infused until a stable level was obtained, when association and dissociation is at equilibrium. The RU obtained at equilibrium was plotted against the protein C concentration, and the curves were used to calculate the Kd's. (D) Membrane binding was determined with light scatter intensity. M2 is the molecular weight of the protein-membrane complex, and M1 is that of the vesicles alone. The concentration of phospholipid was 5 μg/mL, and the protein C concentrations were varied from 0 to 1 μM, yielding protein/phospholipid (P/PL) ratios up to 16. QGNSEDY, (▴); SEDY, (▵); QGN, (■); WT protein C, (●). The means of 2 to 3 different experiments are shown with error bars representing ± SD.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/6/10.1182_blood-2002-06-1691/4/m_h80633972002.jpeg?Expires=1769286629&Signature=3ENIM7aoDr8dhf8httjSB2XGTehv4HOQe-nG9QT6VJgvzWWvY5zOCBcdWsf~0yeE501V8Qv6NNOXcZC2kY0Qtsdf1LXwQVLEdKWNM7U~yzx37gVbWh0069RKebfiRhatw1YKN4BYTZLZOFduFXC3SgnZumhpII9eJQ6ImNOGMlDpaA9CH-sljWim9QEPXFjNrIg5VpvT0ttvRxflvlaOOgoaJQFlWCs-2ojDQ~foiWZkFcIg9qyo9QoJtPMYSyMqx6GKIZcY~06cyi5UXUWzjLP6OkjhoElgRc3sxlo89-OYCgtWajgYgxVcwzg6VxddutwEi2Witp5fbk~oTUy-8g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal