The mi transcription factor (MITF) is a basic-helix-loop-helix leucine zipper transcription factor and is encoded by mi locus. The mi/mi mutant mice showed a significant decrease of skin mast cells in C57BL/6 (B6) genetic background but not in WB genetic background. Kit ligand (KitL) is the most important growth factor for development of mast cells, and the decrease of skin mast cells in B6-mi/mi mice was attributable to the reduced expression of c-kit receptor tyrosine kinase (KIT) that is a receptor for KitL. However, the expression level of KIT in WB-mi/mi mast cells was comparable with that of B6-mi/mi mast cells, suggesting that a factor compensating the reduced expression of KIT was present in WB-mi/mi mice. By linkage analysis, such a factor was mapped on chromosome 10. The mapped position was closely located to the KitL locus. Two alternative spliced forms are known in KitL mRNA: KL-1 and KL-2. Soluble KitL, which is important for development of skin mast cells, is produced more efficiently from KL-1 mRNA than from KL-2 mRNA. The KL-1/KL-2 ratio was higher in WB-mi/mi than in B6-mi/mi mice, suggesting that the larger amount of soluble KitL may compensate for the reduced expression of KIT in WB-mi/mi mice.
Introduction
The mi locus encodes mitranscription factor (MITF), which is a member of basic-helix-loop-helix-leucine zipper protein family of transcription factors.1 Mutant mice of mi/mi genotype were found among the offspring of an irradiated mouse,2 and the mutant gene was introduced into C57BL/6 (B6) genetic background mice (B6-mi/mi mice). The number of skin mast cells in B6-mi/mi mice decreased to one third that of normal (+/+) B6 mice.3 4
We examined the effect of MITF encoded by the mimutant allele (mi-MITF) on the expression of various genes in cultured mast cells (CMCs). CMCs derived from the spleen of B6-mi/mi mice were deficient in the expression of c-kit receptor tyrosine kinase (KIT),5,6 mouse mast cell protease (mMCP)–2,7 –4,8–5,9 –6,10 –7,11–9,7 granzyme B (Gr B),12 and tryptophan hydroxylase (TPH) genes.12 The mi-MITF deletes 1 of 4 consecutive arginines in the basic domain.13,14 Themi-MITF is defective in the DNA binding ability15 and the nuclear localization potential16 and does not transactivate mMCP-2, -4, -6, and -9 genes. Moreover, we found an inhibitory effect of mi-MITF in the transactivation of KIT, TPH, Gr B, and mMCP-7genes by comparing the effect of MITF null mutations.11,17-19 For example, the expression ofmMCP-7 gene was higher in B6-tg/tg CMCs, in which no MITF was produced, than in B6-mi/mi CMCs, indicating that the mi-MITF showed an inhibitory effect on the transcription of mMCP-7 gene.11 MITF binding site was not present in the promoter region that was essential for transcription ofmMCP-7 gene. In contrast, a c-Jun binding motif was present in this region, and the binding of c-Jun to the motif was important for the transactivation. The mi-MITF inhibited the transactivation ability of c-Jun and suppressed the expression level of mMCP-7 gene.11
Because the most important growth factor of mast cells is considered to be Kit ligand (KitL), which is a ligand for KIT,20-23 the decrease in the skin mast cells may be attributable to the deficient signal transduction of the KitL-KIT pathway. In fact, the poor expression of KIT was demonstrated in skin mast cells of B6-mi/mi mice by immunohistochemistry.24
We attempted to investigate the in vivo biologic features of CMCs of the mi/mi genotype by transplantation into tissues of mast cell–deficient (WB × C57BL/6) F1(WBB6F1)–W/Wv mice. To obtain mi/mi CMCs in the same genetic background of WBB6F1-W/Wv recipients, we repeatedly backcrossed the mi mutant gene to the WB strain and obtained WB-mi/mi mice after 8 backcrosses. Unexpectedly, the number of skin mast cells in WB-mi/mi mice did not decrease compared with that of WB-+/+ mice, showing that the effect of mi-MITF on the number of skin mast cells was influenced by the genetic background. The level of KIT expression was reduced in WB-mi/mi CMCs, as in the case of B6-mi/mi CMCs. This suggested that factor(s) compensating for the reduced expression of KIT was present in WB-mi/mimice but not in B6-mi/mi mice. We mapped such a factor on chromosome 10 using microsatellite markers. The mapped position was very closely located to the KitL locus, and there is a possibility that the difference between WB and B6 strains may be attributable to their different splicing patterns of KitL.
Materials and methods
Mice
The original stock of B6-mi/+ mice was purchased from the Jackson Laboratory (Bar Harbor, ME) and was maintained in our laboratory by consecutive backcrosses with our own inbred B6 colony (more than 15 generations at the time of the present experiment). WB-mi/mi mice used in the present study were at the eighth to 10th generations of the repeated backcrosses. WBB6F1-mi/mi mice were obtained by mating between B6-mi/+ and WB-mi/+ mice. (F1 × B6)–mi/mi mice were obtained by mating between WBB6F1-mi/+ and B6-mi/+ mice. Mice of the mi/mi genotype were identified by the white coat color.25
Staining and counting of mast cells
Mice at 2 weeks old were killed by decapitation after ether anesthesia. Pieces of dorsal skin were removed, smoothed onto a piece of filter paper to keep them flat, fixed in Carnoy solution, and embedded in paraffin. Sections (4 μm thick) of skin pieces were stained with alcian blue. The staining method with alcian blue has been described previously.26 Mast cells between epithelium and panniculus carnosus were counted under the microscope, and the number was expressed as mast cells per centimeter of skin.
Cells
Pokeweed mitogen–stimulated spleen cell conditioned medium (PWM-SCM) was prepared according to the method described by Nakahata et al.27 Mice at 2 weeks old were used to obtain CMCs. Mice were killed by decapitation after ether anesthesia, and spleens were removed. Spleen cells were cultured in α-minimal essential medium (α-MEM) (ICN Biomedicals, Costa Mesa, CA) supplemented with 10% PWM-SCM and 10% fetal calf serum (FCS) (Nippon Bio-supp Center, Tokyo, Japan). Half of the medium was replaced every 5 days. More than 95% of cells contained alcian blue–positive granules and were considered to be CMCs 4 weeks after initiation of the culture.
Northern blotting
Each RNA sample was prepared from 1.0 × 107 CMCs by the lithium chloride–urea method.28 Northern blot analysis was performed using mMCP-2,29mMCP-4,30 mMCP-5,31 mMCP-6,32 Gr B,12 KIT,33 TPH,12 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH)34cDNAs labeled with α-[32P]–deoxycytidine triphosphate (dCTP; 10 mCi/mL [370 MBq]; DuPont/NEN Research Products, Boston, MA) by random oligonucleotide priming. After hybridization at 42°C, blots were washed to a final stringency of 0.2 × SSC (1 × SSC is 150 mM NaCl and 15 mM trisodium citrate, pH 7.4) and subjected to autoradiography.
Immunoblotting
The whole-cell extracts of B6-+/+, B6-mi/mi, WB-+/+, and WB-mi/mi CMCs were obtained by the method described previously.16 The extract was suspended in loading buffer, boiled, and analyzed by immunoblot with anti-KIT polyclonal antibody (Santa Cruz Biotech, Santa Cruz, CA) and antitubulin antibody (Santa Cruz Biotech).
MTT assay
To examine the response to recombinant mouse (rm) KitL, an MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma, St Louis, MO) rapid calorimetric assay was carried out. Triplicate aliquots of 3 × 104 CMCs suspended in 100 μL α-MEM supplemented with 10% FCS were cultured in 96-well microtitered plates for 48 hours at 37°C in the presence or absence of various concentrations of rmKitL. MTT (10 μL of 5 mg/mL solution of MTT in phosphate-buffered saline [PBS]) was added to all wells and incubated for 4 hours at 37°C. Acid 2-propanol was added and mixed thoroughly to dissolve the dark blue crystals. The optical density (OD) was then measured on a Micro–enzyme-linked immunosorbent assay plate reader (Corona Electric, Ibaragi, Japan) with a test wavelength of 540 nm and a reference wavelength of 620 nm.
Linkage analysis
Genomic DNA was obtained from the tail of (F1× B6)–mi/mi mice by phenol extraction.35Genotyping for each (F1 × B6)–mi/mi mouse was carried out by polymerase chain reaction (PCR) using genomic DNA as a template, and we determined whether each microsatellite locus was homozygous (B6/B6) or heterozygous (B6/WB). PCR primer sets were purchased from Invitrogen (Carlsbad, CA). Genotyping data on each microsatellite marker were subjected to χ2 analysis.
Sequence analysis of the coding region of KitL
To examine the sequence of the coding region of KitL, PCR was carried out with Pyrobest Taq DNA polymerase (Takara, Kyoto, Japan). The cDNAs of skin of 3 B6-mi/mi mice and 3 WB-mi/mi mice were used as a template. The primers for PCR were 5′-GCTGCCTGGGCTGGATCGCAGCGCTG and 5′-CTGTTACCAGCCACTGTGCGAAGGTAA. The amplified fragment containing the coding region of KitL was cloned into EcoRV site of pBluescript (Stratagene, La Jolla, CA), and the sequence was analyzed with ABI 3100 sequencer (Applied BioSystems, Foster City, CA). The sequence was verified in both directions.
Sequence analysis of introns of KitL gene
The introns 5 and 6 of the KitL gene were amplified by long and accurate (LA)–PCR using the genomic DNA obtained from the tail of 2 B6-+/+ mice and 2 WB-+/+ mice. Intron 5 was amplified using the following primers: 5′-TGGTGGCATCTGACACTAGTGA corresponding to the sequence of exon 5 and 5′-GCTACTGCTGTCATTCCTAAGG corresponding to the sequence of exon 6. Intron 6 was amplified using the following primers: 5′-CCAGAGTCAGTGTCACAAAACC corresponding to the sequence of exon 6 and 5′-CTTCCAGTATAAGGCTCCAAAAGC corresponding to the sequence of exon 7. The amplified fragment was cloned into theEcoRV site of pBluescript, and the sequence was analyzed with the ABI 3100 sequencer.
Semiquantitative reverse transcription (RT)–PCR analysis
Four micrograms of total RNAs was extracted from the dorsal skin of 3 B6-mi/mi and 3 WB-mi/mi mice. The extracted RNAs were subjected to reverse transcription by Superscript (Invitrogen), and the single-strand cDNAs were obtained. One microliter, 0.1 μL, or 0.01 μL of the reaction mixture was added to 25 μL of PCR mixture containing 1.25 units of Taq DNA polymerase (Roche Diagnostics, Mannheim, Germany) and 25 pmol of each of the primers. PCR was carried out to amplify the fragment between exons 7 and 9 of the KitL gene using the primers 5′-AAGACTCGGGCCTACAATGGACAGCCATGG corresponding to the sequence of exon 7 and 5′-CAATGTTGATACGTCCACAATTAC corresponding to the sequence of exon 9.
RT-PCR Southern blotting
The cDNA used for RT-PCR Southern blotting was obtained as described in “Semiquantitative reverse transcription (RT)–PCR analysis.” PCR was carried out to amplify the fragment between exons 5 and 7 of the KitL gene using the primers 5′-TGGTGGCATCTGACACTAGTGA corresponding to the sequence of exon 5 and 5′-CTTCCAGTATAAGGCTCCAAAAGC corresponding to the sequence of exon 7. Fifteen cycles of PCR were performed, and the resulting amplified fragment was electrophoresed in 4% agarose gel. The blotted membrane was hybridized at 67°C with α-[32P]–labeled KitL fragment corresponding to the sequence between exons 5 and 7.
RNase protection assay
The pBluescript containing the coding region of KitL was linearized by SpeI. The α-[32P]–labeled antisense riboprobe was synthesized from the linearized plasmid using T3 RNA polymerase (Invitrogen). RNase protection assay was carried out with an RPA III Kit (Ambion, Austin, TX) according to the manufacturer's instructions. The intensity of the protected band for KL-1 mRNA and that of KL-2 mRNA was quantified using NIH Image (http://rsb.info.nih.gov/nih-image/).
Results
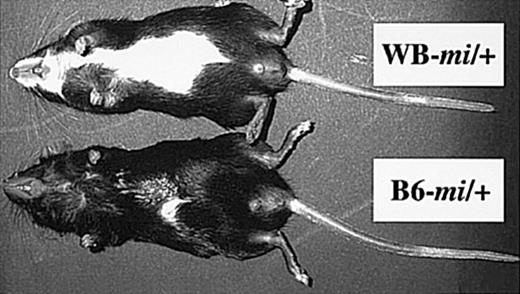
The appearance of WB-mi/mi mice was comparable with that of B6-mi/mi mice: white coat color, microphthalmia, and failure of eruption of incisors. On the other hand, the appearance of WB-mi/+ mice was different from that of B6-mi/+ mice (Figure 1). Abdominal white patches lacking melanocytes were larger in WB-mi/+ mice than in B6-mi/+ mice (Figure 1).
White patches in the abdomen in WB-mi/+ and B6-mi/+ mice.
The white patch was larger in WB-mi/+ mice than in B6-mi/+ mice.
White patches in the abdomen in WB-mi/+ and B6-mi/+ mice.
The white patch was larger in WB-mi/+ mice than in B6-mi/+ mice.
The number of skin mast cells of B6-mi/mi mice decreased to one third that of B6-+/+ mice, but that of WB-mi/mi mice was comparable with that of WB-+/+ mice (Table1). The number of skin mast cells was significantly higher in WB-+/+ mice than in B6-+/+ mice (Table 1). As a result, the number of skin mast cells observed in WB-mi/mimice was higher than the number in B6-+/+ mice, showing the critical effect of genetic backgrounds on the number of skin mast cells.
Effect of mi/mi genotype and strain difference on the number of skin mast cells
| Genotype of mice . | Genetic background . | ||
|---|---|---|---|
| B6 . | WB . | WBB6F1 . | |
| +/+ | 169.5 ± 7.6 (n = 10) | 272.0 ± 9.2 (n = 13)* | 276.6 ± 5.6 (n = 52)* |
| mi/mi | 51.8 ± 2.9 (n = 5)† | 295.2 ± 9.6 (n = 8) | 261.1 ± 6.1 (n = 43) |
| Genotype of mice . | Genetic background . | ||
|---|---|---|---|
| B6 . | WB . | WBB6F1 . | |
| +/+ | 169.5 ± 7.6 (n = 10) | 272.0 ± 9.2 (n = 13)* | 276.6 ± 5.6 (n = 52)* |
| mi/mi | 51.8 ± 2.9 (n = 5)† | 295.2 ± 9.6 (n = 8) | 261.1 ± 6.1 (n = 43) |
P < .01 when compared with the value of B6-+/+ mice by t test.
P < .01 when compared with the value of +/+ mice of the same genetic background by t test.
The expression of genes whose transcription had been shown to be deficient in B6-mi/mi CMCs was compared between WB-mi/mi CMCs and WB-+/+ CMCs by Northern blotting (Figure2). The examined genes were divided into 2 groups: genes showing the normal expression in WB-mi/miCMCs and genes showing the deficient expression in WB-mi/miCMCs, as in the case of B6-mi/mi CMCs. The mMCP-2, mMCP-4, mMCP-5, and Gr B genes belonged to the former group, and the mMCP-6, KIT, and TPH genes belonged to the latter group (Figure 2).
Expression of various genes in CMCs derived from B6-+/+, B6-mi/mi, WB-+/+, and WB-mi/mi mice.
The blot was hybridized with 32P-labeled cDNA probe of mMCP-2, mMCP-4, mMCP-5, Gr B, mMCP-6, KIT, TPH, and GAPDH. Three independent experiments were performed, and comparable results were obtained. A representative experiment is shown.
Expression of various genes in CMCs derived from B6-+/+, B6-mi/mi, WB-+/+, and WB-mi/mi mice.
The blot was hybridized with 32P-labeled cDNA probe of mMCP-2, mMCP-4, mMCP-5, Gr B, mMCP-6, KIT, TPH, and GAPDH. Three independent experiments were performed, and comparable results were obtained. A representative experiment is shown.
Immunoblot demonstrated that the amount of KIT protein was reduced in WB-mi/mi CMCs, and its level was comparable with that of B6-mi/mi CMCs (Figure 3A). When examined with MTT assay, the response to rmKitL was comparable between WB-mi/mi CMCs and B6-mi/mi CMCs (Figure3B).
Immunoblot of KIT and response to rmKitL in CMCs derived from B6-+/+, B6-mi/mi, WB-+/+, and WB-mi/mimice.
(A) Whole cell extracts of various CMCs were immnoblotted with anti-KIT antibody. Three independent experiments were performed, and a representative result is shown. (B) The response of CMCs to rmKitL was examined by MTT assay. The bars represent the means ± standard errors (SEs) of 3 independent experiments. In some cases, the SEs were too small to be shown by bars.
Immunoblot of KIT and response to rmKitL in CMCs derived from B6-+/+, B6-mi/mi, WB-+/+, and WB-mi/mimice.
(A) Whole cell extracts of various CMCs were immnoblotted with anti-KIT antibody. Three independent experiments were performed, and a representative result is shown. (B) The response of CMCs to rmKitL was examined by MTT assay. The bars represent the means ± standard errors (SEs) of 3 independent experiments. In some cases, the SEs were too small to be shown by bars.
It was possible that the decrease of skin mast cells in mice ofmi/mi genotype might be attributable to the reduced expression of KIT.4 However, the number of skin mast cells was normal despite the reduced expression of KIT in WB-mi/mimice. This suggested that factor(s) compensating for the reduced expression of KIT may be present in WB-mi/mi mice but not in B6-mi/mi mice.
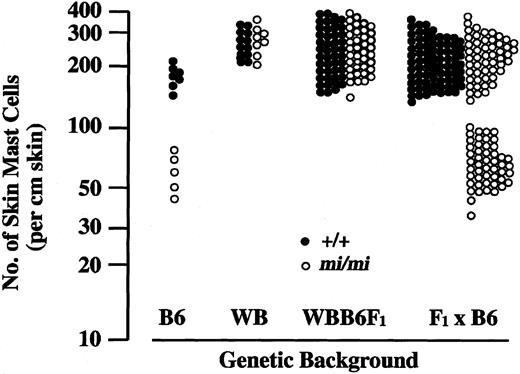
We examined the number of skin mast cells in WBB6F1-mi/mi mice. The number of skin mast cells did not decrease in WBB6F1-mi/mi mice compared with that of WBB6F1-+/+ mice, suggesting that the nonreduction of skin mast cells observed in WB-mi/mi mice was inherited dominantly (Table 1). Next, WBB6F1-mi/+ mice were backcrossed to B6-mi/+ mice, and (F1 × B6)–mi/mi mice were obtained. The number of skin mast cells in (F1 × B6)–mi/mi mice was bimodal (Figure 4). Of 131 (F1× B6)–mi/mi mice, 73 mice showed the high number of skin mast cells (more than 140 per cm skin) that was comparable with the value observed in WB-mi/mi mice, whereas 58 mice showed the low number of skin mast cells (fewer than 100 per cm skin) that was comparable with the value observed in B6-mi/mi mice. We calculated the ratio of mice with the high number of mast cells to mice with the low number of mast cells and examined whether the ratio fitted with the ratio predicted for a single-dominant inheritance. The observed ratio fitted well with the predicted ratio (χ2value, 1.72; P = .19).
Number of skin mast cells in B6, WB, WBB6F1, and (F1 × B6) mice of +/+ or mi/migenotype.
Each point represents the value of an individual mouse.
Number of skin mast cells in B6, WB, WBB6F1, and (F1 × B6) mice of +/+ or mi/migenotype.
Each point represents the value of an individual mouse.
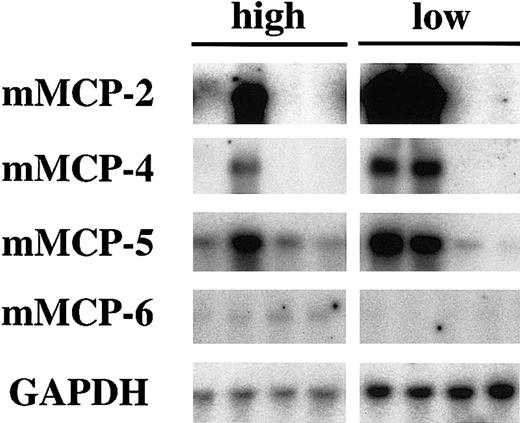
The expression levels of mMCP-2, mMCP-4, and mMCP-5genes did not decrease in WB-mi/mi CMCs (Figure 2). We examined whether the factor that maintained the skin mast cell number in WB-mi/mi mice correlated with the high expression levels of mMCP-2, mMCP-4, and mMCP-5 genes. We obtained CMCs from 4 (F1 × B6)–mi/mi mice with high numbers of skin mast cells and 4 (F1 × B6)–mi/mi mice with low numbers of skin mast cells. One of 4 (F1 × B6)–mi/mi mice with high numbers of skin mast cells showed high expression levels of mMCP-2, mMCP-4, and mMCP-5 genes, but the other 3 mice showed low expression levels (Figure 5). Two of 4 (F1 × B6)–mi/mi mice with low numbers of skin mast cells showed high expression levels of mMCP-2, mMCP-4, and mMCP-5 genes, but the other 2 mice showed low expression levels (Figure 5). The numbers of skin mast cells in (F1 × B6)–mi/mi mice did not correlate with the expression levels of mMCP-2, mMCP-4, andmMCP-5 genes.
Expression of genes in CMCs derived from (F1 × B6)– mi/mi mice with high or low numbers of skin mast cells.
CMCs were obtained from 4 (F1 × B6)–mi/mimice with high number of skin mast cells (indicated as high in the figure) and from 4 (F1 × B6)–mi/mi mice with low numbers of skin mast cells (indicated as low). The blot was hybridized with 32P-labeled cDNA probe of mMCP-2, mMCP-4, mMCP-5, mMCP-6, and GAPDH.
Expression of genes in CMCs derived from (F1 × B6)– mi/mi mice with high or low numbers of skin mast cells.
CMCs were obtained from 4 (F1 × B6)–mi/mimice with high number of skin mast cells (indicated as high in the figure) and from 4 (F1 × B6)–mi/mi mice with low numbers of skin mast cells (indicated as low). The blot was hybridized with 32P-labeled cDNA probe of mMCP-2, mMCP-4, mMCP-5, mMCP-6, and GAPDH.
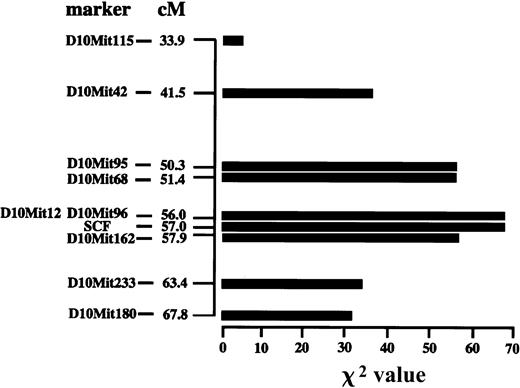
Linkage analysis was carried out using the aforementioned 131 (F1 × B6)–mi/mi mice to map the locus encoding the factor compensating for the reduced expression level of KIT. We selected 70 microsatellite loci that showed apparent differences between B6 and WB strains. The (F1 × B6)–mi/mi mice were typed to be homozygous (B6/B6) or heterozygous (B6/WB) for each microsatellite locus. The ratio of B6/B6 to B6/WB mice was subjected to χ2 analysis. The highest χ2 value of 65.7 was obtained for 2 markers—D10Mit12 and D10Mit96—that are located at 56.0 cM from the centromere of chromosome 10 (Figure 6). Sixty-one markers on 18 other chromosomes did not show significantly different levels of χ2 (less than 8.0; data not shown). No informative marker was found on chromosome 13.
Linkage analysis for the locus encoding a factor that maintained the number of skin mast cells in WB-mi/mimice.
The x-axis represents the scores of χ2 analyses. The y-axis represents the distance of each microsatellite marker from the centromere. The χ2 values against the KitL locus are also shown.
Linkage analysis for the locus encoding a factor that maintained the number of skin mast cells in WB-mi/mimice.
The x-axis represents the scores of χ2 analyses. The y-axis represents the distance of each microsatellite marker from the centromere. The χ2 values against the KitL locus are also shown.
Among the loci reported around the position with the highest χ2 value (56.0 cM), the KitL locus was the only one that may be related to the number of mast cells. The KitL locus was mapped at 57.0 cM from the centromere (http://www.ncbi.nlm.nih.gov/LocusLink/LocRpt.cgi?l=17 311). To retype 131 (F1 × B6)–mi/mi mice against the KitL locus, we searched segments of KitL gene that may be used as a marker. First, the coding region of KitL gene was compared between B6 and WB mice. A single nucleotide alteration resulting in an amino acid change was found; the alteration from G to T caused the change of alanine (B6) to serine (WB) at codon 207 (Figure7). Because this alteration of the coding region was not suitable for the marker, we next compared intron sequences. Three nucleotides altered in intron 5 (data not shown). Four nucleotides altered in intron 6 (Figure8A). In addition, we found a deletion of 42 bases in the intron 6 of WB mice but not in that of B6 mice (Figure8A). Using primers encompassing the deletion as a marker (Figure 8B), we retyped 131 (F1 × B6)–mi/mi mice. The χ2 value of the KitL locus was 65.7; the value was comparable with the value observed for D10Mit12 and D10Mit 96 microsatellite markers (Figure 6).
Comparison of amino acid sequence of KitL between B6 and WB strains.
The altered amino acid (A in B6 and S in WB at aa 207) is boxed.
Comparison of amino acid sequence of KitL between B6 and WB strains.
The altered amino acid (A in B6 and S in WB at aa 207) is boxed.
Comparison of nucleotide sequence of intron 6 of KitL gene between B6 and WB strains and usefulness of the deletion observed in the WB strain as a marker.
(A) The sequence of 450 nucleotides of 3′-splice site (−450 to −1) of intron 6. The altered nucleotides and the deleted part in the WB strain are boxed. The deleted nucleotides in WB strain are shown as dashes. The sequence of the primers used in typing of the KitL locus is indicated by arrows. These sequence data are available from GenBank under accession nos. AF516326 and AF516327. (B) Typing of the KitL locus. Using primers encompassing the deletion as a marker (indicated in panel A), the KitL locus was typed as of either B6/B6 or B6/WB. The band obtained from WB-mi/mi mice is also shown (WB/WB).
Comparison of nucleotide sequence of intron 6 of KitL gene between B6 and WB strains and usefulness of the deletion observed in the WB strain as a marker.
(A) The sequence of 450 nucleotides of 3′-splice site (−450 to −1) of intron 6. The altered nucleotides and the deleted part in the WB strain are boxed. The deleted nucleotides in WB strain are shown as dashes. The sequence of the primers used in typing of the KitL locus is indicated by arrows. These sequence data are available from GenBank under accession nos. AF516326 and AF516327. (B) Typing of the KitL locus. Using primers encompassing the deletion as a marker (indicated in panel A), the KitL locus was typed as of either B6/B6 or B6/WB. The band obtained from WB-mi/mi mice is also shown (WB/WB).
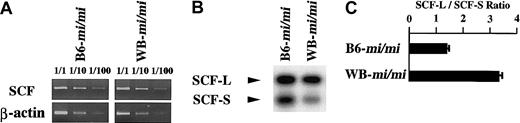
We examined the expression level of KitL mRNA in skin tissues of B6-mi/mi and WB-mi/mi mice by semiquantitative RT-PCR. The expression level was comparable between B6-mi/miand WB-mi/mi mice (Figure 9A). Two isoforms of KitL are known: KL-1 and KL-2.36,37 KL-1 but not KL-2 contains the fragment derived from exon 6 of KitL gene.36,37 Because Besmer and his colleagues reported that KL-1 but not KL-2 was important for development of the skin mast cells,38 we examined the ratio of KL-1 mRNA to KL-2 mRNA (KL-1/KL-2 ratio) in skin tissues. To distinguish KL-1 from KL-2, primers encompassing exon 6 were used in the RT-PCR Southern analysis. The intensity of the band for KL-1 mRNA was comparable with that of KL-2 mRNA in the skin tissues of B6-mi/mi mice (Figure 9B). In contrast, the intensity of the band for KL-1 mRNA was larger than that of KL-2 mRNA in the skin tissue of WB-mi/mi mice (Figure 9A). The KL-1/KL-2 ratio was approximately 3-fold higher in the skin of WB-mi/mi mice than in the skin of B6-mi/mi mice (Figure 9C). Similar results were obtained when the KL-1/KL-2 ratio was compared between B6-+/+ and WB-+/+ mice (data not shown).
Expression level of KitL mRNA and KL-1/KL-2 ratio in skin tissues of B6-mi/mi and WB-mi/mimice.
(A) Semiquantitative RT-PCR to compare the expression level of KitL mRNA. RNAs extracted from skin tissues of B6-mi/mi and WB-mi/mi mice were subjected to RT-PCR. (B) RT-PCR Southern analysis to examine KL-1/KL-2 ratio. (C) KL-1/KL-2 ratio. The intensity of the bands for KL-1 and KL-2 mRNAs was quantified by densitometry, and the ratio was calculated. The bars represent the means ± SEs of 3 independent experiments.
Expression level of KitL mRNA and KL-1/KL-2 ratio in skin tissues of B6-mi/mi and WB-mi/mimice.
(A) Semiquantitative RT-PCR to compare the expression level of KitL mRNA. RNAs extracted from skin tissues of B6-mi/mi and WB-mi/mi mice were subjected to RT-PCR. (B) RT-PCR Southern analysis to examine KL-1/KL-2 ratio. (C) KL-1/KL-2 ratio. The intensity of the bands for KL-1 and KL-2 mRNAs was quantified by densitometry, and the ratio was calculated. The bars represent the means ± SEs of 3 independent experiments.
The KL-1/KL-2 ratio was also evaluated using RNase protection assay. The KL-1/KL-2 ratio was higher in the skin of WB-mi/mi mice than in the skin of B6-mi/mi mice (Table2).
KL-1/KL-2 ratio in skin tissues of B6-mi/mi and WB-mi/mi mice evaluated by RNase protection assay
| . | KL-1/KL-2 ratio of each mouse . | |
|---|---|---|
| Experiment* . | B6-mi/mi . | WB-mi/mi . |
| No. 1 | 0.45 | 1.12 |
| No. 2 | 0.79 | 1.31 |
| . | KL-1/KL-2 ratio of each mouse . | |
|---|---|---|
| Experiment* . | B6-mi/mi . | WB-mi/mi . |
| No. 1 | 0.45 | 1.12 |
| No. 2 | 0.79 | 1.31 |
In 2 individual experiments, the intensity of the protected band for KL-1 mRNA and that of KL-2 mRNA was quantified, and the KL-1/KL-2 ratio was calculated.
Discussion
The B6-mi/mi mice are the most widely used homozygous mutant of mi locus, and the decrease of mast cells in the skin tissue of B6-mi/mi mice was first described by Stechschulte et al.3 Because the skin is the easiest tissue to examine the number of mast cells, mice of themi/mi genotype have been considered to be a mutant with mast cell deficiency. However, the present result showed that the genetic background influenced the number of mast cells in mice of themi/mi genotype. When compared with the wild-type (+/+) mice of the same genetic background, the number of skin mast cells decreased significantly in B6-mi/mi mice but not in WB-mi/mi mice. This is consistent with the old result of Stevens and Loutit.39 They reported that the number of skin mast cells did not decrease in G-mi/mi and (G × CBA) F1-mi/mi mice.
The tissues examined also influenced the comparative numbers of mast cells. In contrast to skin tissue, mast cells did decrease in the gastrointestinal canal and spleen of G-mi/mi and (G × CBA) F1-mi/mi mice.39 Stechschulte et al3 also reported the marked decrease of mast cells in the peritoneal cavity of B6-mi/mi mice. These results were consistent with our unpublished result that practically no mast cells were detectable in the gastrointestinal canal, spleen, and peritoneal cavity of both WB-mi/mi mice and B6-mi/mi mice (T.J. et al, unpublished data, 2002). Mast cell numbers in tissues other than skin did not appear to be influenced by the genetic backgrounds of mi/mi mice.
The WB strain was originally produced by Russell et al to prolong the longevity of the mice with the W/Wgenotype.40,41 In the cross of B6-W/+ males and females, only 4%, rather than the expected 25%, of W/Windividuals were born alive, and the average survival period was 2 days.40 In contrast, when WB-W/+ mice were crossed together, a full 25% live-born W/W offspring was obtained, and the average survival period was 10 days.40This suggested that factor(s) compensating the W/W mutant genotype was present in WB strain. Now the mutant W allele is known to encode KIT without the extracellular domain.42This was consistent with the present study that the factor compensating the reduced expression of KIT in mice with the mi/migenotype was present in WB strain but not in B6 strain.
By linkage analysis, the factor compensating the reduced expression of KIT was mapped near the KitL locus on chromosome 10. There is a possibility that KitL itself is the factor. Between the B6 and WB strains, an amino acid alteration was detected at codon 207 of KitL: alanine in the B6 strain and serine in the WB strain. However, this alteration may not be important, because codon 207 varies among species: serine in rats,43 pigs,44cows,45 and dogs46; proline in human beings47 and cats48; and aspargine in chicken.49 Besides the alteration in the coding region, the ratio of alternatively spliced KitL mRNAs (KL-1/KL-2 ratio) was different between B6 and WB strains.
There are 2 isoforms of KitL protein: membrane-bound form and soluble form.36 37 The latter is produced by proteolytic cleavage from the former. Because the protein translated from KL-2 mRNA lacks the major proteolytic cleavage site, soluble KitL is produced more efficiently from KL-1 mRNA than from KL-2 mRNA. The KL-1/KL-2 ratio was higher in the WB strain than in the B6 strain. The production level of soluble KitL might be higher in the WB strain than in the B6 strain. In fact, the number of skin mast cells was significantly higher in WB-+/+ mice than in B6-+/+ mice.
The number of skin mast cells decreased in the KL-2 gene knock-in mice, in which no KL-1 is produced.38 The number of skin mast cells increased in the transgenic mice of theKL-1 gene but not in the transgenic mice of theKL-2 gene.50 These results suggested that the soluble KitL is critical for the production of normal numbers of skin mast cells.
The development of melanocytes depends on MITF.51 The abdominal white patches lacking melanocytes were larger in WB-mi/+ mice than in B6-mi/+ mice, suggesting that melanocyte progenitors may migrate more easily in the skin of B6-mi/+ mice than in the skin of WB-mi/+ mice. Kunisada et al reported that the increase of melanocytes was observed more markedly in transgenic mice of the KL-2 gene than in transgenic mice of the KL-1 gene.50 This suggested that, in contrast to development of mast cells in the skin, membrane-bound KitL might be more important for the development of melanocytes than soluble KitL. Because the KL-1/KL-2 ratio was lower in B6 strain than in the WB strain, the skin of B6-mi/+ mice may be more suitable for the development of melanocytes than the skin of WB-mi/+ mice.
There is another possibility that the factor compensating for the reduced expression of KIT is not related to KitL. Rhim et al mapped a modifier gene of endothelin receptor B gene near the KitL locus.52 Homozygous mice with mutant endothelin receptor B gene (Ednrbs/Ednrbsmice) show abnormal pigmentation patterns. The genetic background of the mutants affected the pigmentation pattern. C3H-Ednrbs/Ednrbs mice have white patches on the forehead and abdomen, whereas Mayer-Ednrbs/Ednrbs mice have white patches on the back and abdomen. Although the pigmentation patterns ofEdnrbs/Ednrbs mice are influenced by multiple modifier genes, one of them was mapped near the KitL locus. This modifier gene of the Ednrb locus may be consistent with the locus mapped in the present study. On the other hand, either the mutation of Ednrb gene or that of MITF gene causes a similar abnormality (Waardenburg syndrome 453 and Waardenburg syndrome 2, respectively54). The modifier gene of endothelin receptor B and that of KIT might be common.
Taken together, the mutation compensating for the reduced expression of KIT in WB-mi/mi mice was mapped at the position very closely located to the KitL locus. Although the different splicing pattern of KitL between the B6 and WB strains was a plausible explanation, more analysis will be needed to identify such a mutation.
The authors thank Dr H. Hiai for helpful discussions and Ms C. Murakami and Ms T. Sawamura for technical assistance.
Prepublished online as Blood First Edition Paper, October 17, 2002; DOI 10.1182/blood-2002-07-2213.
Supported by grants from the Ministry of Education, Culture, Sports, Science and Technology and from Senri Life Science Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Yukihiko Kitamura, Department of Pathology, Rm C2, Osaka University Medical School, Yamada-oka 2-2, Suita, 565-0871, Japan; e-mail:kitamura@patho.med.osaka-u.ac.jp.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal