The protein Bcl-xL is essential for survival of erythroid progenitor cells, and it increases substantially during late erythrocyte differentiation due to an increase of mRNA. We mapped the transcription start sites of bcl-x mRNA in mouse and human erythroblasts, and we analyzed the function of the mousebcl-x promoter by transient and stable transfection assays in a mouse erythroid cell line using plasmids containing thebcl-x promoter fused to a luciferase reporter gene. In mouse erythroblasts, a cluster of start sites at positions −664, −655, and −644 relative to the ATG initiation codon account for almost all transcripts. Human erythroblasts exhibit a start site at −654 that is homologous to the triplet in the mouse. A short sequence element in the mouse bcl-x promoter that includes nucleotides −1804 through −1734 was identified as very important for transcription. This element also showed strong enhancerlike activity in concert with the SV40 promoter in an enhancer test vector. Analyses of mutations indicated that 2 short sequences within the element, about 15 base pair apart, are necessary for full enhancer activity. Gel shift experiments with oligonucleotides representing these sequences revealed specific binding of nuclear proteins from erythroblasts. Some of these proteins are regulated during the late erythroid differentiation.
Introduction
The bcl-x gene codes for a protein, Bcl-xL, that confers protection from apoptosis to many types of cells. In addition to protecting cells from apoptosis induced by external environmental factors, bcl-x also regulates apoptosis during the development of several tissues. Bcl-x–deficient mice die in utero at about day 13 of gestation, exhibiting extensive apoptosis in the brain, spinal cord, and liver.1 The expression of bcl-x is important for the formation of erythrocytes,2-4 as most erythroblasts die in late stages of differentiation without the protein. Bcl-x also has a role in the function of macrophages5 and in the selective elimination by apoptosis of CD4+, CD8+ (double positive) thymocytes upon engagement of their T-cell receptors.6 7
The expression of bcl-x is regulated in several cell types by cytokines.8-11 Current evidence suggests that this regulation occurs through control of transcription.8-15 Grillot et al16 described several sites in the DNA where transcription is initiated. More recently, evidence was obtained for several different upstream, noncoding exons and thus several different promoters for this gene.17,18 Other studies have suggested functions for particular transcription factors in the control of bcl-xtranscription.9-15 19 However, gene reporter constructs used in some of these last studies lacked the sequences found here to be the major transcription start sites in erythroblasts as well as sequences that strongly enhance transcription. Thus, it is still unclear which are the most important determinants governing transcription of this gene.
We have shown previously that bcl-x transcripts and Bcl-xL protein are greatly increased in erythroblasts during late stages of erythropoiesis.20 We and others have provided evidence that increased transcription of the gene is dependent on erythropoietin (EPO), the cytokine that is essential for survival of late erythroid progenitors.10,12,13,20 Here, we analyze the transcription initiation sites for bcl-x in mouse and human erythroblasts. The start sites that we identify and their relative usages are different in several respects from those reported previously.9,16 18 Using transfection studies in an erythroid cell line, we also identified sequence elements in thebcl-x promoter that have regulatory effects on transcription.
Materials and methods
Cells and tissues
Mouse erythroblasts (FVA cells) were isolated from spleens of 8- to 12-week-old CD2F1 mice that had been infected 2 weeks earlier with the anemia-inducing strain of Friend virus as previously described.21 For isolation of human erythroblasts, erythroid colony-forming cells were prepared as previously described.22 Blood was obtained from healthy volunteers after informed consent. The study was approved by the Vanderbilt University and Department of Veterans Affairs Medical Center (Nashville, TN) institutional review boards. The partially purified human erythroid progenitor cells were cultured in vitro for 10 days to generate differentiating erythroblast (day-10) cells. The mouse erythroid cell line, HCD57,23 was obtained from Dr Stephen Sawyer of the Medical College of Virginia and was maintained in Iscove modified Dulbecco medium containing 20% fetal bovine serum and 5 U/mL of EPO. Mouse fetal livers were isolated from embryos at day 15 of gestation.
RNA isolation, S1 nuclease and RNase protection analyses
RNA was isolated from cells or tissues using the Totally RNA isolation kit (Ambion, Austin, TX). Subsequently, all RNA preparations were treated with 8 units of RNase-free DNase (Promega, Madison, WI) per 100 μg of RNA for 30 minutes at 37°C. The RNAs were re-extracted with phenol/chloroform and precipitated twice with ethanol. DNA products representing specific sections of thebcl-x promoter region were generated by polymerase chain reaction (PCR) or reverse transcriptase–polymerase chain reaction (RT-PCR). The products were ligated into the pGEM-T Easy vector (Promega) and verified by sequencing. Plasmid DNA containing abcl-x promoter insert was linearized for synthesis of a complementary RNA probe using SP6 or T7 RNA polymerase, or for synthesis of a complementary DNA probe using the Klenow fragment ofEscherichia coli DNA polymerase. The Prime-A-Probe and the MAXIscript kits from Ambion were used to synthesize the labeled DNA and RNA probes, respectively. The probes were purified by electrophoresis on a denaturing polyacrylamide gel. For mapping with ribonucleases, 25 μg total RNA was hybridized with 32P-labeled RNA probes (5 × 105 cpm). Hybridization and RNase digestion was done by using the RPA II kit (Ambion). For mapping with S1 nuclease, 25 μg total RNA was hybridized with 32P-labeled DNA probe (5 × 105 cpm) and digested with S1 nuclease using an S1-Assay kit (Ambion). All samples were run on denaturing 6% polyacrylamide sequencing gels. All experiments shown are representative of at least 3 repeated experiments conducted with the probe indicated.
Rapid amplification of cDNA ends
A rapid amplification of cDNA ends (5′-RACE) kit (Gibco BRL, Rockville, MD) was used for synthesis of DNA molecules having one terminus corresponding to the 5′ end of the bcl-x mRNA. The DNA products were ligated into pGEM-T Easy vector (Promega). Clones were isolated, and the DNA inserts were sequenced.
Promoter-luciferase reporter constructs and site-directed mutagenesis
Using PCR from mouse genomic DNA, the downstream end of thebcl-x promoter was obtained, including the BamHI site and the sequence between it and the ATG initiation codon. This sequence was spliced to the Kozak sequence adjacent to the luciferase initiation codon of the pGL3 Basic luciferase reporter vector (Promega). A genomic DNA fragment containing 1.1 kbp ofbcl-x promoter extending upstream from the BamHI site to the XhoI site was then cloned into the above pGL3 construct. Additional upstream promoter sequence was added by inserting PCR fragments generated using mouse genomic DNA. Constructs containing the intact downstream end of the bcl-x promoter sequence but with upstream ends truncated at various points were created by cutting the reporter construct with restriction enzymes having unique sites at the desired truncation points and with an enzyme that cut in the multiple cloning site of the pGL3 vector upstream of thebcl-x promoter sequence. The intervening fragment was removed and the ends fused to achieve the desired deletions. Small deletions at specific locations in the bcl-x promoter sequence of the reporter vectors were created using either the QuikChange XL Site-Directed Mutagenesis Kit or the Chameleon Double-Stranded, Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA). All constructions were verified by sequence determination.
For some promoter constructs, a neomycin resistance (neoR) gene was added to permit selection of stable, transfected cell clones. This was done by placing the 1146 base pair (bp)–XhoI-SalI fragment containing TN5 neoR from pMC1neo Poly A (Stratagene) into the SalI site of the pGL3 promoter constructs. The orientation of the neoR gene was chosen so that the neoR transcripts were made in the same direction as the luciferase transcripts.
Transfection of HCD57 cells and luciferase assays
HCD57 cells (2 × 106) were transfected by the cationic lipid transfection method. 10 μg plasmid DNA and 20 μL transfection reagent DMRIE C (GibcoBRL) were used per transfection. For transfection assays, 9 μg bcl-x promoter construct was added with 1 μg PRL-TK (Promega), a plasmid containing theRenilla luciferase gene controlled by the thymidine kinase (TK) promoter. After transfection, cells were cultured for 24 hours (transient transfection) or 48 hours (stable transfection). For transient transfection, the cells were collected for luciferase assays. For stable transfection, cells were replated in 48-well plates at 2.0 × 104 cells per well in 0.5 mL culture medium plus 0.9 mg/mL G418 for selection. After 2 weeks, wells containing growing cells were harvested and expanded for luciferase assays, genomic DNA analyses, and permanent storage. The Dual Luciferase Reporter Assay Kit (Promega) was used for measurement of luciferase activities.
Electrophoretic mobility shift assays
Nuclear proteins were prepared by the method of Dignam et al.24 Double-stranded oligonucleotides were 5′ end–labeled with γ-32P–adenosine triphosphate (ATP) using the DNA 5′End Labeling Kit (Promega). The32P-labeled probes were mixed with 8 μg nuclear extracts and 4 μg poly(dI-dC) in 20 μL of 20 mM HEPES (N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid) (pH 7.9), 50 mM KCl, 1 mM EDTA (ethylenediaminetetraacetic acid), 1 mM dithiothreitol, 25% (vol/vol) glycerol, and then incubated in the presence or absence of the competitor probes for 30 minutes at room temperature. Complexes of nuclear proteins and 5′ end–labeled oligonucleotides were resolved on 4% polyacrylamide gels in 0.05 M Tris [tris(hydroxymethyl)aminomethane], 0.4 M glycine, 0.002 M EDTA, pH 8.5.25 26
Results
Mapping the transcription start site by nuclease protection analysis
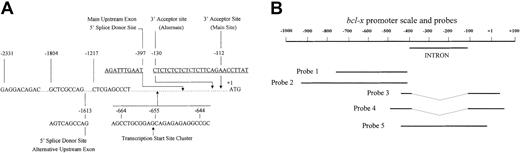
Figure 1A shows a representation of DNA sequences upstream of the protein-coding region of mousebcl-x. We mapped the transcription start sites forbcl-x by nuclease protection analyses, by primer extension analysis (not shown), and by 5′-RACE. Several clones consisting of portions of the bcl-x promoter were isolated and used to generate probes for the nuclease protection assays. Five of the most useful probes are illustrated in Figure 1B. As reported by others16 and as indicated by the structure of our clones isolated by RT-PCR, there is an intron extending from −396 to −112.
The mouse bcl-x promoter and illustration of probes used.
(A) Features of the mouse bcl-x gene upstream of the protein coding sequence. The sequences shown are taken from reference 16 and from the GenBank (accession number AF088904). The indicated splice donor and acceptor sites have been previously published16,17 and were verified here. The indicated cluster of transcription start sites was determined by us to be the predominant site in erythroid cells and corresponds to one of several sites published by others.16 17 Position −1804 is the farthest upstream point of conservation between sequences of the mouse and human genomes in the region of the bcl-x genes (comparison of Genbank sequences AF088904 and AL160175). (B) An illustration of 5 of the most useful probes used in nuclease protection mapping of the transcription start site. Probes 3 and 4 were generated by RT-PCR and thus lack the intron sequence, −396 to −112.
The mouse bcl-x promoter and illustration of probes used.
(A) Features of the mouse bcl-x gene upstream of the protein coding sequence. The sequences shown are taken from reference 16 and from the GenBank (accession number AF088904). The indicated splice donor and acceptor sites have been previously published16,17 and were verified here. The indicated cluster of transcription start sites was determined by us to be the predominant site in erythroid cells and corresponds to one of several sites published by others.16 17 Position −1804 is the farthest upstream point of conservation between sequences of the mouse and human genomes in the region of the bcl-x genes (comparison of Genbank sequences AF088904 and AL160175). (B) An illustration of 5 of the most useful probes used in nuclease protection mapping of the transcription start site. Probes 3 and 4 were generated by RT-PCR and thus lack the intron sequence, −396 to −112.
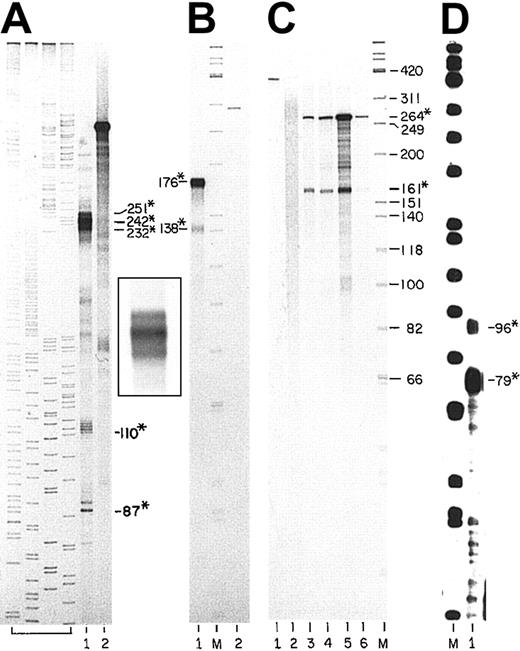
Figure 2A shows an S1 nuclease protection assay using Probe 1 (nucleotides −754 through −412) hybridized to total RNA from late-stage mouse erythroblasts. In lane 1, a group of 3 protected fragments was observed with lengths of 251, 242, and 232 nucleotides. These fragments identify positions −664, −655, and −644 as major transcription start sites. Several faint bands that are larger than the major group identify positions −669, −677, −697, and −725 as minor start sites. In addition, the largest minor band is the length of the full sequence of the probe that is complementary to mouse sequence, indicating that there are rare transcripts initiated upstream of position −754. In Figure 2A and Figure3, bands of 87 and 89 nucleotides in length and a group of bands of 110 to 115 nucleotides in length correspond to cuts at the upstream and downstream boundaries of a stretch of 23 successive thymidylate residues in the bcl-xmRNA. These cuts probably represent altered DNA:RNA duplex structures at those boundaries that the enzymes cut at low frequency rather than transcription start sites.
S1 nuclease protection analyses of the mousebcl-x transcription start sites.
(A) Labeled, single-stranded (ss) DNA probe was the noncoding strand of the region shown in Figure 1B as Probe 1 (−754 through −412). The probe was hybridized to total RNA from mouse erythroblasts (FVA cells) that had been cultured for 36 hours with EPO. Lane 1: the DNA fragments that were protected from S1 nuclease. Lane 2: purified probe, not subjected to digestion (about 7000 cpm). Products of a set of sequencing reactions for a portion of the human coagulation Factor XI gene of known sequence are shown as precise size markers in the first 4 lanes (not numbered). The inset photograph to the right of lane 2 shows the triplet bands of lane 1 at higher resolution. (B) Labeled, ssDNA probe was from the clone shown in Figure 1B as Probe 3 (−433 through +26 lacking the intron). The full length of the bcl-xsequence in Probe 3 is 176 bp. Lane 1: probe hybridized with erythroblast cell RNA (36 hours of culture) and digested with S1 nuclease. Lane M: 5′ end–labeled ΦX174 HinfI fragments. Lane 2: undigested probe (1000 cpm). (C) Labeled, ssDNA probe represented the sequence shown in Figure 1B as Probe 4 (−498 through +49 lacking the intron). Sequence in Probe 4 complementary tobcl-x mRNA is 264 bp. Analyses as for (A) and (B) using S1 nuclease. Lane 1: undigested probe (1000 cpm). Lane 2: probe hybridized with yeast RNA. Lanes 3-6: probe hybridized with total RNAs from thymus, brain, mouse erythroblasts (FVA cells), mouse fetal liver (day 15 gestation). Lane M: as described previously. (D) Labeled, ssDNA was from the clone shown in Figure 1B as Probe 5 (−431 through −34 including the intron). Probe 5 contains 397 bp of sequence of the gene. Lane M: as described in the legend to panel B. Lane 1: probe hybridized to erythroblast RNA (36 hours of culture) and digested with S1 nuclease. Numbers with asterisks are fragments protected from nucleases. Numbers without asterisks are sizes of ΦX174 marker fragments.
S1 nuclease protection analyses of the mousebcl-x transcription start sites.
(A) Labeled, single-stranded (ss) DNA probe was the noncoding strand of the region shown in Figure 1B as Probe 1 (−754 through −412). The probe was hybridized to total RNA from mouse erythroblasts (FVA cells) that had been cultured for 36 hours with EPO. Lane 1: the DNA fragments that were protected from S1 nuclease. Lane 2: purified probe, not subjected to digestion (about 7000 cpm). Products of a set of sequencing reactions for a portion of the human coagulation Factor XI gene of known sequence are shown as precise size markers in the first 4 lanes (not numbered). The inset photograph to the right of lane 2 shows the triplet bands of lane 1 at higher resolution. (B) Labeled, ssDNA probe was from the clone shown in Figure 1B as Probe 3 (−433 through +26 lacking the intron). The full length of the bcl-xsequence in Probe 3 is 176 bp. Lane 1: probe hybridized with erythroblast cell RNA (36 hours of culture) and digested with S1 nuclease. Lane M: 5′ end–labeled ΦX174 HinfI fragments. Lane 2: undigested probe (1000 cpm). (C) Labeled, ssDNA probe represented the sequence shown in Figure 1B as Probe 4 (−498 through +49 lacking the intron). Sequence in Probe 4 complementary tobcl-x mRNA is 264 bp. Analyses as for (A) and (B) using S1 nuclease. Lane 1: undigested probe (1000 cpm). Lane 2: probe hybridized with yeast RNA. Lanes 3-6: probe hybridized with total RNAs from thymus, brain, mouse erythroblasts (FVA cells), mouse fetal liver (day 15 gestation). Lane M: as described previously. (D) Labeled, ssDNA was from the clone shown in Figure 1B as Probe 5 (−431 through −34 including the intron). Probe 5 contains 397 bp of sequence of the gene. Lane M: as described in the legend to panel B. Lane 1: probe hybridized to erythroblast RNA (36 hours of culture) and digested with S1 nuclease. Numbers with asterisks are fragments protected from nucleases. Numbers without asterisks are sizes of ΦX174 marker fragments.
Quantification of transcripts initiated at the main start site during erythroblast differentiation.
Left panel: probe for RNase protection analysis was labeled RNA of complementary sequence to bcl-x mRNA from −930 through −412 (Probe 2, Figure 1B). Lanes labeled C contain samples of probe hybridized with yeast RNA and digested with RNase A and T1. Lanes labeled 0 hours to 48 hours indicate that the RNA used for hybridization was from mouse erythroblast cells cultured for the indicated period in the presence of EPO. Samples were digested with RNases. Lane M is marker, as described in the legend to Figure 2B. Lanes Pr2 contain undigested probe 2 samples (10 000 cpm). Right panel: probe for RNase protection analysis was labeled RNA complementary to bcl-x mRNA sequence from −754 through −412 (Probe 1, Figure 1B). Probe was hybridized with total RNA from mouse erythroblasts cultured for 36 hours and 48 hours in the presence of EPO. Hybridized samples were treated with RNase A and T1. Lane M is ΦX174 marker. Lanes Pr1 contain undigested probe 1.
Quantification of transcripts initiated at the main start site during erythroblast differentiation.
Left panel: probe for RNase protection analysis was labeled RNA of complementary sequence to bcl-x mRNA from −930 through −412 (Probe 2, Figure 1B). Lanes labeled C contain samples of probe hybridized with yeast RNA and digested with RNase A and T1. Lanes labeled 0 hours to 48 hours indicate that the RNA used for hybridization was from mouse erythroblast cells cultured for the indicated period in the presence of EPO. Samples were digested with RNases. Lane M is marker, as described in the legend to Figure 2B. Lanes Pr2 contain undigested probe 2 samples (10 000 cpm). Right panel: probe for RNase protection analysis was labeled RNA complementary to bcl-x mRNA sequence from −754 through −412 (Probe 1, Figure 1B). Probe was hybridized with total RNA from mouse erythroblasts cultured for 36 hours and 48 hours in the presence of EPO. Hybridized samples were treated with RNase A and T1. Lane M is ΦX174 marker. Lanes Pr1 contain undigested probe 1.
Three different probes were used to examine possible transcription start sites between position −412 and the ATG start codon. Two of these probes lacked the intron sequences. Probe 3 (Figure 1B) included the region from −433 through +26 (176 bp of exon sequence), and Probe 4 included the region from −498 through +49 (264 bp of exon sequence). In nuclease protection assays with both probes, the predominant protected species represent the full extents of sequences in the probes that are complementary to the bcl-x promoter; 176 nucleotides in probe 3 (Figure 2B, lane 1) and 264 nucleotides in probe 4 (Figure 2C, lanes 3-6). The transcripts that protect these full-length cloned sequences are undoubtedly transcripts initiated from the start sites identified in Figure 2A and discussed in the preceding paragraph. With each of these 2 probes, however, a second smaller band is present. In Figure 2C, both the longer and shorter protected fragments are evident in other tissues besides erythroid tissues. The smaller species identify protected probe fragments with an upstream end at position −112, the position of the 3′ splice boundary of the intron. For probe 3 in Figure 2B, lane 1, this protected fragment is 138 nucleotides (−112 through +26), and for probe 4 in Figure 2C, lanes 3-6, the smaller protected fragment is 161 nucleotides (−112 through +49). Densitometric analyses of Figure 2B-C (lanes 1 and 5, respectively) and of several other experiments indicate that the molar ratio of the large protected probe fragments to the smaller ones in erythroblasts is about 4 to 1. Thus, using these probes that were prepared by RT-PCR and that span the upstream intron but lack intron sequence, about 20 percent of the protected probe fragments had upstream ends that map at −112; the site of the 3′ splice boundary of the intron. Based on extensive additional data, we conclude that these fragments do not define a transcription start site, but rather are generated because about 20% of transcripts from the start sites further upstream use an alternative 3′ splice acceptor site at −130. The presence of the mixture of spliced RNA transcripts would be expected to lead to a protected probe fragment with an upstream end at −112 because the hybridization of the alternatively spliced transcripts with a probe representative of a single type of splice leads to a mismatch susceptible to nuclease digestion at position −112. We isolated multiple plasmid clones containing DNA inserts generated by RT-PCR with primers that spanned this intron. Sequencing of 30 such clones revealed that the ratio ofbcl-x transcripts spliced at −112 to those spliced at −130 was about 4 to 1. The representation of digestion product fragments in the nuclease protection experiments was as expected from this ratio.
Clones of portions of the bcl-x promoter were isolated that included the intron as well as sequence upstream and downstream. One clone that was used to generate probe 5 of Figure 1B contained the sequence from −431 through −34 (397 bp). Using this probe for an RNase protection assay (Figure 2D), there was no protected fragment corresponding to an unspliced transcript. The vastly predominant protected fragment in the gel was 79 nucleotides long, and there was a moderate amount of a fragment 96 nucleotides in length. These probe fragments correspond to the expected species whose upstream boundaries are the main 3′ splice junction of the intron (−112, the 79 nucleotide fragment) and the alternative 3′ splice site at position −130 (see preceding paragraph). No band longer than 96 nucleotides was seen that corresponds to a transcription start site near −150 that was reported recently by Pecci et al.18
During 48 hours of in vitro culture, there is a great increase in the level of bcl-x mRNA in mouse FVA erythroblasts as they differentiate from early erythroblasts into reticulocytes.20 The level of bcl-x mRNA initiated at the start sites defined in Figure 2 increases in proportion to the total increase in bcl-x mRNA (Figure 3). Thus, transcription from this start site is regulated during erythroblast differentiation and is mainly responsible for the accumulation of total mRNA for Bcl-x.
Verification of start sites by 5′-RACE
To verify that the putative transcription start sites at −664, −655, and −644 represent authentic start sites and not 3′ splice boundaries of an intron, DNA molecules with termini identifying the 5′ end of bcl-x transcripts were generated by 5′-RACE and cloned into the pGEM T-Easy vector. The downstream primer for the PCR step of this procedure extended from −471 through −451. Thirty-seven of fifty clones contained fragments of the size expected to be generated from transcripts initiated at −664, −655, and −644. Sequencing of a randomly chosen group of 15 of these 37 clones showed that each has a terminus at one of those sites. Only one of the isolated clones that had inserts of a size different than expected for those start sites consisted of DNA of bcl-x. That clone contained continuous bcl-x sequence derived from a transcript apparently initiated at position −962.
The transcription start sites in human erythroid progenitors
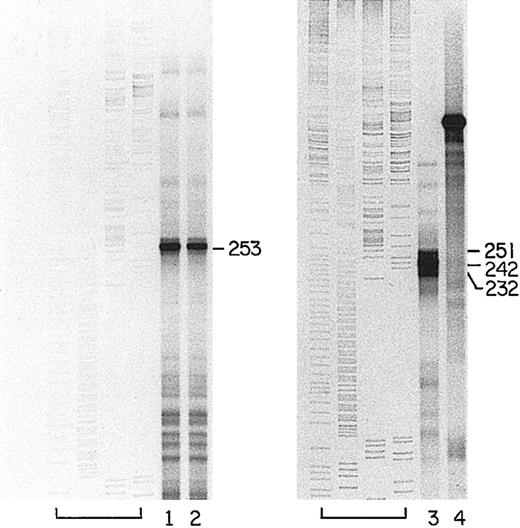
A clone of the human bcl-x promoter region from −858 through −389 was isolated. An S1 nuclease protection assay was performed using a labeled probe from the human clone and RNA purified from human day-10 erythroblasts. A protected fragment of 253 nucleotides in length (Figure 4, lanes 1 and 2) identified a major transcription start site at position −654 that corresponds to the described mouse cluster. This site accounts for about 90% of the specific transcripts detected with this probe. Several minor transcription start sites were located at −789, −711, and −706, and the largest band corresponds to the full length of thebcl-x complementary sequence in the probe, indicating initiation upstream of −858.
Major transcription start site for bcl-x in human erythroblasts.
Left panel: labeled, ssDNA consisting of the noncoding strand of the sequence from −858 through −389 of the human bcl-xsequence (GenBank accession number AL160175) was hybridized to 25 μg total RNA from human erythroblasts derived by 10 days of culture of partially purified erythroid progenitors.22 Lanes 1 and 2 are duplicates showing the major protected fragments after S1 nuclease digestion. Full-length probe (not shown) was 519 nucleotides long. The leftmost lanes of the panel contain labeled sequencing reactions from a portion of the human coagulation Factor XI gene, as in Figure 2A. Right panel is a reproduction of Figure 2A.
Major transcription start site for bcl-x in human erythroblasts.
Left panel: labeled, ssDNA consisting of the noncoding strand of the sequence from −858 through −389 of the human bcl-xsequence (GenBank accession number AL160175) was hybridized to 25 μg total RNA from human erythroblasts derived by 10 days of culture of partially purified erythroid progenitors.22 Lanes 1 and 2 are duplicates showing the major protected fragments after S1 nuclease digestion. Full-length probe (not shown) was 519 nucleotides long. The leftmost lanes of the panel contain labeled sequencing reactions from a portion of the human coagulation Factor XI gene, as in Figure 2A. Right panel is a reproduction of Figure 2A.
Alternative upstream exons
MacCarthy-Morrogh et al17 recently reported the existence of an alternative, noncoding exon upstream of the humanbcl-x coding sequence. They called this exon “exon 1B” and the exon that we have analyzed above “exon 1A.” We examined mouse erythroblasts for transcripts originating from a homologous exon 1B. An upstream primer −1704 through −1687 and a downstream primer +7 through +26 were used for RT-PCR to amplify transcripts. Cloning of the DNA products yielded clones with inserts of 248 bp or 230 bp. Sequencing of multiple clones showed that the 248-bp inserts represented transcripts in which an intron was removed by splicing nucleotide −1613 to nucleotide −130 and the 230-bp inserts represented transcripts in which nucleotide −1613 was spliced to nucleotide −112. When these clones were used to generate probes for use in nuclease protection assays with RNA from FVA erythroblasts, multiple experiments showed no evidence of protection of the exon 1B portion of the sequences (−1704 through −1613), although there was strong protection of fragments representing sequences from −112 (or −130) to +26 (data not shown). Protection of the −112 (or −130) to +26 portion of the probes was undoubtedly achieved by transcripts bearing exon 1A upstream sequences. Thus, while transcripts initiated in exon 1B are present in primary erythroblasts as indicated by cloning of RT-PCR products, they are quantitatively very minor compared with those initiated in exon 1A.
Suggestive evidence for additional upstream exons in the mousebcl-x gene has been presented by Pecci et al.18We conducted RT-PCR reactions using 2 upstream primers in the far upstream exons proposed by Pecci et al18 and a downstream primer in the bcl-x coding sequence. The upstream primers used extended from −3244 through −3225 for the putative P5 exon18 and from −2452 through −2433 for the putative P4 exon.18 The downstream primer contained nucleotides +7 through +26. The RT-PCR reactions (30 PCR cycles with various primer annealing temperatures) yielded reproducible but very faint bands of DNA on electrophoresis that were isolated and cloned. Sequencing of representatives of all the inserted DNAs obtained revealed the primer sequences, but none of the intervening DNAs were related to sequence in the bcl-x gene. Thus, our experiments did not reveal evidence of bcl-x transcripts initiated in the exons proposed by Pecci et al.18
Analysis of promoter function using transient transfection assays
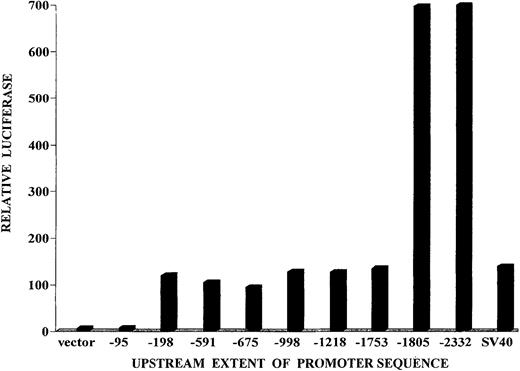
Portions of the mouse bcl-x promoter were placed in the firefly luciferase expression plasmid, pGL3-basic vector. Individual clones extended upstream from the protein initiation codon through positions −94, −197, −590, −674, −997, −1217, −1290, −1385, −1456, −1752, −1804, or −2331. HCD57 cells were transfected with each of these plasmid constructs and analyzed in transient expression assays for firefly luciferase activity. In Figure5, a significant basal level of luciferase activity is seen for constructs having the most downstream 197 bp of bcl-x promoter sequence, but very little activity is seen for the construct containing only the downstream 94 bp of the promoter. A level of luciferase expression that is 5-fold higher than the basal level is gained by inclusion of a sequence from −1752 through −1804. We refer to the sequence responsible for this 5-fold increase as the “upstream element” and the sequence between −94 and −197 responsible for basal expression as the “downstream element.” Further analysis of the downstream element suggested that it is not critical for the majority of transcripts initiated by the full-length promoter (see “Discussion”).
Expression of reporter luciferase directed by portions of the bcl-x promoter in transient transfection assays.
Portions of the bcl-x promoter bearing the complete sequence from −1 (nucleotide preceding A of the ATG codon) to the indicated nucleotide on the horizontal axis were placed in the luciferase reporter vector pGL3 Basic Vector (Promega). The constructs were transfected into HCD57 cells, and the firefly luciferase was measured after 24 hours. A control expression plasmid, pRL-TK, was cotransfected, as described in “Materials and methods.” The ratio of firefly luciferase to Renilla luciferase was determined in the transfected cells. That ratio for the pGL3 luciferase vector without any inserted promoter sequence was adopted as the baseline expression value. Each transfection assay was performed 3 to 5 times, and representative values are shown. For all test plasmids, the range of values obtained in repetitive experiments was less than 15% of the values shown.
Expression of reporter luciferase directed by portions of the bcl-x promoter in transient transfection assays.
Portions of the bcl-x promoter bearing the complete sequence from −1 (nucleotide preceding A of the ATG codon) to the indicated nucleotide on the horizontal axis were placed in the luciferase reporter vector pGL3 Basic Vector (Promega). The constructs were transfected into HCD57 cells, and the firefly luciferase was measured after 24 hours. A control expression plasmid, pRL-TK, was cotransfected, as described in “Materials and methods.” The ratio of firefly luciferase to Renilla luciferase was determined in the transfected cells. That ratio for the pGL3 luciferase vector without any inserted promoter sequence was adopted as the baseline expression value. Each transfection assay was performed 3 to 5 times, and representative values are shown. For all test plasmids, the range of values obtained in repetitive experiments was less than 15% of the values shown.
Analysis of promoter function using stable transfection assays
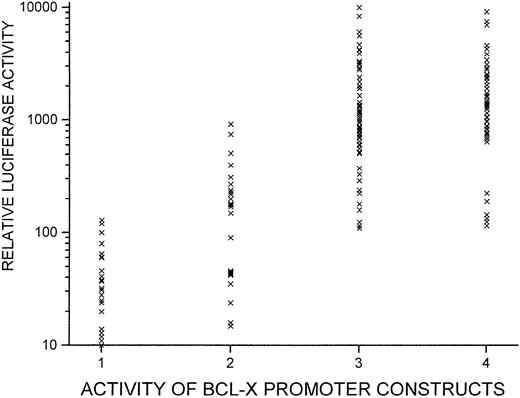
A vector for luciferase expression that also contained a gene for neomycin resistance was prepared. Several of the upstream portions of the bcl-x gene depicted in Figure 5 were placed into this expression plasmid, and clones of HCD57 cells were isolated that contained the plasmids in a stable, integrated state. Thirty to fifty isolated cell clones bearing each construct were analyzed (Figure6). In contrast to the results from transient assays, most clones containing promoter sequences extending upstream to −1217 exhibited higher levels of luciferase expression than clones having promoter sequences only extending through −197. However, constructs bearing the additional sequence between −1217 and −1804 showed a further 5-fold increase in the median expression level, in agreement with the transient assays.
Function of bcl-x promoter sequences in stable transfection assays (integrated constructs).
Several of the bcl-x/reporter constructs depicted in Figure5 were further modified to contain a neomycin resistance gene. These were transfected into HCD57 cells, and clones of cells bearing integrated plasmids were selected by culturing in G418. Approximately 50 luciferase-positive cell clones were isolated for each plasmid construct. Luciferase expression was measured for each cell clone; each “x” on the graph represents a cell clone. Numbers on the x-axis indicate group numbers. Group 1 clones bear a plasmid withbcl-x promoter sequence from −1 through −197; Group 2 have promoter sequence from −1 through −1217; Group 3 have sequence from −1 through −1804; and Group 4 have sequence from −1 through −2331. Relative luciferase units are standardized on a per 0.5 × 106 cells basis.
Function of bcl-x promoter sequences in stable transfection assays (integrated constructs).
Several of the bcl-x/reporter constructs depicted in Figure5 were further modified to contain a neomycin resistance gene. These were transfected into HCD57 cells, and clones of cells bearing integrated plasmids were selected by culturing in G418. Approximately 50 luciferase-positive cell clones were isolated for each plasmid construct. Luciferase expression was measured for each cell clone; each “x” on the graph represents a cell clone. Numbers on the x-axis indicate group numbers. Group 1 clones bear a plasmid withbcl-x promoter sequence from −1 through −197; Group 2 have promoter sequence from −1 through −1217; Group 3 have sequence from −1 through −1804; and Group 4 have sequence from −1 through −2331. Relative luciferase units are standardized on a per 0.5 × 106 cells basis.
Analysis of the mouse upstream element
The upstream boundary of the regulatory upstream element (position −1804) is the furthest upstream extent of sequence homology between the mouse and human bcl-x genes (Figure 1). The upstream element from −1734 through −1804 or a larger sequence from −1217 through −1801 was placed in the enhancer test vector pGL3-Promoter Vector (Promega). Each of those sequences caused a 3-fold increase of expression directed by the SV40 promoter in that enhancer test vector when introduced transiently into HCD57 cells (Figure7B). In experiments using the larger sequence, the increased expression required that the segment be placed upstream of the SV40 promoter and was greatest when it was placed in the same orientation as it is in the bcl-x promoter. It did enhance luciferase activity about 40% when placed upstream but in the reverse orientation (Figure 7B). The upstream element also functioned as a strong enhancer when transfected into mouse fibroblasts (NIH 3T3 cells, data not shown), indicating that its function is not restricted to erythroid cells.
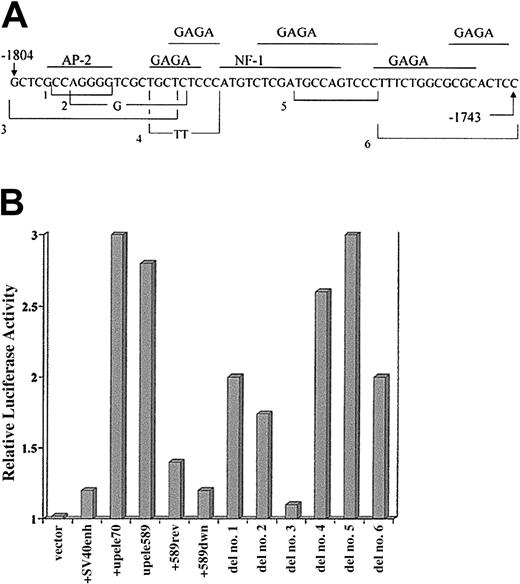
Analysis of regulatory sequences of the upstream element.
(A) The sequence of the upstream element of the bcl-xpromoter that has strong enhancerlike activity. Also depicted are the structures of several mutations created in that sequence that were analyzed for functional activity. Mutants 1 through 6 are indicated by brackets encompassing a sequence deleted from the normal sequence. Bases shown within a bracket indicate that those bases were substituted for the deleted sequence. The transcription factors indicated above the sequence were retrieved by the Transcription Element Search Software (www.cbil.upenn.edu/tess), based on consensus sequence motifs found by that program. The motifs of the factors shown are present in homologous sequences of the mouse and human bcl-x genes. GAGA factor has great importance for transcription in Drosophila, but no homolog of it has been identified in mammalian cells. (B) Results of transient transfection assays in HCD57 cells using constructs of the enhancer test vector, PGL3-Promoter Vector (Promega). This vector contains the SV40 promoter. The plasmids represented by the histogram columns contain, from left to right: vector alone; vector containing the SV40 enhancer; vector with bcl-x promoter sequence from −1804 through −1734 (upele70), forward orientation; vector withbcl-x promoter −1801 through −1213 (upele589), forward orientation; vector with upele589 in reverse orientation; vector with upele 589 placed downstream of luciferase gene. Other columns labeled as “del” represent experiments with upele70, mutated as indicated in (A), inserted upstream of the luciferase gene and in the normal orientation. Test plasmids were cotransfected with the control plasmid, pRL-TK, as an internal control for experimental variation. The activity of the unmodified PGL3-Promoter Vector was chosen as 1. Transfection experiments for each test plasmid were repeated 3 to 5 times. In all cases, the range of the determined values was equal to or less than 13% of the values shown.
Analysis of regulatory sequences of the upstream element.
(A) The sequence of the upstream element of the bcl-xpromoter that has strong enhancerlike activity. Also depicted are the structures of several mutations created in that sequence that were analyzed for functional activity. Mutants 1 through 6 are indicated by brackets encompassing a sequence deleted from the normal sequence. Bases shown within a bracket indicate that those bases were substituted for the deleted sequence. The transcription factors indicated above the sequence were retrieved by the Transcription Element Search Software (www.cbil.upenn.edu/tess), based on consensus sequence motifs found by that program. The motifs of the factors shown are present in homologous sequences of the mouse and human bcl-x genes. GAGA factor has great importance for transcription in Drosophila, but no homolog of it has been identified in mammalian cells. (B) Results of transient transfection assays in HCD57 cells using constructs of the enhancer test vector, PGL3-Promoter Vector (Promega). This vector contains the SV40 promoter. The plasmids represented by the histogram columns contain, from left to right: vector alone; vector containing the SV40 enhancer; vector with bcl-x promoter sequence from −1804 through −1734 (upele70), forward orientation; vector withbcl-x promoter −1801 through −1213 (upele589), forward orientation; vector with upele589 in reverse orientation; vector with upele 589 placed downstream of luciferase gene. Other columns labeled as “del” represent experiments with upele70, mutated as indicated in (A), inserted upstream of the luciferase gene and in the normal orientation. Test plasmids were cotransfected with the control plasmid, pRL-TK, as an internal control for experimental variation. The activity of the unmodified PGL3-Promoter Vector was chosen as 1. Transfection experiments for each test plasmid were repeated 3 to 5 times. In all cases, the range of the determined values was equal to or less than 13% of the values shown.
Deletion mutations of the upstream element (Figure 7A) were tested in the enhancer test vector (Figure 7B). Deletions that eliminated a portion of the middle of this element showed no loss of enhancer activity (an example shown is del no. 5 that eliminates the putative nuclear factor [NF]–1 binding site). However, deletion of the most upstream 20 bp of the sequence (mutant no. 3, Figure 7) caused loss of 85% of the enhancing activity, indicating that this section has a role in the mechanism of regulation. Additionally, deletion of 17 bases near the downstream end of this element (mutant no. 6, Figure 7) reduced the enhancer activity by 50%. All of these mutations also have been created and tested in the full-length bcl-xpromoter/luciferase construct, and the pattern and magnitude of the effects of the mutations on expression directed by the completebcl-x promoter (not shown) were identical to those caused in the enhancer test vector (Figure 7B).
Binding of nuclear proteins to sequences in the upstream element
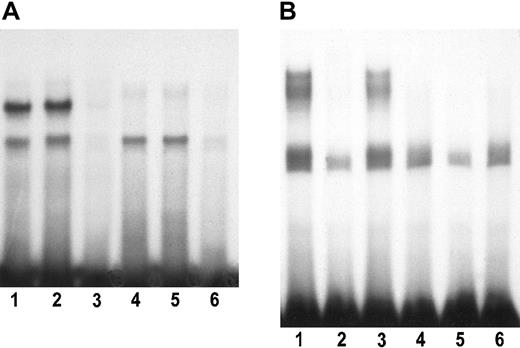
We synthesized double-stranded oligonucleotides that spanned the regions shown by mutations to affect enhancer function of the upstream element. One oligonucleotide consisted of the most upstream 24 nucleotide pairs, positions −1804 through −1781 (sequence A), and the second oligonucleotide consisted of nucleotide pairs from position −1759 through −1743 (sequence B). Figure8A shows that 2 specific complexes are formed between proteins from nuclei of FVA erythroblasts and the sequence A oligonucleotide. Activity of one complex (upper band) declines during the differentiation of the erythroblasts, whereas the second remains constant or increases somewhat. A nonhomologous competitor DNA has no effect on these complexes. In Figure 8B, several specific complexes are seen in experiments with the more downstream sequence B oligonucleotide. All of the specific complexes involving sequence B decline as the erythroblasts differentiate (missing by 24 hours of culture). We have tested antibodies to all of the following transcription factors in supershift assays with both of the above labeled oligonucleotides: AP-2 (all isoforms), Brn-3α, PU-1, GATA-1, YY1, PPAR-α, PPAR-γ, PPAR-δ, E47, and SP1. None of the antibodies eliminated or shifted any of the bands.
Binding of nuclear proteins to the upstream element.
(A) The end-labeled, double-stranded oligonucleotide had a sense strand sequence 5′-GCTCGCCAGGGGTCGCTGCTCTCCCATG-3′, which includes the most upstream 24 bp of the upstream element (Figure 7A). Lanes 1-3 and lanes 4-6 represent complexes formed by extracts from mouse splenic erythroblasts (FVA cells) cultured for 0 hours (freshly isolated cells) or 24 hours, respectively. Lanes 1 and 4, no unlabeled competitor oligonucleotide; lanes 2 and 5, a 100-fold excess of a heterologous competitor oligonucleotide bearing a binding sequence for the transcription factor TAL1 (5′-CTCCCAGCAGCTGGCCTAGGAGATAGCAGCAG-3′); lanes 3 and 6, a 100-fold excess of unlabeled homologous competitor. (B) The end-labeled, double-stranded oligonucleotide had a sense strand sequence 5′-GTCCCTTTCTGGCGCGCACTCCTTTTGC-3′, which includes the sequence of the upstream element deleted in mutant 6 depicted in Figure 7A-B. Lanes 1-3 represent complexes formed with nuclear extracts of 0-hour splenic erythroblasts (FVA cells), and lanes 4-6 represent complexes formed with extracts from FVA cells cultured for 24 hours. Lanes 1 and 4, no unlabeled competitor oligonucleotide; lanes 2 and 5, a 100-fold excess of unlabeled homologous competitor oligonucleotide; lanes 3 and 6, a 100-fold excess of unlabeled heterologous competitor (TAL1 binding sequence as in Figure 8A).
Binding of nuclear proteins to the upstream element.
(A) The end-labeled, double-stranded oligonucleotide had a sense strand sequence 5′-GCTCGCCAGGGGTCGCTGCTCTCCCATG-3′, which includes the most upstream 24 bp of the upstream element (Figure 7A). Lanes 1-3 and lanes 4-6 represent complexes formed by extracts from mouse splenic erythroblasts (FVA cells) cultured for 0 hours (freshly isolated cells) or 24 hours, respectively. Lanes 1 and 4, no unlabeled competitor oligonucleotide; lanes 2 and 5, a 100-fold excess of a heterologous competitor oligonucleotide bearing a binding sequence for the transcription factor TAL1 (5′-CTCCCAGCAGCTGGCCTAGGAGATAGCAGCAG-3′); lanes 3 and 6, a 100-fold excess of unlabeled homologous competitor. (B) The end-labeled, double-stranded oligonucleotide had a sense strand sequence 5′-GTCCCTTTCTGGCGCGCACTCCTTTTGC-3′, which includes the sequence of the upstream element deleted in mutant 6 depicted in Figure 7A-B. Lanes 1-3 represent complexes formed with nuclear extracts of 0-hour splenic erythroblasts (FVA cells), and lanes 4-6 represent complexes formed with extracts from FVA cells cultured for 24 hours. Lanes 1 and 4, no unlabeled competitor oligonucleotide; lanes 2 and 5, a 100-fold excess of unlabeled homologous competitor oligonucleotide; lanes 3 and 6, a 100-fold excess of unlabeled heterologous competitor (TAL1 binding sequence as in Figure 8A).
Discussion
Our nuclease protection analyses indicate that the location of transcription initiation for greater than 90% of bcl-xtranscripts in mouse erythroblasts is a cluster of sites at −664, −655, and −644 relative to the initiation codon. The demonstration (Figure 3) that transcripts initiated at this site are strongly increased during differentiation indicates that this site cluster is involved in regulation of Bcl-x levels during erythroid differentiation. In human erythroblasts a homologous site was observed at −654. Most of the remaining rare bcl-x transcripts in erythroid cells appear to be initiated further upstream but to represent alternative start sites to the predominant cluster for the same 5′ noncoding exon. Grillot et al16 published results of primer extension studies that identified a transcription start site at −655 in mouse thymus, spleen, liver, heart, and kidney, although these authors provided no quantitative data about its usage relative to the usage of other sites that they proposed. Based on primer extension analysis, these authors also reported a bcl-x transcription start site in the vicinity of position −100.16 We also did extensive preliminary studies using primer extension analyses, and the findings are summarized in Table 1. We found evidence by that method for a start site at −101 as well as others upstream of −655. Because no site at −101 was observed by nuclease protection analyses with numerous probes, we believe that this result of the primer extension method is an artifact or else that −101 is a very minor start site. Also, while some of the additional sites found by primer extension upstream of position −664 may be real, they must be minor sites in erythroblasts or there should have been much more prominent protection of probe fragments larger than 251 nucleotides from probes 1 and 2 in the nuclease protection experiments (Figures 2-3). Primer extension analysis is not a quantitative method. Although putative start sites can be identified, the relative importance of such sites cannot be determined through its use.
Summary of primer extension studies
| Ends of primers (positions) . | Start sites identified . |
|---|---|
| 27 | −101 |
| −493 | −655 |
| −493 | −831 |
| −602 | −644 |
| −756 | −872 |
| Ends of primers (positions) . | Start sites identified . |
|---|---|
| 27 | −101 |
| −493 | −655 |
| −493 | −831 |
| −602 | −644 |
| −756 | −872 |
In mouse erythroblasts, we found evidence by RT-PCR and subsequent isolation of clones that some transcription was initiated at an alternative upstream exon, exon 1B, reported for human bcl-xby MacCarthy-Morrogh et al.17 However, by RNase protection assays the latter transcripts were not detected, indicating that transcription from exon 1B is not quantitatively significant in the erythroblasts. Pecci et al18 proposed additional upstream exons based on evidence from RT-PCR. However, the downstream primers that they used to support their conclusions were far upstream of the coding sequence of the gene, within the purported exons themselves. We attempted to demonstrate transcripts that contained the suggested exons connected to the coding sequence of bcl-x by RT-PCR using upstream primers in the proposed exons and a downstream primer in the bcl-x coding sequence. Using various primer annealing conditions, we obtained reproducible but extremely low levels of discrete RT-PCR products. However, all isolated plasmid clones containing those products contained DNA unrelated to thebcl-x gene. We thus suggest that further evidence of spliced transcripts containing both the proposed exons and bcl-xcoding sequence is required to prove authenticity of these exons. The suggested exons are upstream of any sequence homology between mouse and human DNA in the vicinity of the bcl-x gene.
In studies of regulation of bcl-x transcription, investigators have implicated various transcription factors as important: in cardiac myocytes, it was Stat1;9 in macrophage, Ets 2;11 in erythroid cells, Stat5;12,13 and in neuronal cells, NF-kappaB15 and Brn-3a.19 In several studies,12,13 the reporter constructs used in transient transfection assays of promoter function lacked the main transcription start site for exon 1A observed in the current study and contained only sequences downstream of this site. Other studies used constructs that contained the start sites and extended sequence upstream. An additional complicating factor in interpreting the results is that an alternative upstream, noncoding exon (exon 1B) has been documented for humanbcl-x17 and mouse bcl-x (our data). This alternative first exon has an additional transcription start site and possibly additional regulatory elements. Thus, understanding ofbcl-x transcription regulation in a particular cell type requires determination of which upstream exon(s) are used and analyses of multiple potential regulatory sequences in the context of the complete, upstream promoter region.
In transfection assays of promoter function (Figures 5-7) in the mouse erythroid cell line HCD57, we discovered that the most powerful regulatory sequence in the bcl-x promoter is located from −1734 through −1804. By making mutations in this upstream element, we have shown that 2 short sequences within it are critical: the most upstream 20 base pairs of the element, positions −1785 through −1804 (sequence A), and a second sequence revealed by a deletion from −1743 through −1759 (sequence B). Significantly, sequence A is the most upstream sequence that has homology between the mouse and humanbcl-x genes. In contrast to our results, Grillot et al16 found no evidence of a positive regulatory element upstream of −600. However, Glasgow et al15 showed in transient transfections into PC12 cells that 3.2 kbp of upstream promoter had about 8 times the promoter activity as the most downstream 0.6 kbp. MacCarthy-Morrogh et al17 and Sugars et al19 also showed evidence for a strong regulatory element in a 683-bp sequence spanning the region of the human gene from −2362 through −1680. The homologous sequence to our mouse upstream element is the human sequence from −1827 through −1751. Despite its location far upstream of the start sites for exon 1A, the upstream element may regulate transcription at these start sites. In primary erythroblasts, we found that transcripts of bcl-x initiated at exon 1B are very rare, and strong regulation of transcripts initiated at exon 1A was observed. Our transfection studies were done in a mouse erythroid progenitor cell line, HCD57, because we have been unable to find conditions suitable for transfection of primary erythroblasts. We have used RT-PCR to obtain a semiquantitative estimate of the relative usage of exon 1A versus exon 1B for luciferase reporter transcripts in stable, transfected HCD57 cell clones with the full 1804 bp of promoter sequence. The results indicate that exon 1A is represented several-fold more frequently than exon 1B in such transcripts (data not shown). This indication that exon 1A is preferentially used in these lines, together with the data showing that the upstream element causes a 5-fold enhancement of luciferase expression in these lines (Figures 5-6), suggests that the upstream element does regulate transcription of exon 1A.
In the present study and in others,16 a basal promoter activity was observed in reporter plasmids containing as little as 197 bp of sequence immediately upstream of the bcl-x protein initiation codon (Figure 5). In constructs containing only 197 bp of downstream promoter sequence, we showed that this minimal promoter activity in transient transfection assays was reduced by 66% by deletion of the 7 repeats of CT from −116 through −130 (data not shown). However, in constructs containing the full 1804 bp ofbcl-x promoter, this deletion reduced the activity by only 20%, indicating that these repeats are not highly critical for function in the context of the full promoter. Because we found no transcription start site within the most downstream 200 base pairs of the promoter, we cannot presently interpret the significance of the basal transcription in transfection experiments nor of the physiologic importance of CT repeats in regulating transcription of the endogenousbcl-x gene.
Electrophoretic mobility shift assays (Figure 8) reveal that both of the important sequences of the upstream element (sequences A and B) bind specifically to nuclear proteins in mouse erythroblasts. Two major complexes are formed with the oligonucleotide representing sequence A (Figure 8A). One complex is down-regulated during the time in whichbcl-x transcripts accumulate in late erythroblasts, and the second complex remains constant or is slightly increased. These data are consistent with the former complex being a repressor and the latter a stimulator. The function of the upstream element is not restricted to erythroid cells, as our observation of its function in NIH 3T3 cells has shown. However, the changes in the nuclear proteins that bind the element occurring during erythroid differentiation do suggest that this element may be specifically regulated in erythroid cells. Specific complexes also were demonstrated between sequence B and nuclear proteins of erythroblasts. In this case the proteins forming these complexes diminish during erythroid differentiation (Figure 8B).
It has been reported that EPO specifically causes up-regulation ofbcl-x transcription by activating Stat5 that binds to sites in the first intron.12 13 However, plasmid reporter constructs used in those studies contained only about 650 bp of thebcl-x promoter sequence immediately upstream of the initiation codon. We have not been able to show any specific effect of EPO on transcription of bcl-x promoter constructs, either in transient assays with HCD57 cells or in clones of HCD57 cells containing stable integrated reporter plasmids. In all experiments, the activities of reporter genes under the control of the TK or SV40 promoters in control plasmids rose and fell in parallel with the firefly luciferase reporter gene driven by the bcl-xpromoter upon EPO addition or withdrawal. These results do not prove that EPO does not regulate bcl-x directly, but only that any specific effect was masked by a broader effect of EPO on transcription in these cells. Because of the functional importance of the upstream element, we suspect that it plays a role in regulating bcl-xtranscription during erythroid differentiation. However, it will be necessary to determine what proteins operate on this element and to study the regulation of those proteins in primary erythroid cells before we can understand this regulation and the possible role of EPO in it. Testing of multiple antibodies in supershift assays in gel mobility shift experiments has so far failed to identify any known protein involved in the interactions.
Prepublished online as Blood First Edition Paper, October 31, 2002; DOI 10.1182/blood-2002- 04-1217.
Supported by a Veterans Health Administration Merit Review Grant (M.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maurice Bondurant, Veterans Affairs Medical Center, ACRE Building, Rm F411, 1310 24th Ave South, Nashville, TN 37212; e-mail:maurice.c.bondurant@vanderbilt.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal