Human β-globin transgenes regulated by the locus control region (LCR) express at all integration sites in transgenic mice. For such LCR activity at ectopic sites, the 5′HS3 element requires the presence of the AT-rich region (ATR) in β-globin intron-2. Here, we examine the dependence of 5′HS3 LCR activity on transcription factor binding sites in the ATR. In vitro DNaseI footprint analysis and electrophoretic mobility shift assays of the ATR identified an inverted double Gata-1 site composed of 2 noncanonical sequences (GATT and GATG) and an Oct-1 consensus site. Mutant Oct-1, Gata-1, or double mutant sites were created in the ATR of the BGT50 construct composed of a 5′HS3 β/γ-globin hybrid transgene. Transgenes with double mutant sites expressed at all sites of integration, but mean expression levels in transgenic mice were reduced from 64% per copy (BGT50) to 37% (P < .05). Mutation of the inverted double Gata-1 site had no effect at 61% per copy expression levels. In contrast, mutation of the Oct-1 site alone reduced per-copy expression levels to 31% (P < .05). We conclude that the ability of 5′HS3 to activate expression from all transgene integration sites is dependent on sequences in the ATR that are not bound at high affinity by transcription factors. In addition, the Oct-1 site in the ATR is required for high-level 5′HS3 β/γ-globin transgene expression and should be retained in LCRβ-globin expression cassettes designed for gene therapy.
Introduction
The development of small globin gene constructs that express to high levels is a prerequisite for successful retrovirus or lentivirus-mediated gene therapy of sickle-cell anemia.1-3 Extensive analysis from many laboratories has shown that a locus control region (LCR) located upstream of the human β-globin gene cluster is capable of directing high-level β-globin transgene expression at all ectopic sites in transgenic mice.4-8 The LCR is composed of multiple DNaseI hypersensitive sites (HS),9-11 of which only 4 (5′HS1 to 5′HS4) are required together to obtain full expression levels from single-copy β-globin transgenes.12,13 These large 5′HS1-4 LCR cassettes range from 4-kb to 6.5-kb in length, compromising their use in gene therapy vectors. A smaller 3.0-kb 5′HS2-4 cassette directs 45% expression levels from single-copy β-globin transgenes,14 and similar 5′HS2-4 cassettes have been successfully used in lentivirus vectors to cure β-thalassemia and sickle cell anemia in mice.15-17 Despite their tremendous success in validating the principles of gene therapy in mouse models, expression from existing 5′HS2-4 globin gene therapy vectors remains suboptimal, resulting in protein expression of approximately 15% or requiring multiple integrations to be effective.15-17
To design smaller LCR cassettes that express globins to higher levels, we have shown that the 850-bp 5′HS3 element alone can activate β-globin expression to mean levels of 70% at all single-copy integration sites.13,18 However, 5′HS3 cannot reproducibly activate expression when linked to γ-globin transgenes,19-21 indicating that 5′HS3 LCR activity is dependent on gene proximal regulatory elements within the β-globin gene. As γ-globin protein has strong antisickling properties that are desirable for treating sickle cell anemia,22 it is important to define these β-globin proximal elements and link them to γ-globin coding sequences and minimal LCR elements such as 5′HS3.
There are several β-globin gene proximal elements that could influence LCR activity by 5′HS3, including the β-globin promoter,23,24 intron-2 matrix attachment region (MAR),25,26 intron-2 enhancer, or 3′ enhancer.23,27-29 Through the use of hybrid β/γ-globin transgenes it was shown that 5′HS3-directed LCR activity requires β-globin intron-2 and the 260-bp 3′ enhancer region, but not the β-globin promoter.19 These hybrid β/γ-globin transgenes can be stably transmitted through lentivirus vectors with high efficiency.30 However, deletion of the 372-bp AT-rich region (ATR) that colocalizes with the intron-2 MAR resulted in lower mean expression levels from 5′HS3 β/γ-globin transgenes with undetectable expression at some integration sites.19Hence, 5′HS3 LCR activity is dependent on sequences located within the β-globin intron-2 ATR. Although it is evident that high expression at all ectopic sites requires the ATR, it has proved difficult to incorporate the ATR in retrovirus vectors because it reduces titer by introducing premature polyadenylation signals, cryptic splice sites, and deletions into viral genomic RNA.31 32
The function of the ATR may be to bind transcription factors that cooperate with LCR elements. Jackson et al have reported several transcription factor binding sites dispersed through the β-globin intron-2.33 Here, we identify a novel noncanonical double Gata-1 site with unique factor-binding characteristics and confirm the presence of an Oct-1 consensus site in the ATR. In vivo functional analysis of mutant sites in 5′HS3 β/γ-globin transgenes in mice shows that LCR activity at all ectopic integration sites is dependent on sequences in the ATR that are not bound at high affinity by transcription factors. The Oct-1 consensus site is required for high-level expression from 5′HS3 β/γ-globin transgenes, while the inverted double Gata-1 site has no discernable role in this assay. These findings strongly suggest that the Oct-1 site should be retained in the intron-2 of β-globin or β/γ-globin cassettes designed for gene therapy.
Materials and methods
In vitro DNaseI footprint analysis
The β-globin AT-rich region was isolated as a 521-bpBamHI-DraI fragment, corresponding to the in vitro–characterized MAR,25 including the 372-bp ATR.31,32 The fragment was cloned into theBamHI site of Litmus 38 plasmid vector, usingBamHI-linkers to produce the ATR construct. The footprint probe was produced using a modified protocol previously described27 using restriction enzymes in the flanking polylinkers to radiolabel single DNA strands with32P(α)dCTP (Amersham Pharmacia, Baie d'Urfe, Quebec, Canada) using Klenow fragment (Roche, Laval, Quebec, Canada). Seven to ten thousand disintegrations per minute (dpm) of probe was mixed with 2 ng poly-dIdC, MEL cell nuclear extract, and digested with 4 U of DNaseI (Roche) for 120 seconds at 4°C in a DNaseI buffer (20 mM Hepes [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7.9, 1 mM MgCl2, 50 mM KCl, 250 μM phenylmethylsulfonyl fluoride [PMSF], 100 μM EDTA [ethylenediaminetetraacetic acid], 1.5 mM DTT). Equal counts of probe (3000-5000 dpm) were electrophoresed on a 6% sequencing gel. The footprinted samples were coelectrophoresed with dideoxyNTP sequencing ladders produced using a T7 Sequenase Kit (Amersham Pharmacia). The ATR vector (1 μg) was used as a template, and the FTspe1 primer (5′-GTG GGG CCC GTG CAA TTG AAG CC-3′) was used to sequence from the 5′ end, and FTnhe1 primer (5′-GCG AAT TCG TGG ATC CAA AAT TA-3′) was used to sequence from the 3′ end. All primers were obtained from Invitrogen (Carlsbad, CA).
Electrophoretic mobility shift assay
Oligonucleotide probes for electrophoretic mobility shift assay (EMSA) were annealed, radiolabeled, and bound to nuclear extract as described.27 An aliquot of each sample was electrophoresed on a 5% native acrylamide gel. Competitor oligonucleotides were used at 50-fold excess of the EMSA probe. Factor specific antibodies were used by preincubating the nuclear extract with the antibody for 45 minutes or 16 hours at 4°C before addition of labeled EMSA probe. Consensus oligonucleotides and antibodies for Oct-1, Gata-1, and Sp1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The sense strands of the other EMSA probes used are FT1 5′-ACG AAT GAT TGC ATC AGT GTG GAA GT-3′, FT1B 5′-ACG AAT TTT TGC ATC AGT GTG GAA GT-3′, FT1C 5′-ACG AAT GAT TGC AAA AGT GTG GAA GT-3′, FT1D 5′-ACG AAT TTT TGC AAA AGT GTG GAA GT-3′, FT2 5′-GTG TGC TTA TTT GCA TAT TCA T-3′, and FT2B 5′-GTG TGCTTG CTT GCA TAT TCA T-3′.
PCR-based site-specific mutagenesis
The template for the mutant Oct-1 AT-rich region (mOct-1) and the mutant Gata-1 AT-rich region (mGata-1) constructs was the BGT50 construct.19 The template for the mutant Gata-1/mutant Oct-1 (mGata-1/mOct-1) construct was the mOct-1 construct, and the mutation was introduced at the Gata-1 factor-binding site. The primers used to introduce the mutation as described34 at the Oct-1 or Gata-1 sites are the same oligonucleotides used for EMSA: FT1D (S) and (AS) for the mGata-1 and mGata-1/mOct-1 constructs; and FT2B (S) and (AS) for the mOct-1 construct. The first set of polymerase chain reactions (PCR) was separately amplified; the sequence between a 5′ primer named B50 (S) (5′-CTT TGC CCA GCT GAG TGA ACT GC-3′) that lies in exon 2 and the antisense mutant oligonucleotide; and the sequence between the sense mutant oligonucleotide and a 3′ primer named B50 (AS) (5′-GCC CTG AAA GAA AGA GAT TAG GG-3′) that lies in intron-2. The first PCR reactions were performed using 50-ng template, 10 pmole each primer, deoxynucleoside triphosphate (dNTP) mix (10 nmole each; Invitrogen), and 10 U Pfx DNA polymerase (Invitrogen). The 2 PCR products were gel purified. The second PCR reaction was performed by using 1 μg each of the products from the first PCR reaction, 500 pmole of B50 (S) and B50 (AS), dNTP mix, and 10 U of Pfx DNA polymerase. The product of this PCR reaction contained the entire AT-rich region (BamHI-DraI) with site-specific mutations at the targeted factor-binding site. The 521-bp mutant BamHI-DraI fragment was isolated and cloned into Litmus 38 as described above and sequence verified before subcloning into the BGT50 5′HS3 β/γ-globin construct.19
Generation of transgenic mice
Transgene DNA was isolated as 3.9-kbClaI-ClaI fragments and purified using Elutip-d columns (Schleicher and Schuell, Dassell, Germany), ethanol precipitated, and resuspended in injection buffer (10 mM Tris [tris(hydroxymethyl)aminomethane]-HCl pH 7.5, 0.2 mM EDTA) and diluted to 0.1-0.5 ng/μL. Injected FVB mouse eggs were transferred into recipient CD1 females and fetuses dissected at day 15.5. Fetal heads were collected for identification of transgenic animals by slot blot hybridization to a β-globin intron-2 probe, and fetal liver split in 2 for subsequent DNA and RNA analysis from frozen samples.
DNA analysis
Southern transfer and hybridization were by standard procedures. Copy-number determination was performed using a Molecular Dynamics PhosphorImager (Sunnyvale, CA). Single-copy animals showed a single random-sized end-fragment in BamHI andEcoRI digests hybridized with a β-globin intron-2 probe. With multicopy animals, the intensity of the end-fragment was defined as 1 copy and was used to calculate the copy number of the multicopy junction-fragment in the same lane. The intactness of the transgene in the DNA sample was verified by Southern blot analysis using PstI digests hybridized to a β-globin intron-2 probe and an endogenous mTHY-1 probe. Nonintact transgenes were not included in the calculation of copy number. A loading control of bred single copy transgenic mouse DNA included in the PstI Southerns permitted mosaic analysis as described.14 Mice that were highly mosaic were excluded from study after demonstration that < 20% of the fetal liver cells contained intact transgenes.
RNA analysis
Fetal liver (embryonic day 15.5) RNA was extracted using Trizol Reagent (Invitrogen), 1 μg was hybridized to32P(α)dATP-labeled (Amersham Pharmacia) double-stranded 3′ DNA probe (the EcoRI fragment containing exon 3) for detection of human γ-globin.35 A32P(γ)ATP-labeled (Amersham Pharmacia) double-stranded 5′DNA probe was used for mouse βmajor-globin detection as a loading control.23 RNA/DNA hybrids were subsequently digested with 75 units of S1 nuclease (Roche) and run on a 6% sequencing gel as described.23 Probe excess was demonstrated by including a sample that contains 3 μg of fetal liver RNA. The protected 170 nucleotide (nt) Huγ and 95 nt Moβ bands were quantified on a Molecular Dynamics PhosphorImager and the percentage expression levels calculated according to the formula (Huγ/Moβ) × 100%. The percentage expression levels were corrected for specific activities of the probe preparations by normalization using T225 fetal liver RNA isolated from a heterozygous BGT50 line, previously shown to express Huγ at 40% per Moβ copy.19 Expression per transgene was calculated as (2 Moβ genes/number of Huγ transgenes) × (% expression)/(% transgenic cells) × 100%.
Statistical analysis
To determine if mean expression from a mutant transgene is statistically different from the mean expression level of the BGT50 construct, the Wilcoxon rank-sum test was performed. Since at least 8 transgenic mice were generated for each transgene, a normal approximation can be made using the test statistic:
Results
In vitro DNaseI footprint analysis of the AT-rich region
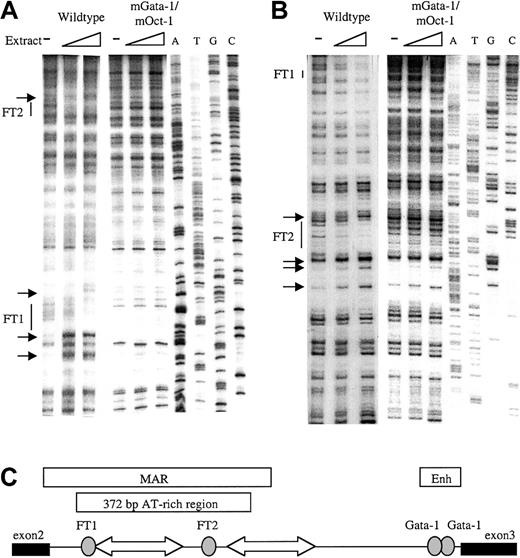
To identify factor-binding sites in the β-globin intron-2 ATR, in vitro DNaseI footprint analysis was performed. The 521BamHI-DraI fragment containing the 372-bp ATR19,31 32 was cloned into the Litmus 38 vector. Radiolabeled sense and antisense ATR fragments were incubated with mouse erythroleukemia (MEL) cell nuclear extract, briefly digested by DNaseI, and separated adjacent to a sequence ladder. Two footprinted regions were detected on the sense strand (Figure1A); a 5′ footprinted region (FT1) downstream of 2 strong hypersensitive bases and a 3′ footprinted region (FT2). Strong footprints on the antisense strand also colocalized with the same FT1 and FT2 sequences (Figure 1B). Location of the 2 strong footprints within β-globin intron-2 is illustrated in Figure 1C.
In vitro DNaseI footprint analysis of the β-globin intron-2 AT-rich region (ATR).
(A) Footprint of ATR sense strand probes protected by MEL cell nuclear extract showing footprints FT1 and FT2 with associated hypersensitive bases (small arrows) on the wild-type ATR fragment but not on the mGata-1/mOct-1 ATR fragment. ATGC indicates dideoxy sequencing ladders. (B) Footprint of the ATR antisense strand shows FT1 and FT2 on the wild-type ATR fragment but not the mGata-1/mOct-1 ATR fragment. (C) Location of FT1 and FT2 footprints in β-globin intron-2 aligned beside the reported AT-rich region and MAR. Gray ovals indicate footprints; block arrows, tracts of adenosine/thymidine–rich sequence; thin line, intron-2; thick lines, exons.
In vitro DNaseI footprint analysis of the β-globin intron-2 AT-rich region (ATR).
(A) Footprint of ATR sense strand probes protected by MEL cell nuclear extract showing footprints FT1 and FT2 with associated hypersensitive bases (small arrows) on the wild-type ATR fragment but not on the mGata-1/mOct-1 ATR fragment. ATGC indicates dideoxy sequencing ladders. (B) Footprint of the ATR antisense strand shows FT1 and FT2 on the wild-type ATR fragment but not the mGata-1/mOct-1 ATR fragment. (C) Location of FT1 and FT2 footprints in β-globin intron-2 aligned beside the reported AT-rich region and MAR. Gray ovals indicate footprints; block arrows, tracts of adenosine/thymidine–rich sequence; thin line, intron-2; thick lines, exons.
FT1 is an inverted noncanonical double Gata-1 site
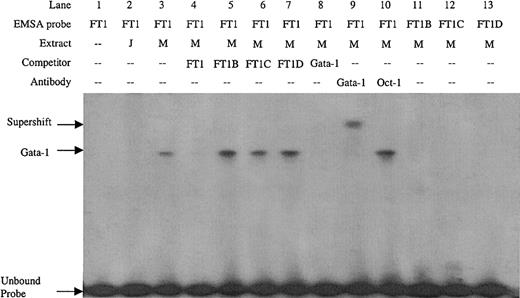
Double-stranded DNA oligonucleotides were created covering the FT1 footprint. Sequence inspection of the FT1 region revealed an inverted noncanonical double Gata-1 site (5′-GATTGCATC-3′). EMSA with the FT1 probe detected a factor present in MEL cell nuclear extract but not Jurkat cell nuclear extract (Figure 2, lanes 2 and 3), consistent with the erythroid specificity of Gata-1. Preincubation of MEL extract with Gata-1 antibody, but not Oct-1 antibody, supershifted this complex (Figure 2, lanes 9 and 10), which also was competed with Gata-1 consensus sites, confirming its identity as Gata-1. To determine which of the inverted double Gata-1 sites are bound, mutations of GA to TT were introduced in either the 5′, 3′, or both noncanonical Gata-1 sites (FT1B, FT1C, and FT1D, respectively). The mutant oligonucleotides did not compete the FT1 complex, nor were they bound by factors in MEL nuclear extracts when used as EMSA probes (Figure 2, lanes 5-7 and 11-13). These data demonstrate that Gata-1 binding requires the entire inverted double site, and mutation of either or both noncanonical sites results in loss of Gata-1 binding.
EMSA on the FT1 footprint of the ATR bound by Gata-1 factor present in MEL extract but not Jurkat extract.
Competitors and probes with mutations that remove the 5′, 3′, or both noncanonical Gata-1 sites (FT1B, FT1C, and FT1D, respectively) and a consensus Gata-1 competitor are indicated, as are antibodies used in supershift experiments. M indicates MEL extract; J, Jurkat extract.
EMSA on the FT1 footprint of the ATR bound by Gata-1 factor present in MEL extract but not Jurkat extract.
Competitors and probes with mutations that remove the 5′, 3′, or both noncanonical Gata-1 sites (FT1B, FT1C, and FT1D, respectively) and a consensus Gata-1 competitor are indicated, as are antibodies used in supershift experiments. M indicates MEL extract; J, Jurkat extract.
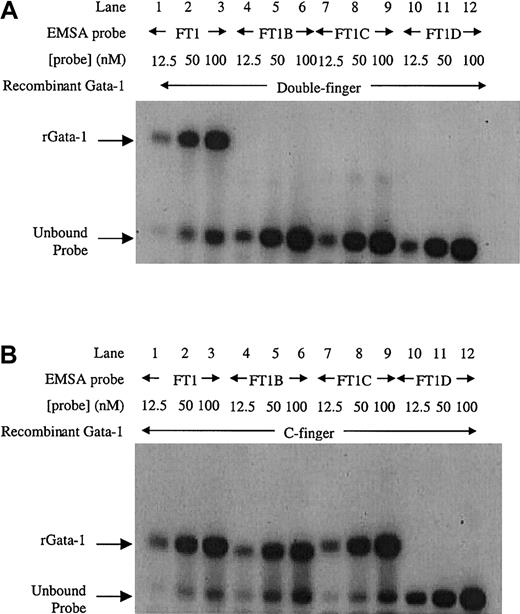
To further investigate Gata-1 binding to this inverted double site, EMSA using recombinant Gata-1 proteins36 was performed. Increasing amounts of FT1 probe for the inverted double site was bound by the double finger recombinant protein consisting of both C-terminal and N-terminal zinc fingers (Figure 3A, lanes 1-3). In contrast, the mutant sites (FT1B, FT1C, and FT1D) were unable to bind recombinant Gata-1 protein (Figure 3A, lanes 4-12). These data confirm that the entire inverted double site is required to bind Gata-1. A recombinant C-finger Gata-1 protein consisting of only the C-terminal zinc finger bound to the inverted double site (Figure3B, lanes 1-3). The C-finger protein also bound to probes that contained only one noncanonical site (Figure 3B, lanes 4-9), but not to the probe in which both noncanonical sites were mutated (Figure 3B, lanes 10-12). These results show that the C-finger protein is capable of binding to either noncanonical site. Furthermore, the C-finger protein bound to the inverted double site comigrated with the complex formed at single mutant Gata-1 sites, indicating that only one C-finger protein molecule can bind to the inverted double site at a time. These data are consistent with Gata-1 factor binding FT1 as a monomer that simultaneously contacts both the noncanonical sites in the inverted double site through its pair of zinc fingers.
EMSA with recombinant Gata-1 proteins on the inverted noncanonical double site.
(A) EMSA with a recombinant Gata-1 (rGata-1) molecule containing both C- and N-terminal zinc fingers (double-finger) binds FT1 probe containing both noncanonical Gata-1 sites, but not probes with the 5′, 3′, or both sites mutated (FT1B, FT1C, and FT1D, respectively). (B) EMSA performed with a recombinant Gata-1 protein containing only the C-terminal zinc finger (C-finger) binds all 3 probes that contain at least one noncanonical site (FT1, FT1B, and FT1C). Each probe was assayed with 10 mL 12.5, 50, and 100 nM DNA concentration.
EMSA with recombinant Gata-1 proteins on the inverted noncanonical double site.
(A) EMSA with a recombinant Gata-1 (rGata-1) molecule containing both C- and N-terminal zinc fingers (double-finger) binds FT1 probe containing both noncanonical Gata-1 sites, but not probes with the 5′, 3′, or both sites mutated (FT1B, FT1C, and FT1D, respectively). (B) EMSA performed with a recombinant Gata-1 protein containing only the C-terminal zinc finger (C-finger) binds all 3 probes that contain at least one noncanonical site (FT1, FT1B, and FT1C). Each probe was assayed with 10 mL 12.5, 50, and 100 nM DNA concentration.
FT2 is an Oct-1 site
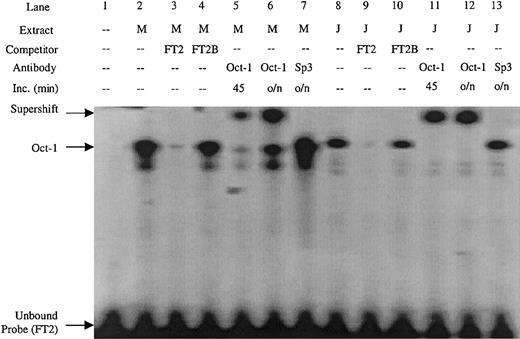
Double-stranded DNA oligonucleotides were created covering the FT2 footprint. Sequence inspection of the FT2 region revealed an Oct-1 consensus site. An FT2 EMSA probe detected a complex in both MEL and Jurkat nuclear extract (Figure 4, lanes 2 and 8) that was competed by excess unlabeled probe, but not by a mutant Oct-1 competitor containing an AT to GC substitution (FT2B) (Figure 4, lanes 3-4 and 9-10). The complex was also supershifted by Oct-1 antibody in both MEL and Jurkat extracts, but not with Sp3 antibody (Figure 4, lanes 5-7 and 11-14). These data show that FT2 is bound by the ubiquitous Oct-1 factor.
EMSA performed with the FT2 footprint of the ATR is bound by Oct-1 factor present in MEL and Jurkat extracts.
The FT2B competitor contains a mutation in the Oct-1 consensus site, antibodies used in supershift experiments are indicated. M indicates MEL extract; J, Jurkat extract; Inc, length (in minutes) of incubation of extract and antibody prior to addition of EMSA probe; o/n, overnight.
EMSA performed with the FT2 footprint of the ATR is bound by Oct-1 factor present in MEL and Jurkat extracts.
The FT2B competitor contains a mutation in the Oct-1 consensus site, antibodies used in supershift experiments are indicated. M indicates MEL extract; J, Jurkat extract; Inc, length (in minutes) of incubation of extract and antibody prior to addition of EMSA probe; o/n, overnight.
Creation of mutant AT-rich regions
PCR-based site-specific mutagenesis was used to create 3 mutant ATRs: 1) the inverted double Gata-1 site mutated at both noncanonical sites (mGata-1 ATR); 2) the Oct-1 site mutated (mOct-1 ATR); and 3) both the inverted double Gata-1 site and the Oct-1 site mutated (mGata-1/mOct-1 ATR). To demonstrate that the mutant sites were not bound by any factors, DNaseI footprint analysis with MEL nuclear extract was performed. Footprint analysis of the mGata-1 ATR region detected a footprint at the Oct-1 site in the FT2 region but no footprint at the FT1 region as expected (data not shown). Conversely, analysis of the mOct-1 ATR region detected the Gata-1 footprint in FT1, but no footprint in FT2 region (data not shown). Finally, analysis of the mGata-1/mOct-1 ATR region did not detect any footprints on the sense or antisense strands (Figure 1A-B).
Functional analysis in transgenic mice
To test the functional importance of the Gata-1 and Oct-1 sites, the mGata-1/mOct-1 ATR was cloned into intron-2 of the BGT50 construct yielding BGT133 (Figure 5, bottom). BGT50 contains a 5′HS3 LCR element linked to the 815-bp β-globin promoter, γ-globin exon-1/intron-1/exon-2, β-globin intron-2, γ-globin exon-3, and the 260-bp β-globin 3′ enhancer (Figure 5, top). BGT50 transgenes in mice express human γ-globin (Huγ) mRNA at all integration sites tested at 64% ± 11% (SE) per copy.19Transgenic animals were created with the BGT133 construct, and fetuses collected at day 15.5. Fetal head DNA was screened by slot blot hybridization to identify transgenic animals. Southern blot analysis was performed on fetal head and liver DNA of positive animals to determine copy number, transgene intactness, and mosaicism in fetal liver as described (data not shown). Highly mosaic animals containing < 20% transgenic cells in the fetal liver were excluded.
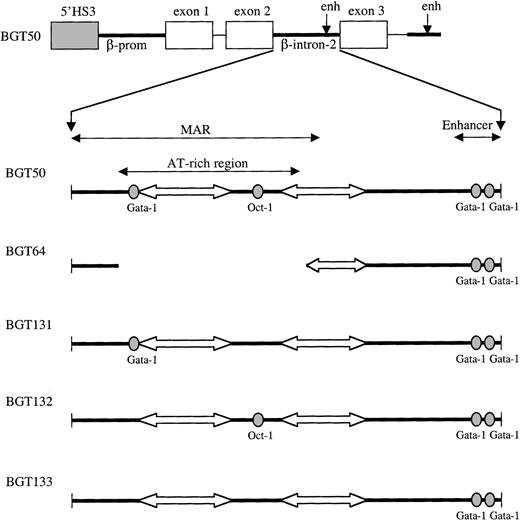
Map of 5′HS3 β/γ-globin transgene constructs designed to test LCR dependence on footprints present in the ATR.
Gray boxes indicate the 0.85-kb 5′HS3; white boxes, γ-globin exons; thin lines, γ-globin intron-1 and downstream sequences; and thick lines, β-globin sequences. Within β-globin intron-2, wild-type factor-binding sites are depicted as ovals and A/T tracts as block arrows.
Map of 5′HS3 β/γ-globin transgene constructs designed to test LCR dependence on footprints present in the ATR.
Gray boxes indicate the 0.85-kb 5′HS3; white boxes, γ-globin exons; thin lines, γ-globin intron-1 and downstream sequences; and thick lines, β-globin sequences. Within β-globin intron-2, wild-type factor-binding sites are depicted as ovals and A/T tracts as block arrows.
Mutation of inverted double Gata-1 and Oct-1 sites reduces transgene expression
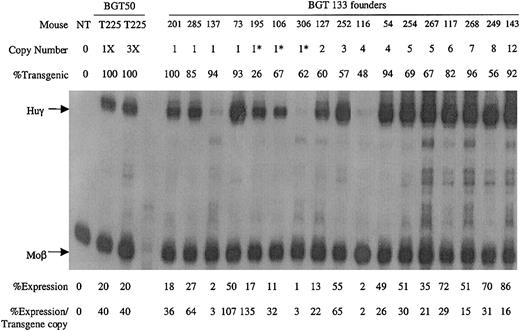
A total of 17 transgenic animals were identified with intact BGT133 transgenes, including 7 single-copy animals. Expression was determined by S1 nuclease protection assay with Huγ and mouse βmajor-globin (Moβ) probes (Figure6). RNA from a single-copy BGT50 bred line (T225)19 that expressed Huγ at 40% per Moβ copy (the low end of the BGT50 expression range), and a nontransgenic (NT) animal served as controls. All single-copy and multicopy BGT133 animals express Huγ mRNA to a mean level of 37% ± 8% (SE). These data demonstrate that the inverted double Gata-1 and Oct-1 sites in the ATR are not essential for activating LCR-dependent transgene expression at ectopic sites of integration. However, mutation of the inverted double Gata-1 and Oct-1 sites reduced mean transgene expression levels from 64% to 37%.
Expression of globin mRNA in transgenic mice containing the BGT133 (mGata-1/mOct-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the Huγ-globin transgene in 7/7 single-copy and 10/10 multicopy animals, to a mean of 37% of each endogenous mouse βmajor-globin gene. NT, nontransgenic; Huγ, human γ-globin protected probe fragment; Moβ, mouse βmajor-globin protected probe fragment; BGT50 T225, a heterozygous BGT50 transgenic line; 3X, probe excess control using 3 μg of fetal liver RNA; copy number = 1*, one intact and one partial copy of the transgene.
Expression of globin mRNA in transgenic mice containing the BGT133 (mGata-1/mOct-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the Huγ-globin transgene in 7/7 single-copy and 10/10 multicopy animals, to a mean of 37% of each endogenous mouse βmajor-globin gene. NT, nontransgenic; Huγ, human γ-globin protected probe fragment; Moβ, mouse βmajor-globin protected probe fragment; BGT50 T225, a heterozygous BGT50 transgenic line; 3X, probe excess control using 3 μg of fetal liver RNA; copy number = 1*, one intact and one partial copy of the transgene.
Mutation of inverted double Gata-1 site does not affect transgene expression
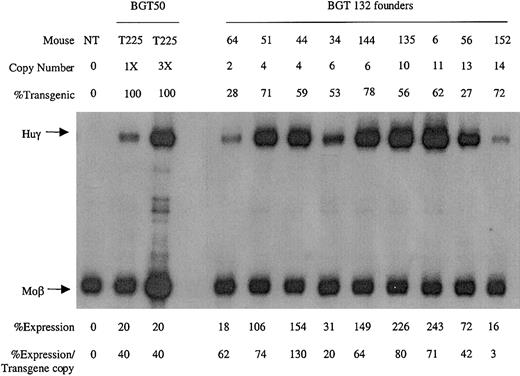
To identify which mutation was responsible for the reduction in transgene expression levels, transgenic animals were generated with the BGT132 (mGata-1 ATR) and BGT131 (mOct-1 ATR) constructs. Nine multicopy transgenic animals were identified with intact BGT132 transgenes. S1 protection assays demonstrate that all the animals express Huγ to a mean level of 61% ± 12% (Figure 7), suggesting that mutation of the inverted double Gata-1 site does not reduce transgene expression levels.
Expression of globin mRNA in transgenic mice containing the BGT132 (mGata-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the γ-globin transgene in 9/9 multicopy animals to a mean of 61% of each endogenous mouse βmajor-globin gene. Abbreviations are as described in the legend to Figure 6.
Expression of globin mRNA in transgenic mice containing the BGT132 (mGata-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the γ-globin transgene in 9/9 multicopy animals to a mean of 61% of each endogenous mouse βmajor-globin gene. Abbreviations are as described in the legend to Figure 6.
Mutation of the Oct-1 site reduces transgene expression
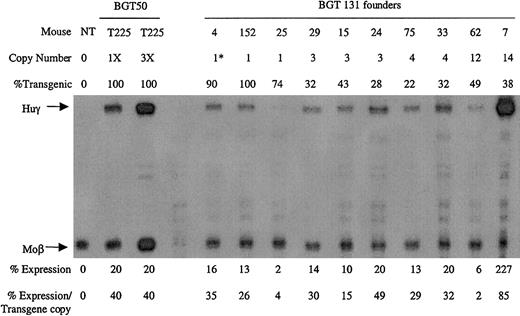
Ten transgenic animals were identified with intact BGT131 (mOct-1 ATR) transgenes, including 3 single-copy animals. All single-copy and multicopy BGT131 animals express the Huγ transgene to a mean level of 31% ± 8% when assayed by S1 protection assay (Figure8). These data demonstrate that mutation of the Oct-1 site alone reduces transgene expression to levels equivalent to the BGT133 mice (mGata-1/mOct-1 ATR) described above. Transgenic mice expressing BGT64 (in which the entire ATR is deleted) also express to this same level (31% ± 2%),19indicating that the absence of the Oct-1 site can fully account for reduced expression levels by BGT64.
Expression of globin mRNA in transgenic mice containing the BGT131 (mOct-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the γ-globin transgene 3/3 single-copy and 7/7 multicopy animals to a mean of 31% of each endogenous mouse βmajor-globin gene. Abbreviations are as described in the legend to Figure 6.
Expression of globin mRNA in transgenic mice containing the BGT131 (mOct-1 ATR) construct.
S1 nuclease protection analysis on day 15.5 fetal liver RNA reveals expression of the γ-globin transgene 3/3 single-copy and 7/7 multicopy animals to a mean of 31% of each endogenous mouse βmajor-globin gene. Abbreviations are as described in the legend to Figure 6.
Discussion
LCR activity by 5′HS3 at ectopic transgene integration sites requires a functional interaction with elements in the β-globin gene that include the intron-2 AT-rich region (ATR).19 Here we characterize 2 strong footprints in the ATR. FT1 is an inverted double Gata-1 site composed of 2 noncanonical binding sites bound by a single Gata-1 molecule. FT2 is an Oct-1 consensus site. Functional analysis of the 2 footprinted regions using mutations that destroy Gata-1 and Oct-1 binding demonstrate that these sites are not required for 5′HS3 to activate expression at ectopic sites in transgenic mice. However, LCR-regulated transgene expression levels are dependent on the Oct-1 site in the ATR. These conclusions have important implications for the design of β-globin gene therapy cassettes.
Novel inverted double Gata-1 site
EMSA on the FT1 region revealed binding of Gata-1 to an inverted repeat of noncanonical Gata-1 sites. Although previous analysis suggested that Gata-1 bound a single noncanonical site at this location,33 our experiments show that both noncanonical sites are required for Gata-1 complex formation and this unique binding profile is mediated by simultaneous interactions with both Gata-1 zinc fingers. The existence of canonical and noncanonical double sites with different apparent activities36 could suggest an unusual function for Gata-1 bound to the ATR.
Oct-1 binds FT2
Analysis of the FT2 footprinted region by EMSA identified a complex competed by an Oct-1 site, supershifted by an Oct-1 antibody, and which failed to bind probes in which the Oct-1 consensus was mutated. These data are consistent with the presence of an Oct-1 site in β-globin intron-2 as reported.33 Oct-1 is an ubiquitous transcription factor with activator37 and repressor38 functions in transcriptional regulation that binds enhancers39 and promoters.38
Mutant Oct-1 site reduces 5′HS3β/γ-globin transgene expression levels
To determine the importance of the Oct-1 and Gata-1 sites within the ATR, point mutations that destroyed these sites were introduced into the ATR of the 5′HS3 β/γ-globin construct (BGT50) and introduced into transgenic mice. If the LCR is dependent on the Gata-1 or Oct-1 sites in the ATR, then mutations removing one or both sites could result in lack of expression at some integration sites and/or a decrease in mean expression levels per transgene.
Analysis of BGT133 (mGata-1/mOct-1 ATR) fetal liver RNA demonstrated expression in 7/7 single-copy and 12/12 multicopy animals at a mean expression level of 37% ± 8% (SE) per transgene copy (Table 1). BGT133 expression is significantly different (P < .05) from BGT50 expression of 64% ± 11% (Table 1), indicating that mutation of one or both sites reduces transgene expression levels. To identify the binding site responsible for reduced expression, transgenic animals were created containing mutations at either the Gata-1 or the Oct-1 sites. BGT132 (mGata-1 ATR) transgenic animals expressed in 9/9 multicopy animals with a mean expression of 61% ± 12%, indicating that mutation of the inverted double Gata-1 site alone does not significantly reduce (P > .50) transgene expression levels (Table 1). The lack of an effect for the mutant Gata-1 site may reflect redundancy between it and other Gata-1 sites present in the transgene including 5′HS3,40 β-globin promoter,23,24 intron-2 enhancer, and 3′ enhancer.23,27-29 Alternatively, the inverted double site may bind Gata-1 in a conformation that fails to contribute to overall expression levels41 or perturb nucleosome structure.42 This is consistent with the lack of an HS at this location in intron-2,9-11 indicating that significant nucleosome remodeling does not occur at the inverted double Gata-1 site in vivo.
Description of AT-rich regions present in each 5′HS3 β/γ-globin hybrid transgene and a summary of transgenic animals and transgene expression levels
| Transgene . | AT-rich region . | Mice expressing single-copy . | Mice expressing total . | Mean expression* ± standard error (SE) . | Difference from BGT50‡ . |
|---|---|---|---|---|---|
| BGT50 | Wild-type | 7/7 | 8/8 | 64 ± 11 | NA |
| BGT64 | ΔAT-rich region | 2/4 | 13/17 | 31† ± 2 | P < .01 |
| BGT131 | mOct-1 | 3/3 | 10/10 | 31 ± 8 | P< .05 |
| BGT132 | mGata-1 | — | 9/9 | 61 ± 12 | P> .50 |
| BGT133 | mGata-1/mOct-1 | 7/7 | 18/18 | 37 ± 8 | P< .05 |
| Transgene . | AT-rich region . | Mice expressing single-copy . | Mice expressing total . | Mean expression* ± standard error (SE) . | Difference from BGT50‡ . |
|---|---|---|---|---|---|
| BGT50 | Wild-type | 7/7 | 8/8 | 64 ± 11 | NA |
| BGT64 | ΔAT-rich region | 2/4 | 13/17 | 31† ± 2 | P < .01 |
| BGT131 | mOct-1 | 3/3 | 10/10 | 31 ± 8 | P< .05 |
| BGT132 | mGata-1 | — | 9/9 | 61 ± 12 | P> .50 |
| BGT133 | mGata-1/mOct-1 | 7/7 | 18/18 | 37 ± 8 | P< .05 |
NA indicates not applicable; —, no single copy transgenic mice obtained.
Mean expression listed as the mean of the percentage of human γ-globin transcripts to endogenous mouse βmajor-globin transcripts on a per copy basis of each mouse.
Calculated from expression levels of the 13 expressing mice.
Calculated by the Wilcoxon rank sum method.
Analysis of BGT131 (mOct-1 ATR) transgenic animals demonstrated expression in 3/3 single-copy and 7/7 multicopy animals at a mean level of 31% ± 8%. These data show that mutation of the Oct-1 site alone significantly reduces (P < .05) mean expression levels (Table 1). This reduction is similar to the decreased expression (P < .01) observed when the ATR is deleted in BGT6419 at a mean expression of 31% ± 2% (Table 1), indicating that Oct-1 can fully account for this reduction. The strong effect on expression levels of the Oct-1 mutation is surprising, but as the other known Oct-1 site in the β-globin locus is located in the γ-globin promoter,43 it implies that there would be no redundant sites for Oct-1 in the hybrid β/γ-globin genes used here. These data are the first functional data demonstrating that a gene proximal Oct-1 site plays an important role in controlling expression levels from LCR-regulated β-globin transgenes at ectopic integration sites.
Mutant ATR constructs express detectable levels at ectopic sites
We previously showed that 5′HS3 β/γ-globin transgenes containing the ATR (BGT50) express at all integration sites tested, whereas some integration sites are silent when the AT-rich region is deleted (BGT64).19 The 3 ATR mutations of the BGT50 construct tested here express detectable levels at all integration sites. These results show that sequences within the ATR other than the strong transcription factor binding sites ensure expression at all 5′HS3 β/γ-globin transgene integration sites. The remarkable AT-richness of the sequence raises 2 models that are testable. First, the ATR contains a MAR that may tether the β/γ-globin transgene to the nuclear matrix.25,26 This effect may contribute to LCR activity by extending open chromatin, similar to the chromatin modifications44,45 extending from MARs that surround the immunoglobulin-μ enhancer.46 A report using higher concentrations of nuclear extract in footprint experiments indicated the presence of a weak SATB1 site downstream of the Oct-1 site at FT2.33 SATB1 is expressed primarily in thymocytes where it binds MARs and acts as a repressor, but the phenotype in SATB1 knockout mice is limited to effects on T cells.47 Despite its normal silencing function in T cells, weak SATB1 binding in erythroid cells could contribute to intron-2 MAR activity. Second, periodic runs of A or T can position nucleosomes48,49 that can be anchored to their positions by adjacent transcription factor binding sites.50 If Gata-1 and Oct-1 are bound to linker regions, this could anchor a positioned nucleosome on the intervening 160 bp of AT rich sequence (Figure 1C) without creating a detectable HS. An array of phased nucleosomes across intron-2 might result, and these could increase Gata-1 accessibility to sites at the intron-2 enhancer, facilitating HS formation at this location and the spread of open chromatin. Sequences that position nucleosomes within β-globin loci have been reported.51 The Oct-1 site may be more important than the Gata-1 site for elevating expression levels directed by the intron-2 enhancer in this scenario, but the AT sequences alone could be sufficient to establish weaker unanchored nucleosome positioning and permit detectable expression at all transgene integration sites.
Use of the ATR in gene therapy
The intron-2 ATR is deleterious for high titer production of globin retrovirus vectors, but the Oct-1 site in the ATR is important for high-level expression in transgenic mice. Hence, deletion of the ATR in globin retrovirus vectors improves virus titer31,32but compromises mean expression levels.19 Our analysis of RNA extracted from transient transgenic mice precludes single-cell analysis to study possible variegation effects. However, mean expression levels of 64% from 5′HS3-regulated β/γ-globin hybrid transgenes suggests that most cells are expressing therapeutically effective levels of globin RNA that are higher than the 31% levels obtained from ATR deleted cassettes. These findings suggest that components of the ATR, including the Oct-1 site, should be retained to create the ideal LCR β-globin cassette for gene therapy capable of expressing high mean levels at all ectopic integration sites. Future experiments designed to test LCR dependence on MAR activity or nucleosome positioning in the ATR may define minimal sequences that can perform this function and be transmitted through gene therapy vectors at high titer.
Thanks to J. Rubin, D. Pannell, and L. Wei for technical advice and support, and to J. Omichinski for the peptides used in this study.
Prepublished online as Blood First Edition Paper, September 26, 2002; DOI 10.1182/blood-2002-07-2086.
Supported by a grant from the Canadian Institutes of Health Research (CIHR) (J.E.) and a Hospital for Sick Children Research Training Centre Graduate Studentship (R.R.B.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Note added in proof
SATB1 has recently been shown to regulate nucleosome positioning.52
Author notes
James Ellis, Elm Building, Room 8154, Hospital for Sick Children, 555 University Ave, Toronto, ON, Canada M5G 1X8; e-mail: jellis@sickkids.ca.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal