Abstract
To identify genes linked to normal plasma cell (PC) differentiation and to classify multiple myeloma (MM) with respect to the expression patterns of these genes, we analyzed global mRNA expression in CD19-enriched B cells (BCs) from 7 tonsils, CD138-enriched PCs from 11 tonsils, 31 normal bone marrow samples, and 74 MM bone marrow samples using microarrays interrogating 6800 genes. Hierarchical clustering analyses with 3288 genes clearly segregated the 4 cell types, and chi-square and Wilcoxin rank sum tests (P < .0005) identified 359 and 500 previously defined and novel genes that distinguish tonsil BCs from tonsil PCs (early differentiation genes [EDGs]), and tonsil PCs from bone marrow PCs (late differentiation genes [LDGs]), respectively. MM as a whole was found to have dramatically variable expression of EDGs and LDGs, and one-way analysis of variance (ANOVA) was used to identify the most variable EDGs (vEDGs) and LDGs (v1LDG and v2LDG). Hierarchical cluster analysis with these genes revealed that previously defined MM gene expression subgroups (MM1-MM4) could be linked to one of the 3 normal cell types. Clustering with 30 vEDGs revealed that 13 of 18 MM4 cases clustered with tonsil BCs (P = .000 05), whereas 14 of 15 MM3 cases clustered with tonsil PCs when using 50 v1LDG (P = .000 008), and 14 of 20 MM2 cases clustered with bone marrow PCs when using 50 v2LDG (P = .000 09). MM1 showed no significant linkage with normal cell types studied. Thus, genes whose expression is linked to distinct transitions in late-stage B-cell differentiation can be used to classify MM.
Introduction
Although many of the steps in B-cell (BC) development have been elucidated, the final stages of plasma cell (PC) differentiation are not well understood. PCs generated during primary humoral immune responses begin their differentiation in the light zones of the germinal centers of lymph nodes or in the red pulp of spleen and have a life span of only a few days.1-3 During secondary humoral immune responses, long-lived PCs migrate from the secondary lymphoid tissues into the bone marrow or into the lamina propria of the mucosa where they survive and secrete large amounts of immunoglobulin for at least 3 weeks.4,5 Despite morphologic and functional similarities, PCs isolated from different organs exhibit distinct differences. For example, life spans of tonsil- and bone marrow–derived PCs are different1,6,7 and differences in somatic hypermutation of complementarity-determining regions (CDRs) of IGV genes is evident.8,9 Although most of these observations have been derived from rodent models, accumulating evidence suggests that human PCs follow a similar progressive developmental process.10-17 Recent studies, using alterations in the expression of a panel of CD markers and the transcription factors BSAP and PRDI-BF1, showed that human PCs follow a gradient of increasing maturity in the direction of tonsil to peripheral blood to bone marrow.18
PC differentiation is marked by the loss or down-regulation of several molecules including major histocompatibility complex (MHC) class II, CD19, CD20, CD22, CD44, CD45, as well as transcription factors CIITA19 and BSAP/Pax-5.20-23 On the other hand, PCs turn on or up-regulate the transcription factors PRDI-BF1 (Blimp-1)24,25 and MUM1/IRF4,26 the rough endoplasmic reticulum-associated antigen Vs38c, and cell-surface molecules CD138 and CD38.27 Recently, XBP-1 has been identified as the only transcription factor known to be required for the terminal differentiation of PCs.28 For a comprehensive examination of the molecular events in PC development, see Calame's review.27
Multiple myeloma (MM) is a tumor of terminally differentiated PCs that home to and expand in the bone marrow.29 Although MM appears to originate in a postgerminal center cell, as suggested by the presence of somatic hypermutation,30,31 much speculation exists concerning the exact cell in which this malignant transformation occurs. The hypoproliferative nature of MM, with labeling indexes in the clonal PCs rarely exceeding 1%,32 has led to the hypothesis that MM is a tumor arising from a transformed precursor cell that proliferates and differentiates giving rise to the clonal expansion of terminally differentiated PCs. Indeed, the bone marrow of patients with multiple myeloma contains BC populations at different stages of differentiation that are clonally related to the malignant PCs.33 Corradini and colleagues have shown that bone marrow BCs, transcribing the MM PC-derived VDJ gene joined to immunoglobulin M (IgM) sequence in IgG- and IgA-secreting MM, can exist.34 Other investigations have shown that the clonogenic cell in MM originates from a preswitched but somatically mutated BC that lacks intraclonal variation.30,31Detection of a high frequency of circulating BCs that share clonotypic Ig heavy-chain VDJ rearrangements with MM PCs using single-cell in situ reverse transcriptase–polymerase chain reaction (RT-PCR) has furthered this hypothesis.35
Microarray analysis of global gene expression patterns has become a powerful means of identifying clinical subgroups of hematopoietic neoplasms.36-42 Alizadeh et al used gene expression profiling to identify distinct clinical entities of diffuse large B-cell lymphoma (DLBCL) related to either pregerminal or postgerminal center B cells with the pregerminal center–like group having a poorer clinical course.37 The conclusions of these studies were exquisitely dependent on the ability to put DLBCL gene expression in the context of cells representing different stages of normal BC differentiation.
Here we show that comparative microarray profiling of CD19-enriched tonsil BCs with CD138-enriched PCs from tonsil and bone marrow allowed the identification of previously defined and novel genes differentially expressed during late stage BC development. Hierarchical clustering of MM and normal samples using these genes revealed that subsets of MM could be linked to the 3 normal cell types studied. These MM clusters were found to be consistent with previously defined unsupervised gene expression defined subgroups, such that MM4, MM3, and MM2 have tonsil BC–like, tonsil PC–like, or bone marrow PC–like expression features, respectively.
Materials and methods
Cell isolation and analysis
Tonsils were obtained from patients undergoing tonsillectomy for chronic tonsillitis. Tonsil tissues were minced, softly teased, and filtered. The mononuclear cell fraction from tonsil preparations and bone marrow aspirates was separated by a standard Ficoll-Hypaque gradient (Pharmacia Biotech, Piscataway, NJ). The cells in the light density fraction (specific gravity [SG] ∼ 1.077) were resuspended in cell culture media and 10% fetal bovine serum, red blood cell (RBC) lysed, and several phosphate-buffered saline (PBS) wash steps were performed. Tonsil and bone marrow PC enrichment was performed using directly conjugated monoclonal mouse anti-CD138 microbeads (Miltenyi Biotec, Auburn, CA) and an immunomagnetic bead selection process as previously described.42 B-lymphocyte isolation was performed using directly conjugated monoclonal mouse anti–human CD19 microbeads (Miltenyi-Biotec). All samples were obtained with appropriate consent according to University of Arkansas for Medical Sciences guidelines.
Cytospin preparations were prepared following microbead enrichment and cells fixed and stained using DiffQuick (Dade Diagnostics, Aguada, Puerto Rico). Both CD19- and CD138-enriched cells were subjected to immunofluorescence microscopy for cytoplasmic immunoglobulin light chain (cIg) expression. The analysis was performed essentially as described.43 Briefly, cytospin preparations of cells were fixed in 100% ethanol and stained with 100 μL of a 1:20 dilution of AMCA (7-amino-4-methylcourmarin-3-acitic acid) conjugated goat anti–human-kappa immunoglobulin light chain (Vector Laboratories, Burlingame, CA), washed 2 times in 1 × PBD (1 × PBS + 0.1% NP-40), then 100 μL of a 1:100 dilution of fluorescein isothiocyanate (FITC)–conjugated goat anti–human-lambda immunoglobulin light chain (Vector Laboratories) washed and stained with propidium iodide at 0.1 μg/mL in 1 × PBS for 5 minutes, washed in 1 × PBD, and antifade added (Molecular Probes, Eugene, OR). Cells were visualized using an Olympus BX60 epifluorescence microscope (Olympus, Melville, NY) equipped with appropriate filters.
Fluorescence-activated cell sorting (FACS) analysis was performed on unpurified mononuclear cells and CD19-enriched or CD138-enriched cells using FITC-labeled CD20, phycoerythrin (PE)–labeled CD38, FITC- or PE + Texas Red–labeled CD45, PE- or PE + Cy5–labeled CD138, and isotype-matched control G1 antibodies (Beckman Coulter, Miami, FL). For detection of CD138 after CD138 microbead enrichment, we employed an indirect detection strategy using an FITC-labeled rabbit anti–mouse IgG antibody (Beckman Coulter). Cells were taken after Ficoll Hypaque gradient or after microbead enrichment, washed in PBS, and stained at 4°C with antibodies. After staining, cells were resuspended in 1 × PBS and analyzed using an Epics XL-MCL flow cytometry system (Beckman Coulter).
RNA purification and microarray hybridization and analysis
Detailed protocols for RNA purification, cDNA synthesis, cRNA preparation, and hybridization to the Affymetrix HuGeneFL GeneChip microarray have been described.42 The HuGenFL gene expression data for all samples described in this study can be obtained from our web site (http://lambertlab.uams.edu).
Gene expression data analysis
Hierarchical clustering of average linkage clustering with the centered correlation metric was employed.44 A total of 3228 genes were scanned across 7 cases each of tonsil BCs, tonsil PCs, bone marrow PCs, and MM PCs. The 3228 genes were derived from 6800 by filtering out all control genes, all genes with absent absolute calls, and genes not fulfilling the test of Max-Min greater than 3.0 (3.0 being the natural log of the average difference call).
Gene expression profiles of 7 CD19-enriched BCs were compared with those from 11 CD138-enriched tonsil PC samples. The genes differentiating these 2 groups were defined as early differentiation genes (EDGs). Gene expression profiles of the same 11 tonsil PC samples were compared with those of 31 CD138-enriched bone marrow PCs. The most significantly differentially expressed genes in this comparison were defined as late differentiation genes (LDGs). The first test applied was a chi-square test (χ2) of the absolute call (an Affymetrix algorithm-based parameter of whether the gene is absent or present). In each comparison, genes with χ2 values more than 3.84 (P < .05) or having “present” absolute calls in more than half of the samples in each group were retained. In this way, 2662 and 2549 genes discriminated between the tonsil BCs and tonsil PCs and tonsil PCs and bone marrow PCs, respectively.
To compare gene expression levels, the nonparametric Wilcoxin rank sum (WRS) test (P < .0005) was applied to natural log transformed average difference call (an Affymetrix algorithm-based quantitative measure of gene expression). In this analysis, 496 and 646 discriminated between tonsil BCs and tonsil PCs and tonsil PCs and bone marrow PCs, respectively. By combining the χ2 and WRS data, 359 EDGs and 500 LDGs were identified. To define whether expression changes were up or down in one group compared with the other, the nonparametric Spearman correlation test of the average difference call was employed.
To classify MM with respect to EDGs and LDGs, 74 newly diagnosed cases of MM and the tonsil BC, tonsil PC, and bone marrow PC samples were tested for variance across the 359 EDGs and 500 LDGs using a one-way analysis of variance (ANOVA) test. The top 50 EDGs showing the most significant variance across all samples were defined as variable EDGs (vEDGs); likewise, the top 50 LDGs showing the most significant variance were defined as variable 1 LDGs (v1LDG). Subtracting the v1LDGs from the 500 LDGs and then applying one-way ANOVA to the remaining genes was used to identify variable 2 LDGs (v2LDG). Hierarchical clustering was applied to all samples using the vEDGs, v1LDGs, and v2LDGs. Of the 50 vEDGs, 20 were left out of the clustering analysis because these genes generally showed no variability across the MM sample group and thus could not be used to distinguish MM subgroups. These genes were filtered out by applying the Max-Min greater than 2.5 test.
Results
Cell analysis of CD19-enriched tonsil BCs and CD138-enriched tonsil and bone marrow PCs
FACS analysis of the tonsil preparations before CD19 microbead enrichment showed that approximately 70% of the cells had a CD20hi/CD38lo immunophenotype (Figure1A). After anti-CD19 immunomagnetic bead enrichment, the CD20hi/CD38lo cells were enriched to 98% (Figure 1B) and were essentially void of CD138+ cells. A mean of 95% (SD 3%) of the cells in the 7 tonsil-derived CD19-enriched samples used for gene expression profiling had a CD20hi/CD38+/locell surface phenotype. Morphologic analysis (Figure 1B, top right) and FACS suggested that the CD19-enriched cells consisted of a combination of follicular mantle and subepithelial B lymphocytes (CD38lo, small cells) and germinal center centroblasts and centrocytes (CD38+, larger cells). Immunofluoresence staining for cIg light chain expression revealed that CD19-enriched fractions contained less than 5% cIg staining cells, indicating only a minor contamination with PCs (Figure1B, bottom right).
Cell analyses of representative normal samples before and after immunomagnetic bead enrichment.
2-color dot plots or histogram plot of FACS analysis of the tonsil mononuclear fractions prior to CD19 selection (A), tonsil mononuclear fractions prior to CD138 selection (C), and bone marrow mononuclear cells prior to CD138 selection (E). 2-color dot plots or histogram plot of dual antibody FACS analysis of cell surface phenotype, light microscopy of morphology, and immunofluoresence microscopy of cytoplasmic immunoglobulin light chain expression in CD19-enriched tonsil BCs (B), CD138-enriched tonsil PC (D), and CD138-enriched bone marrow PCs (F). Antibodies used in individual FACS analyses are indicated to the left and bottom of each FACS plot. The percentage of cells in each gate window is indicated. Original magnification, × 60. Stains are described in “Materials and methods.”
Cell analyses of representative normal samples before and after immunomagnetic bead enrichment.
2-color dot plots or histogram plot of FACS analysis of the tonsil mononuclear fractions prior to CD19 selection (A), tonsil mononuclear fractions prior to CD138 selection (C), and bone marrow mononuclear cells prior to CD138 selection (E). 2-color dot plots or histogram plot of dual antibody FACS analysis of cell surface phenotype, light microscopy of morphology, and immunofluoresence microscopy of cytoplasmic immunoglobulin light chain expression in CD19-enriched tonsil BCs (B), CD138-enriched tonsil PC (D), and CD138-enriched bone marrow PCs (F). Antibodies used in individual FACS analyses are indicated to the left and bottom of each FACS plot. The percentage of cells in each gate window is indicated. Original magnification, × 60. Stains are described in “Materials and methods.”
FACS for the cells with a PC phenotype in the tonsil mononuclear fractions revealed that CD38hi/CD45−cells represented between 1% and 2% and CD138hi/CD45− cells represented less than 1% (Figure 1C). CD138 selection dramatically enriched cells with a PC immunophenotype (Figure 1D). A mean of 89% (SD 4%) of the cells in the 11 CD138-enriched tonsil samples had a CD38hi/CD45lo/− immunophenotype. Due to the nature of the selection process, CD138lo/−/CD38+ PCs were lost during CD138 enrichment. The CD138-selected cells had PC morphology with an increased cytoplasmic-to-nuclear ratio and prominent endoplasmic reticulum (Figure 1D, top right) and more than 90% were cIg-positive (Figure 1D, bottom right).
CD38/CD45 and CD138/CD45 dual-color FACS analysis of the bone marrow mononuclear cell samples from healthy donors revealed that between 0.5% and 2% of the population appeared to be PCs (Figure 1E). The percentage of CD38hi/CD45− cells in the preselected bone marrow mononuclear cells was less than that seen in the tonsil. FACS analysis after CD138 enrichment showed that more than 95% of the cells had a PC immunophenotype (Figure 1F). A mean of 92% (SD 4%) of the 31 normal bone marrow–derived and 95% (SD 3%) of the 74 MM bone marrow–derived CD138-enriched cells (data not shown) had a CD38hi/CD45lo/− cell surface phenotype. A comparison of the FACS analysis of the CD138-enriched cells from the tonsil and bone marrow revealed that the percentages of CD38+/CD45+ and CD38+/CD20+ cells were greatly reduced in the bone marrow cells compared with tonsil cells. As with the tonsil PC, the bone marrow CD138-enriched cells had PC morphology (Figure 1F, top right) and more than 95% were cIg positive (Figure 1F, bottom right).
Comparative gene expression profiling of CD19- and CD138-enriched B-cell populations
The expression patterns of approximately 6800 genes were determined for CD19-enriched tonsil BCs, CD138-enriched tonsil PCs, and bone marrow PCs using Affymetrix high-density oligonucleotide microarrays. The mean average difference call for a panel of CD markers and transcription factors known to change during PC development was compared across the 3 normal cell types (Table1). Genes for CD45,CD20, CD79B, CD52, CD19, CD22, CD83, and CD72showed high expression in tonsil BCs, intermediate levels in tonsil PCs, and low or absent expression in bone marrow PCs.CD21 showed no significant differences in the tonsil BC to tonsil PC comparison, but showed a significant reduction in bone marrow PCs. Conversely, CD138, CD38, and CD63were absent or weakly expressed on tonsil BCs, with intermediate levels on tonsil PCs, and high in bone marrow PCs. To our knowledge, this is the first indication that CD63 may be differentially regulated during PC differentiation. CD27 showed significant up-regulation in the comparison of tonsil BCs to tonsil PCs; however, the tonsil and bone marrow PCs showed no significant differences.
Microarray-derived expression levels of genes differentially expressed during PC development
| Accession no. . | Symbol . | TBCs . | TPCs . | BPCs . |
|---|---|---|---|---|
| Y00062 | CD45 | 11 495 ± 2 198 | 4 979 ± 2 522 | 1 385 ± 706 |
| M27394 | CD20 | 23 860 ± 5 494 | 3 799 ± 2 977 | 289 ± 358 |
| M89957 | CD79B | 14 758 ± 3 348 | 4 696 ± 2 440 | 1 243 ± 1 357 |
| X62466 | CD52 | 14 576 ± 2 395 | 4 348 ± 2 074 | 2 831 ± 1 002 |
| M84371 | CD19 | 12 339 ± 1 708 | 6 174 ± 1 345 | 2 852 ± 852 |
| M26004 | CD21 | 8 909 ± 1 640 | 5 434 ± 4 053 | 458 ± 140 |
| X59350 | CD22 | 10 349 ± 1 422 | 5 356 ± 1 610 | 1 929 ± 612 |
| Z11697 | CD83 | 9 201 ± 1 900 | 2 380 ± 1 087 | 392 ± 403 |
| M54992 | CD72 | 6 177 ± 1 620 | 865 ± 554 | 454 ± 548 |
| Z48199 | CD138 | 719 ± 519 | 9 935 ± 3545 | 24 643 ± 6 206 |
| D84276 | CD38 | 3 122 ± 967 | 9 833 ± 3 419 | 14 836 ± 3 462 |
| X62654 | CD63 | 2 310 ± 431 | 6 815 ± 1 582 | 16 878 ± 3 305 |
| M63928 | CD27 | 6 235 ± 1 736 | 15 937 ± 6 691 | 16 714 ± 4 442 |
| M31627 | XBP1 | 12 978 ± 1 676 | 54 912 ± 13 649 | 49 558 ± 10 798 |
| U52682 | IRF4 | 1 863 ± 630 | 8 422 ± 3 061 | 11 348 ± 3 118 |
| U00115 | BCL6 | 7 979 ± 1 610 | 3 303 ± 2 070 | 618 ± 335 |
| X74301 | CIITA | 1 553 ± 263 | 236 ± 217 | 113 ± 82 |
| U16031 | STAT6 | 1 314 ± 512 | 386 ± 335 | 191 ± 187 |
| S76617 | BLK | 3 654 ± 1 551 | 388 ± 592 | 95 ± 86 |
| X68149 | CXCR5 | 3 381 ± 1 173 | 183 ± 299 | 92 ± 183 |
| U29680 | BCL2A1 | 3 290 ± 1 073 | 1 121 ± 817 | 483 ± 209 |
| L00058 | MYC | 1 528 ± 474 | 348 ± 239 | 2 103 ± 903 |
| L06797 | CXCR4 | 11 911 ± 2 093 | 6 673 ± 3 508 | 18 033 ± 5 331 |
| Accession no. . | Symbol . | TBCs . | TPCs . | BPCs . |
|---|---|---|---|---|
| Y00062 | CD45 | 11 495 ± 2 198 | 4 979 ± 2 522 | 1 385 ± 706 |
| M27394 | CD20 | 23 860 ± 5 494 | 3 799 ± 2 977 | 289 ± 358 |
| M89957 | CD79B | 14 758 ± 3 348 | 4 696 ± 2 440 | 1 243 ± 1 357 |
| X62466 | CD52 | 14 576 ± 2 395 | 4 348 ± 2 074 | 2 831 ± 1 002 |
| M84371 | CD19 | 12 339 ± 1 708 | 6 174 ± 1 345 | 2 852 ± 852 |
| M26004 | CD21 | 8 909 ± 1 640 | 5 434 ± 4 053 | 458 ± 140 |
| X59350 | CD22 | 10 349 ± 1 422 | 5 356 ± 1 610 | 1 929 ± 612 |
| Z11697 | CD83 | 9 201 ± 1 900 | 2 380 ± 1 087 | 392 ± 403 |
| M54992 | CD72 | 6 177 ± 1 620 | 865 ± 554 | 454 ± 548 |
| Z48199 | CD138 | 719 ± 519 | 9 935 ± 3545 | 24 643 ± 6 206 |
| D84276 | CD38 | 3 122 ± 967 | 9 833 ± 3 419 | 14 836 ± 3 462 |
| X62654 | CD63 | 2 310 ± 431 | 6 815 ± 1 582 | 16 878 ± 3 305 |
| M63928 | CD27 | 6 235 ± 1 736 | 15 937 ± 6 691 | 16 714 ± 4 442 |
| M31627 | XBP1 | 12 978 ± 1 676 | 54 912 ± 13 649 | 49 558 ± 10 798 |
| U52682 | IRF4 | 1 863 ± 630 | 8 422 ± 3 061 | 11 348 ± 3 118 |
| U00115 | BCL6 | 7 979 ± 1 610 | 3 303 ± 2 070 | 618 ± 335 |
| X74301 | CIITA | 1 553 ± 263 | 236 ± 217 | 113 ± 82 |
| U16031 | STAT6 | 1 314 ± 512 | 386 ± 335 | 191 ± 187 |
| S76617 | BLK | 3 654 ± 1 551 | 388 ± 592 | 95 ± 86 |
| X68149 | CXCR5 | 3 381 ± 1 173 | 183 ± 299 | 92 ± 183 |
| U29680 | BCL2A1 | 3 290 ± 1 073 | 1 121 ± 817 | 483 ± 209 |
| L00058 | MYC | 1 528 ± 474 | 348 ± 239 | 2 103 ± 903 |
| L06797 | CXCR4 | 11 911 ± 2 093 | 6 673 ± 3 508 | 18 033 ± 5 331 |
Accession numbers are GenBank accession numbers for the Affymetrix probe set. Symbols are Human Genome Organization (HUGO)–approved gene symbols for the Affymetrix probe set. The numbers in the columns under the tonsil BC (TBC), tonsil PC (TPC), and bone marrow PC (BPC) headings represent the mean of the average difference call plus or minus the standard deviation for the given gene. Differences in expression across comparisons were significant (P < .01) unless the data cell is shaded.
Expression of the transcription factor IRF4 was significantly elevated in tonsil PCs compared with tonsil BCs and was higher in bone marrow PCs than in tonsil PCs. XBP1 showed a more than 4-fold increase in expression in the comparison of tonsil BCs to tonsil PCs, but no significant difference between tonsil and bone marrow PCs. On the other hand, CTIIA, STAT6, andBLK (a direct target of BSAP orPAX5,20-23) and the BCL2 homologueBCL2A1 were down-regulated in the tonsil BC to tonsil PC transition, whereas BCL6 was down-regulated in the tonsil PC to bone marrow PC transition. Although not present on the HuGeneFL GeneChip, recent studies using the U95Av2 GeneChip, have revealed that Blimp-1 (PRDM1) expression is significantly elevated in both tonsil and bone marrow PCs compared with tonsil BCs (our unpublished data, May 2002). Interestingly, whereasMYC showed significant down-regulation in the tonsil BC to tonsil PC transition, it was reactivated in bone marrow PCs to levels higher than those in the tonsil BCs. Whereas the chemokine receptorsCXCR4 and CXCR5 showed down-regulation in the tonsil BC to tonsil PC transition, CXCR4, likeMYC, was reactivated in bone marrow PCs.
We next used χ2 and WRS analysis to identify 359 and 500 genes whose mRNA expression was significantly different (P < .000 05) in comparisons of tonsil BCs to tonsil PCs and tonsil PCs to bone marrow PCs, respectively. Genes that were significantly differentially expressed in the tonsil BC to tonsil PC transition were referred to as EDGs and those differentially expressed in the tonsil PC to bone marrow PC transition were referred as LDGs. A total of 235 of the 359 EDGs (65%) and 43 of the top 50 EDGs (86%) were down-regulated in tonsil PCs compared with tonsil BCs. The 50 most significantly differentially expressed EDGs are listed in Table2. The largest group of genes in the top 50 EDGs encode transcription factors. Of 16 transcription factors, only 3 (XBP1, IRF4, and BMI1), were up-regulated. There were 4 ets domain–containing proteins (ETS1, SPIB, SPI1, and ELF1) that were found to be down-regulated EDGs. Other transcription factors included the repressors EED and ID3, as well as the activatorsRUNX3, ICSBP1, REL, ERG3, and FOXM1. MYC and CIITA were also among the down-regulated transcription factors. The IRF family member, interferon consensus sequence binding protein, ICSBP1, which is a lymphoid-specific negative regulator, was the only gene that was expressed at a 3-plus level in tonsil BCs and shut down in tonsil PCs (and bone marrow PCs). Genes coding for proteins involved in signaling represented the second-most abundant class of EDGs.CASP10 represented the only signaling protein up-regulated in tonsil PCs. GBP1, the Rho family members ARHGand ARHH, and the proto-oncogene HRAS were down-regulated GTP-binding EDGs. There were two members of the tumor necrosis factor family, TNF and lymphotoxin beta(LTB), as well as the TNF receptor binding proteinTRAF5, that were identified. The IL4R and the cxc receptor, CXCR5 (BLR1), represented the only receptors in the list. MNK1 and the B lymphocyte–restricted kinaseBLK (direct target of BSAP) were the only kinases in the top 50 EDGs. Adhesion molecules ITGA6 andPECAM1 were up-regulated EDGs and represented the only adhesion genes in the EDG class. These genes also showed an up-regulation in the comparison of tonsil PC to bone marrow PC, andPECAM1 was also identified in the top 50 LDGs (Table3). Other multiple-member classes of down-regulated EDGs included cell cycle (CCNF, CCNG2, and CDC20) or DNA repair/maintenance (TERF2, LIG1, MSH2, RPA1) genes.
Early differentiation genes: top 50 differentially expressed genes in comparison of CD19-enriched tonsil BCs and CD138-enriched tonsil PCs
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | ||
|---|---|---|---|---|---|
| TBCs . | TPCs . | BPCs . | |||
| U60519 | CASP10 | Apoptosis | − | + | ++ |
| X53586 | ITGA6 | Adhesion | − | + | ++ |
| U04735 | STCH | Chaperone | + | ++ | ++ |
| L13689 | BMI1 | Transcription; repressor; PcG | + | ++ | +++ |
| L34657 | PECAM1 | Adhesion | + | ++ | +++ |
| U52682 | IRF4 | Transcription; IRF family | + | +++ | +++ |
| M31627 | XBP1 | Transcription; bZip family | +++ | ++++ | ++++ |
| AB000410 | OGG1 | DNA glycosylase | + | − | − |
| D87432 | SLC7A6 | Solute transporter | + | − | − |
| J04101 | ETS1 | Transcription; etsfamily | + | − | − |
| L38820 | CDID | Immunity | + | − | − |
| M28827 | CDIC | Immunity | + | − | − |
| M55542 | GBP1 | Signaling; GTP binding | + | − | − |
| M81182 | ABCD3 | ABC transporter | + | − | − |
| M85085 | CSTF2 | mRNA cleavage stimulating factor | + | − | − |
| U74612 | FOXM1 | Transcription; fork-head family | + | − | − |
| U84720 | RAE1 | RNA export | + | − | − |
| V00574 | HRAS | Signaling; GTP-binding protein | + | − | − |
| X02910 | TNF | Signaling; TNFα | + | − | − |
| X63741 | EGR3 | Transcription;egr family | + | − | − |
| X93512 | TERF2 | Telomere repeat binding protein | + | − | − |
| Z36714 | CCNF | Cell cycle; cyclin F | + | − | − |
| AB000409 | MNK1 | Signaling; kinase | + | − | + |
| M33308 | VCL | Cytoskeleton | + | − | ++ |
| D16480 | HADHA | Mitochondrial oxidation | ++ | − | − |
| M63488 | RPA1 | DNA replication/repair | ++ | − | − |
| U03911 | MSH2 | DNA repair | ++ | − | − |
| U69108 | TRAF5 | Signaling; TNFR-associated protein | ++ | − | − |
| X12517 | SNRPC | mRNA splicing | ++ | − | − |
| X52056 | SPI1 | Transcription; ets family | ++ | − | − |
| X68149 | BLR1 | Signaling; cxc receptor | ++ | − | − |
| X74301 | CIITA | Transcription; adaptor | ++ | − | − |
| X75042 | REL | Transcription; rel/dorsal family | ++ | − | − |
| L00058 | MYC | Transcription; bHLHZip | ++ | − | ++ |
| M36067 | LIG1 | DNA ligase | ++ | + | + |
| M82882 | ELF1 | Transcription; ets family | ++ | + | + |
| S76617 | BLK | Signaling; kinase | ++ | + | + |
| U47414 | CCNG2 | Cell cycle; cyclin G | ++ | + | + |
| U61167 | SH3D1B | Unknown; SH3-containing protein | ++ | + | + |
| X61587 | ARHG | Signaling; Rho G | ++ | + | + |
| Z35278 | RUNX3 | Transcription; contains runt domain | ++ | + | + |
| U89922 | LTB | Signaling; TNF-c | ++++ | + | + |
| M34458 | LMNB1 | Cytoskeletal matrix | +++ | + | − |
| U90651 | EED | Transcription; repression; PcG | +++ | + | + |
| X69111 | ID3 | Transcription; repression; bHLH | +++ | + | + |
| X52425 | IL4R | Signaling; cytokine receptor | +++ | ++ | − |
| Z35227 | ARHH | Signaling; Rho H | +++ | ++ | + |
| U89922 | LTB | Signaling; TNF-c | ++++ | + | + |
| U05340 | CDC20 | Cell cycle; activator of APC | ++++ | ++ | − |
| X66079 | SPIB | Transcription;ets family | ++++ | ++ | − |
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | ||
|---|---|---|---|---|---|
| TBCs . | TPCs . | BPCs . | |||
| U60519 | CASP10 | Apoptosis | − | + | ++ |
| X53586 | ITGA6 | Adhesion | − | + | ++ |
| U04735 | STCH | Chaperone | + | ++ | ++ |
| L13689 | BMI1 | Transcription; repressor; PcG | + | ++ | +++ |
| L34657 | PECAM1 | Adhesion | + | ++ | +++ |
| U52682 | IRF4 | Transcription; IRF family | + | +++ | +++ |
| M31627 | XBP1 | Transcription; bZip family | +++ | ++++ | ++++ |
| AB000410 | OGG1 | DNA glycosylase | + | − | − |
| D87432 | SLC7A6 | Solute transporter | + | − | − |
| J04101 | ETS1 | Transcription; etsfamily | + | − | − |
| L38820 | CDID | Immunity | + | − | − |
| M28827 | CDIC | Immunity | + | − | − |
| M55542 | GBP1 | Signaling; GTP binding | + | − | − |
| M81182 | ABCD3 | ABC transporter | + | − | − |
| M85085 | CSTF2 | mRNA cleavage stimulating factor | + | − | − |
| U74612 | FOXM1 | Transcription; fork-head family | + | − | − |
| U84720 | RAE1 | RNA export | + | − | − |
| V00574 | HRAS | Signaling; GTP-binding protein | + | − | − |
| X02910 | TNF | Signaling; TNFα | + | − | − |
| X63741 | EGR3 | Transcription;egr family | + | − | − |
| X93512 | TERF2 | Telomere repeat binding protein | + | − | − |
| Z36714 | CCNF | Cell cycle; cyclin F | + | − | − |
| AB000409 | MNK1 | Signaling; kinase | + | − | + |
| M33308 | VCL | Cytoskeleton | + | − | ++ |
| D16480 | HADHA | Mitochondrial oxidation | ++ | − | − |
| M63488 | RPA1 | DNA replication/repair | ++ | − | − |
| U03911 | MSH2 | DNA repair | ++ | − | − |
| U69108 | TRAF5 | Signaling; TNFR-associated protein | ++ | − | − |
| X12517 | SNRPC | mRNA splicing | ++ | − | − |
| X52056 | SPI1 | Transcription; ets family | ++ | − | − |
| X68149 | BLR1 | Signaling; cxc receptor | ++ | − | − |
| X74301 | CIITA | Transcription; adaptor | ++ | − | − |
| X75042 | REL | Transcription; rel/dorsal family | ++ | − | − |
| L00058 | MYC | Transcription; bHLHZip | ++ | − | ++ |
| M36067 | LIG1 | DNA ligase | ++ | + | + |
| M82882 | ELF1 | Transcription; ets family | ++ | + | + |
| S76617 | BLK | Signaling; kinase | ++ | + | + |
| U47414 | CCNG2 | Cell cycle; cyclin G | ++ | + | + |
| U61167 | SH3D1B | Unknown; SH3-containing protein | ++ | + | + |
| X61587 | ARHG | Signaling; Rho G | ++ | + | + |
| Z35278 | RUNX3 | Transcription; contains runt domain | ++ | + | + |
| U89922 | LTB | Signaling; TNF-c | ++++ | + | + |
| M34458 | LMNB1 | Cytoskeletal matrix | +++ | + | − |
| U90651 | EED | Transcription; repression; PcG | +++ | + | + |
| X69111 | ID3 | Transcription; repression; bHLH | +++ | + | + |
| X52425 | IL4R | Signaling; cytokine receptor | +++ | ++ | − |
| Z35227 | ARHH | Signaling; Rho H | +++ | ++ | + |
| U89922 | LTB | Signaling; TNF-c | ++++ | + | + |
| U05340 | CDC20 | Cell cycle; activator of APC | ++++ | ++ | − |
| X66079 | SPIB | Transcription;ets family | ++++ | ++ | − |
Quantitative gene expression (QGE) was defined as being at 1 of 5 levels. A gene was “−” if all the samples in the group had an absent absolute call (AC), “+” when all samples had present AC and average difference call (AD) < 1000, “++” when all samples had present AC and AD between 1000 and 5000, “+++” when all samples had present AC and AD between 5000 and 10 000, and “++++” when all samples had present AC and AD > 10 000. Up-regulated LDGs (n = 7) in the tonsil BC to tonsil PC transition are listed first. A hierarchy in the listing of genes was established such that QGE (5 expression levels) increases from top to bottom and from left to right for up-regulated genes and increases from top to bottom and decreases from left to right for down-regulated genes. Although the expression patterns of bone marrow PCs were not used to derive the list of EDGs, the gene expression levels in these cells are provided for reference.
Accession numbers are GenBank accession numbers corresponding to the Affymetrix GeneChip probe set for the given gene. Symbols are HUGO-approved gene symbols. TBCs indicates CD19-enriched tonsil BCs; TPCs, CD138-enriched tonsil PCs; BPCs, CD138-enriched bone marrow PCs.
Late differentiation genes: top 50 differentially expressed genes in comparison of CD138-enriched tonsil PCs and CD138-enriched bone marrow PCs
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |
|---|---|---|---|---|
| TPCs . | BPCs . | |||
| U32114 | CAV2 | Signaling; membrane caveolae | − | + |
| U60115 | FHL1 | Signaling; LIM domain | − | + |
| U73936 | JAG1 | Signaling; Notch ligand | − | + |
| X57025 | IGF1 | Signaling; growth factor | − | + |
| Z32684 | XK | Membrane transport | − | + |
| D10511 | ACAT1 | Metabolism; ketone | − | ++ |
| Y08999 | ARPC1A | Actin polymerization | − | ++ |
| M14745 | BCL2 | Signaling; antiapoptosis | − | ++ |
| M24486 | P4HA1 | Collagen synthesis | − | ++ |
| M60315 | BMP6 | Signaling; TGF family | − | ++ |
| U25956 | SELPLG | Adhesion | − | ++ |
| X16983 | ITGA4 | Adhesion | − | ++ |
| Z18951 | CAV1 | Signaling; membrane caveolae | − | ++ |
| M60092 | AMPD1 | Metabolism; energy | − | +++ |
| U15932 | DUSP5 | Signaling; phosphatase | − | ++++ |
| U95626 | CCR2 | Signaling; chemokine receptor | − | ++++ |
| D78132 | RHEB2 | Signaling; ras homolog | + | ++ |
| L41887 | SFRS7 | mRNA splicing factor | + | ++ |
| M23161 | LOC90411a | Unknown | + | ++ |
| M37721 | PAM | Metabolism; hormone amidation | + | ++ |
| M69023 | TSPAN-3a | Unknown | + | ++ |
| U02556 | TCTE1L | Dynein homolog | + | ++ |
| U41060 | LIV-1a | Unknown | + | ++ |
| U44772 | PPT1 | Lysosome enzyme | + | ++ |
| U70660 | ATOX1 | Metabolism; antioxidant | + | ++ |
| X92493 | PIP5K1B | Signaling; kinase | + | ++ |
| M23254 | CAPN2 | Cysteine protease | + | +++ |
| J02763 | S100A6 | Signaling; calcium binding | ++ | +++ |
| L13689 | BMI1 | Transcription; repressor; PcG | ++ | +++ |
| L34657 | PECAM1 | Adhesion | ++ | +++ |
| M23294 | HEXB | Metabolism; hexoaminidase | ++ | +++ |
| M64098 | HLDBP | Metabolism; sterol | ++ | ++++ |
| U52101 | EMP3 | Adhesion | ++ | ++++ |
| X66087 | MYBL1 | Transcription; myb-like | + | − |
| X54942 | CKS2 | Cell cycle; kinase regulator | ++ | − |
| X73568 | SYK | Signaling; kinase | ++ | − |
| L08177 | EBI2 | Signaling; receptor | ++ | − |
| M25629 | KLK1 | Protease; serine | ++ | − |
| U00115 | BCL6 | Transcription; Zn-finger | ++ | − |
| U23852 | LCK | Signaling; kinase | ++ | − |
| U60975 | SORL1 | Endocytosis | ++ | − |
| X63380 | MEF2B | Transcription; MADs box | ++ | − |
| L25878 | EPXH1 | Metabolism; epoxide hydrolase | ++ | + |
| Z35227 | ARHH | Signaling; Rho C | ++ | + |
| X89986 | BIK | Signaling; apoptosis | +++ | − |
| M13792 | ADA | Metabolism; purine | +++ | + |
| U10485 | LRMP | ER membrane protein | +++ | + |
| M81601 | TCEA1 | Transcription; elongation | +++ | ++ |
| X70326 | MACMARCK | Actin binding | ++++ | + |
| X56494 | PKM2 | Metabolism; energy | ++++ | + |
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |
|---|---|---|---|---|
| TPCs . | BPCs . | |||
| U32114 | CAV2 | Signaling; membrane caveolae | − | + |
| U60115 | FHL1 | Signaling; LIM domain | − | + |
| U73936 | JAG1 | Signaling; Notch ligand | − | + |
| X57025 | IGF1 | Signaling; growth factor | − | + |
| Z32684 | XK | Membrane transport | − | + |
| D10511 | ACAT1 | Metabolism; ketone | − | ++ |
| Y08999 | ARPC1A | Actin polymerization | − | ++ |
| M14745 | BCL2 | Signaling; antiapoptosis | − | ++ |
| M24486 | P4HA1 | Collagen synthesis | − | ++ |
| M60315 | BMP6 | Signaling; TGF family | − | ++ |
| U25956 | SELPLG | Adhesion | − | ++ |
| X16983 | ITGA4 | Adhesion | − | ++ |
| Z18951 | CAV1 | Signaling; membrane caveolae | − | ++ |
| M60092 | AMPD1 | Metabolism; energy | − | +++ |
| U15932 | DUSP5 | Signaling; phosphatase | − | ++++ |
| U95626 | CCR2 | Signaling; chemokine receptor | − | ++++ |
| D78132 | RHEB2 | Signaling; ras homolog | + | ++ |
| L41887 | SFRS7 | mRNA splicing factor | + | ++ |
| M23161 | LOC90411a | Unknown | + | ++ |
| M37721 | PAM | Metabolism; hormone amidation | + | ++ |
| M69023 | TSPAN-3a | Unknown | + | ++ |
| U02556 | TCTE1L | Dynein homolog | + | ++ |
| U41060 | LIV-1a | Unknown | + | ++ |
| U44772 | PPT1 | Lysosome enzyme | + | ++ |
| U70660 | ATOX1 | Metabolism; antioxidant | + | ++ |
| X92493 | PIP5K1B | Signaling; kinase | + | ++ |
| M23254 | CAPN2 | Cysteine protease | + | +++ |
| J02763 | S100A6 | Signaling; calcium binding | ++ | +++ |
| L13689 | BMI1 | Transcription; repressor; PcG | ++ | +++ |
| L34657 | PECAM1 | Adhesion | ++ | +++ |
| M23294 | HEXB | Metabolism; hexoaminidase | ++ | +++ |
| M64098 | HLDBP | Metabolism; sterol | ++ | ++++ |
| U52101 | EMP3 | Adhesion | ++ | ++++ |
| X66087 | MYBL1 | Transcription; myb-like | + | − |
| X54942 | CKS2 | Cell cycle; kinase regulator | ++ | − |
| X73568 | SYK | Signaling; kinase | ++ | − |
| L08177 | EBI2 | Signaling; receptor | ++ | − |
| M25629 | KLK1 | Protease; serine | ++ | − |
| U00115 | BCL6 | Transcription; Zn-finger | ++ | − |
| U23852 | LCK | Signaling; kinase | ++ | − |
| U60975 | SORL1 | Endocytosis | ++ | − |
| X63380 | MEF2B | Transcription; MADs box | ++ | − |
| L25878 | EPXH1 | Metabolism; epoxide hydrolase | ++ | + |
| Z35227 | ARHH | Signaling; Rho C | ++ | + |
| X89986 | BIK | Signaling; apoptosis | +++ | − |
| M13792 | ADA | Metabolism; purine | +++ | + |
| U10485 | LRMP | ER membrane protein | +++ | + |
| M81601 | TCEA1 | Transcription; elongation | +++ | ++ |
| X70326 | MACMARCK | Actin binding | ++++ | + |
| X56494 | PKM2 | Metabolism; energy | ++++ | + |
Please see Table 2 note.
A total of 310 of 500 (62%) LDGs were up-regulated or turned on in the tonsil PC to bone marrow PC transition. This is in contrast to the EDG where a majority of the genes were turned off or down-regulated in the tonsil BC to tonsil PC transition. The 50 most significantly differentially expressed EDGs are listed in Table3. Although 16 EDGs were transcription factors, only 5 LDGs belonged to this class. The BMI1 gene, which was an up-regulated EDG, was also an up-regulated LDG, indicating that the gene undergoes a progressive increase in expression during differentiation. BMI1 was the only up-regulated transcription factor. The genes MYBL1, MEF2B, andBCL6 were shut down in bone marrow PCs and the transcription elongation factor TCEA1 was also down-regulated. The largest class of LDGs (n = 16; 11 up-regulated and 5 down-regulated) coded for proteins involved in signaling. The LIM-containing protein with both nuclear and focal adhesion localization, FHL1; and the secreted protein, JAG1, a ligand for Notch, IGF1; and BMP6 were up-regulated. The dual-specific phosphataseDUSP5 and the chemokine receptor CCR2 represented genes with the most dramatically altered expression and were turned on to extremely high levels in bone marrow PCs while absent in tonsil PCs. Additional up-regulated LDGs included CAV1 andCAV2, plasma membrane proteins important in transportation of materials and organizing numerous signal transduction pathways.45 There were 4 adhesion molecules (SELPG, ITGA4, PECAM1, and EMP3) up-regulated in bone marrow PCs. As seen in the EDGs, no LDG adhesion genes were down-regulated.ARHH, which was down-regulated in tonsil PCs, also showed a significant decrease in bone marrow PCs compared with tonsil PCs. The lymphocyte-specific kinases SYK and LCK were shut off in bone marrow PCs. Consistent with a role in regulating longevity of bone marrow PCs, the antiapoptotic BCL2 was up-regulated and the proapoptotic BIK was down-regulated. A lymphoid-restricted, integral endoplasmic reticulum membrane proteinLRMP (JAW1), was a down-regulated LDG, a finding consistent with previous studies showing down-regulation of this gene at the PC stage of BC development.46
Identification of genes with similar expression between MM and cells at different stages of B-cell development
To provide a comprehensive assessment of the distinctions between the samples under study, we performed a hierarchical cluster analysis with 3288 genes on 7 tonsil BC, 7 tonsil PC, 7 bone marrow PC, and 7 MM PC samples (Figure 2). As expected, this analysis revealed a major division between the CD19-enriched tonsil BC samples and all the CD138-enriched PC samples, with the exception of one tonsil PC sample being clustered with tonsil BCs. The CD138-enriched PC branch was further subdivided into 2 distinct subbranches, one containing the tonsil and bone marrow PCs and the other containing the MM PCs. The tonsil and bone marrow PCs were separated on separate subbranches.
A 2-dimensional hierarchical cluster analysis of 7 tonsil BC (TBC), 7 tonsil PC (TPC), 7 bone marrow PC (BPC), and 7 MM PC samples clustered based on the correlation of experimental expression profiles of 3288 probe sets.
The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are adjacent to each other. Experimental samples are arranged the same way along the horizontal axis; those with the most similar patterns of expression across all genes are adjacent to each other. Sample groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean.
A 2-dimensional hierarchical cluster analysis of 7 tonsil BC (TBC), 7 tonsil PC (TPC), 7 bone marrow PC (BPC), and 7 MM PC samples clustered based on the correlation of experimental expression profiles of 3288 probe sets.
The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are adjacent to each other. Experimental samples are arranged the same way along the horizontal axis; those with the most similar patterns of expression across all genes are adjacent to each other. Sample groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean.
Although hierarchical clustering with 3288 genes distinguished MM from the other normal tissues, we recognized that MM also exhibited a high degree of variability in expression of EDGs and LDGs, with some MM having tonsil BC– or tonsil PC–like patterns for these genes. Thus, to determine the extent of this variability and to see if it could be used to classify MM, a one-way ANOVA analysis of the EDGs and LDGs was performed across the normal cell types and MM. The 50 most-variable EDG (vEDGs) are listed in Table 4. This list consists of 18 up-regulated and 32 down-regulated EDGs that exhibit tonsil BC–like expression in all MM. The cyclin-dependent kinase 8 (CDK8), which was undetectable (“−,” absent absolute call) in tonsil BCs, was up-regulated to a “++++” level in both tonsil and bone marrow PCs, but was either absent or at a “+” or “+ +” level in all MM cases, representing one of the most dramatic examples of a vEDG. Of the 50 v1LDGs (Table 5), 34 were LDGs that exhibited up-regulation in the tonsil PC to bone marrow PC transition and had tonsil PC–like expression in all or a subset of MM. The remaining 16 v1LDGs had the reverse pattern. Unlike vEDGs, only 15 of the top 50 v1LDGs showed tonsil PC–like patterns in all MM. The cxc chemokines SDF1, PF4, and PPBPrepresented the most dramatic v1LDGs. Expression of these genes was undetectable in both tonsil PCs and MM PCs, yet these genes were expressed at 3-plus (SDF1) or 4-plus (PF4 and PPBP) in bone marrow PCs. Results forSDF1 were validated by the fact that 2 separate and distinct probe sets interrogating different regions of SDF1(accession numbers L36033 and U19495) showed identical patterns across the samples.
vEDGs: EDGs with similar expression patterns in tonsil BCs and all or subsets of MM
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |||
|---|---|---|---|---|---|---|
| TBCs . | TPCs . | BPCs . | MM . | |||
| D28364 | ANXA2 | Annexin family | − | + | + | −/+ |
| U81787 | WNT10B | Signaling; ligand | − | + | ++ | −/++ |
| U88898 | LOC51581a | Unknown | − | + | + | −/+ |
| X12451 | CTSL | Protease; cysteine | − | ++ | ++ | − |
| Z25347 | CDK8 | Cell cycle; kinase | − | ++++ | ++++ | −/++ |
| D38548 | KIAA0076a | Unknown | + | ++ | ++ | +/++ |
| D86479 | AEBP1 | Extracellular matrix | + | ++ | ++ | + |
| U04689 | OR1D2 | Signaling; receptor | + | ++ | + | + |
| M31328 | GNB3 | Signaling; G protein | + | ++ | ++ | + |
| U13395 | WWOX | Metabolism; oxidoreductase | + | ++ | ++ | + |
| X14675 | BCR | Signaling; GTPase for RAC | + | ++ | ++ | + |
| X16665 | HOXB2 | Transcription; homeobox domain | + | ++ | ++ | −/+ |
| Z11899 | POU5F1 | Transcription; homeobox domain | + | ++ | ++ | + |
| Z36531 | FGL2 | Secreted fibrinogenlike | + | ++ | ++ | + |
| X80907 | PIK3R2 | Signaling; kinase adaptor | + | +++ | +++ | ++ |
| D31846 | AQP2 | Aquaporin | ++ | +++ | +++ | ++ |
| L18983 | PTPRN | Phosphatase; membrane | ++ | ++++ | ++++ | ++ |
| M23323 | CD3E | Signaling; T-cell receptor partner | ++ | ++++ | ++++ | ++ |
| D83781 | K1AA0197a | Unknown | + | − | − | + |
| HT4824 | CBS | Metabolism; cystathionine-beta-synthase | + | − | − | −/++ |
| S78873 | RABIF | Signaling; GTP-releasing factor | + | − | − | +/++ |
| U32645 | ELF4 | Transcription;ets domain | + | − | − | −/+ |
| X97630 | EMK1 | Signaling; kinase; ELK domain | + | − | − | + |
| Z24724 | UNKNOWNa | Cell cycle | + | − | − | +/++ |
| D16480 | HADHA | Mitochondrial oxidation | ++ | − | − | −/++ |
| L77701 | COX17 | Mitochondrial oxidation | ++ | − | − | −/++ |
| M90356 | BTF3L2 | Transcription; NAC domain | ++ | − | − | ++ |
| U08815 | SF3A3 | Spliceosome | ++ | − | − | +/++ |
| U53225 | SNX1 | Intracellular trafficking | ++ | − | − | +/++ |
| M25753 | CCNB1 | Cell cycle | ++ | + | − | −/++ |
| D87448 | TOPBP1a | Topoisomerase II binding protein | ++ | + | + | +/++ |
| L38810 | PSMC5 | 26S proteasome subunit 5 | ++ | + | + | ++ |
| M29551 | PPP3CB | Signaling; calcium-dependent phosphatase | ++ | + | + | +/++ |
| M32886 | SRI | Signaling; calcium binding | ++ | + | + | ++ |
| U24704 | PSMD4 | 26S proteasome subunit 4 | ++ | + | + | ++ |
| U25165 | FXR1 | RNA binding protein | ++ | + | + | ++ |
| U37022 | CDK4 | Cell cycle; kinase | ++ | + | + | ++ |
| U53003 | C21orf33 | Unknown; highly conserved | ++ | + | + | ++ |
| X89985 | BCL7B | Actin crosslinking | ++ | + | + | ++ |
| D49738 | CKAP1 | Tubulin folding | +++ | + | + | ++ |
| D43950 | CCT5 | Chaperonin | +++ | ++ | ++ | +++ |
| D82348 | ATIC | Metabolism; purine biosynthesis | +++ | ++ | ++ | +++ |
| D86550 | DYRK1A | Signaling; kinase | +++ | ++ | ++ | +++ |
| L06132 | VDAC1 | Anion channel | +++ | ++ | ++ | ++/+++ |
| L43631 | SAFB | Nuclear scaffold factor | +++ | ++ | ++ | ++/+++ |
| M30448 | CSNK2B | Signaling; casein kinase regulation | +++ | ++ | ++ | ++/++++ |
| X76013 | QARS | Metabolism; glutaminyl tRNA synthetase | +++ | ++ | ++ | ++/+++ |
| D83735 | CNN2 | Actin binding | ++++ | ++ | ++ | ++/++++ |
| M86667 | NAP1L1 | Nucleosome assembly | ++++ | ++ | ++ | +++ |
| X04828 | GNAI2 | Signaling; G protein | ++++ | ++ | ++ | ++/+++ |
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |||
|---|---|---|---|---|---|---|
| TBCs . | TPCs . | BPCs . | MM . | |||
| D28364 | ANXA2 | Annexin family | − | + | + | −/+ |
| U81787 | WNT10B | Signaling; ligand | − | + | ++ | −/++ |
| U88898 | LOC51581a | Unknown | − | + | + | −/+ |
| X12451 | CTSL | Protease; cysteine | − | ++ | ++ | − |
| Z25347 | CDK8 | Cell cycle; kinase | − | ++++ | ++++ | −/++ |
| D38548 | KIAA0076a | Unknown | + | ++ | ++ | +/++ |
| D86479 | AEBP1 | Extracellular matrix | + | ++ | ++ | + |
| U04689 | OR1D2 | Signaling; receptor | + | ++ | + | + |
| M31328 | GNB3 | Signaling; G protein | + | ++ | ++ | + |
| U13395 | WWOX | Metabolism; oxidoreductase | + | ++ | ++ | + |
| X14675 | BCR | Signaling; GTPase for RAC | + | ++ | ++ | + |
| X16665 | HOXB2 | Transcription; homeobox domain | + | ++ | ++ | −/+ |
| Z11899 | POU5F1 | Transcription; homeobox domain | + | ++ | ++ | + |
| Z36531 | FGL2 | Secreted fibrinogenlike | + | ++ | ++ | + |
| X80907 | PIK3R2 | Signaling; kinase adaptor | + | +++ | +++ | ++ |
| D31846 | AQP2 | Aquaporin | ++ | +++ | +++ | ++ |
| L18983 | PTPRN | Phosphatase; membrane | ++ | ++++ | ++++ | ++ |
| M23323 | CD3E | Signaling; T-cell receptor partner | ++ | ++++ | ++++ | ++ |
| D83781 | K1AA0197a | Unknown | + | − | − | + |
| HT4824 | CBS | Metabolism; cystathionine-beta-synthase | + | − | − | −/++ |
| S78873 | RABIF | Signaling; GTP-releasing factor | + | − | − | +/++ |
| U32645 | ELF4 | Transcription;ets domain | + | − | − | −/+ |
| X97630 | EMK1 | Signaling; kinase; ELK domain | + | − | − | + |
| Z24724 | UNKNOWNa | Cell cycle | + | − | − | +/++ |
| D16480 | HADHA | Mitochondrial oxidation | ++ | − | − | −/++ |
| L77701 | COX17 | Mitochondrial oxidation | ++ | − | − | −/++ |
| M90356 | BTF3L2 | Transcription; NAC domain | ++ | − | − | ++ |
| U08815 | SF3A3 | Spliceosome | ++ | − | − | +/++ |
| U53225 | SNX1 | Intracellular trafficking | ++ | − | − | +/++ |
| M25753 | CCNB1 | Cell cycle | ++ | + | − | −/++ |
| D87448 | TOPBP1a | Topoisomerase II binding protein | ++ | + | + | +/++ |
| L38810 | PSMC5 | 26S proteasome subunit 5 | ++ | + | + | ++ |
| M29551 | PPP3CB | Signaling; calcium-dependent phosphatase | ++ | + | + | +/++ |
| M32886 | SRI | Signaling; calcium binding | ++ | + | + | ++ |
| U24704 | PSMD4 | 26S proteasome subunit 4 | ++ | + | + | ++ |
| U25165 | FXR1 | RNA binding protein | ++ | + | + | ++ |
| U37022 | CDK4 | Cell cycle; kinase | ++ | + | + | ++ |
| U53003 | C21orf33 | Unknown; highly conserved | ++ | + | + | ++ |
| X89985 | BCL7B | Actin crosslinking | ++ | + | + | ++ |
| D49738 | CKAP1 | Tubulin folding | +++ | + | + | ++ |
| D43950 | CCT5 | Chaperonin | +++ | ++ | ++ | +++ |
| D82348 | ATIC | Metabolism; purine biosynthesis | +++ | ++ | ++ | +++ |
| D86550 | DYRK1A | Signaling; kinase | +++ | ++ | ++ | +++ |
| L06132 | VDAC1 | Anion channel | +++ | ++ | ++ | ++/+++ |
| L43631 | SAFB | Nuclear scaffold factor | +++ | ++ | ++ | ++/+++ |
| M30448 | CSNK2B | Signaling; casein kinase regulation | +++ | ++ | ++ | ++/++++ |
| X76013 | QARS | Metabolism; glutaminyl tRNA synthetase | +++ | ++ | ++ | ++/+++ |
| D83735 | CNN2 | Actin binding | ++++ | ++ | ++ | ++/++++ |
| M86667 | NAP1L1 | Nucleosome assembly | ++++ | ++ | ++ | +++ |
| X04828 | GNAI2 | Signaling; G protein | ++++ | ++ | ++ | ++/+++ |
Please see Table 2 note.
The range of QGE levels for genes with variable expression in MM are given as lowest level/highest level.
v1LDGs: LDGs showing similar expression patterns in tonsil PCs and all or subsets of MM
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | ||
|---|---|---|---|---|---|
| TPCs . | BPCs . | MM . | |||
| U90902 | 23612a | Unknown; related to TIAM1 | − | + | −/++ |
| D12775 | AMPD3 | Metabolism; AMP deaminase | − | + | −/++ |
| U37546 | BIRC3 | Signaling; antiapoptosis | − | ++ | −/++ |
| Z11793 | SEPP1 | Metabolism; selenium transport | − | +++ | −/+ |
| L36033 | SDF1 | Signaling; cxc chemokine | − | +++ | − |
| U19495 | SDF1 | Signaling; cxc chemokine | − | +++ | − |
| M27891 | CST3 | Protease inhibitor | − | ++++ | −/++++ |
| M26602 | DEFA1 | Immunity | − | ++++ | −/++++ |
| M25897 | PF4 | Signaling; cxc chemokine | − | ++++ | − |
| M54995 | PPBP | Signaling; cxc chemokine | − | ++++ | − |
| U79288 | KIAA0513a | Unknown | + | ++ | +/++ |
| M59465 | TNFAIP1 | Signaling; antiapoptosis | + | ++ | +/++++ |
| X53586 | ITGA6 | Adhesion | + | ++ | +/++ |
| D50663 | TCTEL1 | Dynein light chain | + | ++ | +/++ |
| U40846 | NAGLU | Metabolism; heparan sulfate degradation | + | ++ | +/++ |
| M80563 | S100A4 | Signaling; calcium binding | + | ++ | +/++++ |
| X04085 | CAT | Metabolism; catalase | + | ++ | +/++ |
| L02648 | TCN2 | Metabolism; vitamin B12 transport | + | ++ | + |
| L35249 | ATP6B2 | Lysosome; vacuolar proton pump | + | ++ | + |
| L09209 | APLP2 | Amyloid beta precursor like | + | ++ | + |
| L41870 | RB1 | Cell cycle | + | ++ | +/++ |
| X76732 | NUCB2 | Signaling; calcium binding | + | +++ | +/+++ |
| D29805 | 5B4GALT1 | Adhesion | + | +++ | + |
| M29877 | FUCA1 | Lysosome; fucosidase | + | +++ | +/++ |
| M32304 | TIMP2 | Metalloproteinase 2 inhibitor | + | +++ | +/++++ |
| D10522 | MACS | Actin crosslinking | + | ++++ | −/++ |
| L38696 | RALYa | RNA binding | ++ | +++ | ++ |
| U05875 | IFNGR2 | Signaling; interferon gamma receptor | ++ | +++ | ++/+++ |
| U78095 | SPINT2 | Protease inhibitor; blocks HGF | ++ | +++ | −/+++ |
| L13977 | PRCP | Lysosomal; angiotensinase C | ++ | ++++ | ++ |
| U12255 | FCGRT | IgG Fc receptor | ++ | ++++ | −/+++ |
| L06797 | CXCR4 | Signaling; SDF1 receptor | ++ | ++++ | ++/++++ |
| D82061 | FABGL | Metabolism | ++ | ++++ | ++/+++ |
| Y00433 | GPX1 | Oxidation protection | +++ | ++++ | ++/+++ |
| M60752 | H2AFA | Histone; nucleosome | + | − | −/++ |
| U18300 | DDB2 | DNA repair | + | − | + |
| X63692 | DNMT1 | DNA methyltransferase | + | − | + |
| D11327 | PTPN7 | Signaling; phosphatase | ++ | − | ++ |
| X54942 | CKS2 | Cell cycle; kinase regulator | ++ | − | +/+++ |
| D14874 | ADM | Adrenomedullin | ++ | + | +/++++ |
| D86976 | KIAA0223a | Minor histocompatability antigen | ++ | + | +/+++ |
| X52979 | SNRPB | mRNA splicing | ++ | + | +/++ |
| Z49254 | MRPL23 | Mitochondrial ribosomal protein | ++ | + | ++ |
| U66464 | HPK1 | Signaling; kinase | ++ | + | +/++ |
| U91903 | FRZB | Signaling; WNT antagonists | ++ | + | +/++ |
| D87453 | MRPS27 | Mitochondrial ribosomal protein | ++ | + | +/++ |
| X59932 | CSK | Signaling; kinase | +++ | ++ | ++ |
| L17131 | HMG1Y | Transcription; high mobility group | ++++ | + | ++/++++ |
| L19779 | H2AFO | Histone; nucleosome | ++++ | ++ | ++++ |
| U70439 | SSP29a | Unknown | ++++ | +++ | +++/++++ |
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | ||
|---|---|---|---|---|---|
| TPCs . | BPCs . | MM . | |||
| U90902 | 23612a | Unknown; related to TIAM1 | − | + | −/++ |
| D12775 | AMPD3 | Metabolism; AMP deaminase | − | + | −/++ |
| U37546 | BIRC3 | Signaling; antiapoptosis | − | ++ | −/++ |
| Z11793 | SEPP1 | Metabolism; selenium transport | − | +++ | −/+ |
| L36033 | SDF1 | Signaling; cxc chemokine | − | +++ | − |
| U19495 | SDF1 | Signaling; cxc chemokine | − | +++ | − |
| M27891 | CST3 | Protease inhibitor | − | ++++ | −/++++ |
| M26602 | DEFA1 | Immunity | − | ++++ | −/++++ |
| M25897 | PF4 | Signaling; cxc chemokine | − | ++++ | − |
| M54995 | PPBP | Signaling; cxc chemokine | − | ++++ | − |
| U79288 | KIAA0513a | Unknown | + | ++ | +/++ |
| M59465 | TNFAIP1 | Signaling; antiapoptosis | + | ++ | +/++++ |
| X53586 | ITGA6 | Adhesion | + | ++ | +/++ |
| D50663 | TCTEL1 | Dynein light chain | + | ++ | +/++ |
| U40846 | NAGLU | Metabolism; heparan sulfate degradation | + | ++ | +/++ |
| M80563 | S100A4 | Signaling; calcium binding | + | ++ | +/++++ |
| X04085 | CAT | Metabolism; catalase | + | ++ | +/++ |
| L02648 | TCN2 | Metabolism; vitamin B12 transport | + | ++ | + |
| L35249 | ATP6B2 | Lysosome; vacuolar proton pump | + | ++ | + |
| L09209 | APLP2 | Amyloid beta precursor like | + | ++ | + |
| L41870 | RB1 | Cell cycle | + | ++ | +/++ |
| X76732 | NUCB2 | Signaling; calcium binding | + | +++ | +/+++ |
| D29805 | 5B4GALT1 | Adhesion | + | +++ | + |
| M29877 | FUCA1 | Lysosome; fucosidase | + | +++ | +/++ |
| M32304 | TIMP2 | Metalloproteinase 2 inhibitor | + | +++ | +/++++ |
| D10522 | MACS | Actin crosslinking | + | ++++ | −/++ |
| L38696 | RALYa | RNA binding | ++ | +++ | ++ |
| U05875 | IFNGR2 | Signaling; interferon gamma receptor | ++ | +++ | ++/+++ |
| U78095 | SPINT2 | Protease inhibitor; blocks HGF | ++ | +++ | −/+++ |
| L13977 | PRCP | Lysosomal; angiotensinase C | ++ | ++++ | ++ |
| U12255 | FCGRT | IgG Fc receptor | ++ | ++++ | −/+++ |
| L06797 | CXCR4 | Signaling; SDF1 receptor | ++ | ++++ | ++/++++ |
| D82061 | FABGL | Metabolism | ++ | ++++ | ++/+++ |
| Y00433 | GPX1 | Oxidation protection | +++ | ++++ | ++/+++ |
| M60752 | H2AFA | Histone; nucleosome | + | − | −/++ |
| U18300 | DDB2 | DNA repair | + | − | + |
| X63692 | DNMT1 | DNA methyltransferase | + | − | + |
| D11327 | PTPN7 | Signaling; phosphatase | ++ | − | ++ |
| X54942 | CKS2 | Cell cycle; kinase regulator | ++ | − | +/+++ |
| D14874 | ADM | Adrenomedullin | ++ | + | +/++++ |
| D86976 | KIAA0223a | Minor histocompatability antigen | ++ | + | +/+++ |
| X52979 | SNRPB | mRNA splicing | ++ | + | +/++ |
| Z49254 | MRPL23 | Mitochondrial ribosomal protein | ++ | + | ++ |
| U66464 | HPK1 | Signaling; kinase | ++ | + | +/++ |
| U91903 | FRZB | Signaling; WNT antagonists | ++ | + | +/++ |
| D87453 | MRPS27 | Mitochondrial ribosomal protein | ++ | + | +/++ |
| X59932 | CSK | Signaling; kinase | +++ | ++ | ++ |
| L17131 | HMG1Y | Transcription; high mobility group | ++++ | + | ++/++++ |
| L19779 | H2AFO | Histone; nucleosome | ++++ | ++ | ++++ |
| U70439 | SSP29a | Unknown | ++++ | +++ | +++/++++ |
Please see Table 2 note.
The range of QGE levels for genes with variable expression in MM are given as lowest level/highest level.
Having identified tonsil PC–like MM genes in v1LDGs, we sought to identify a subset of LDGs that had bone marrow PC–like expression in MM. By subtracting the v1LDGs from the 500 LDGs we were able to eliminate the genes with tonsil PC–like expression from the list of LDGs. Applying one-way ANOVA to the remaining genes allowed the identification of so-called variable 2 LDGs (v2LDGs). Unlike the vEDGs and v1LDGs, whose expression in subsets of MM resembled that seen in tonsil BCs and tonsil PCs, respectively, v2LDGs tended to show similar expression levels between bone marrow PCs and subsets of MM (Table 6). All v2LDGs showed variability within MM and the variability could be dramatic. For example, whereas expression of the apoptosis-inducer BIK was absent in bone marrow PCs, the expression ranged from negative to 4-plus in MM. A large class of v2LDGs represented genes coding for enzymes involved in metabolism with a majority involved in glucose metabolism. Metabolism genes were not a predominant class in vEDGs and v1LDGs.
v2LDGs: LDGs showing similar expression patterns in bone marrow PC and subsets of MM
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |
|---|---|---|---|---|
| BPCs . | MM . | |||
| U61145 | EZH2 | Transcription; SET domain | − | −/+ |
| HT4000 | SYK | Signaling; lymphocyte kinase | − | −/++ |
| X89986 | BIK | Signaling; apoptosis inducer | − | −/++++ |
| D85181 | SC5DL | Metabolism; sterol-C5-desaturase | + | −/+ |
| M98045 | FPGS | Metabolism; folylpolyglutamate synthase | + | −/++ |
| L41559 | PCBD | Transcription; enhances TCF1 activity | + | −/++ |
| L25876 | CDKN2 | Cell cycle; CDK inhibitor; phosphatase | + | +/++ |
| U76638 | BRAD1 | Transcription; BRCA1 heterodimer | + | +/++ |
| L05072 | IRF1 | Transcription; IRF family | + | +/++ |
| D87440 | KIAA0252a | Unknown | + | +/++ |
| U02680 | PTK9 | Tyrosine kinase | + | +/++ |
| U28042 | DDX10 | Oncogene; ATP-dependent RNA helicase | + | +/++ |
| L20320 | CDK7 | Cell cycle; kinase | + | +/++ |
| X56494 | PKM2 | Metabolism; pyruvate kinase | + | +/++++ |
| M12959 | TCRA | Signaling; T-cell receptor | ++ | −/++ |
| HT3981 | INSL3 | Signaling; insulinlike peptide; IGF family | ++ | −/++ |
| U21931 | FBP1 | Metabolism; fructose bisphophatase | ++ | −/++++ |
| Z48054 | PXR1 | Metabolism; peroxisome biogenesis | ++ | +/++ |
| D84145 | WS-3a | Dynatin 6 | ++ | +/++ |
| D14661 | KIAA0105a | Transcription; WT1-associating protein | ++ | +/++ |
| X77548 | NCOA4 | Transcription; nuclear receptor coactivator | ++ | +/++ |
| M90696 | CTSS | Cysteine protease | ++ | +/++ |
| D11086 | IL2RG | Cytokine receptor | ++ | +/++ |
| U70426 | RGS16 | Signaling; GTPase activating protein | ++ | +/+++ |
| X14850 | H2AX | Histone; required for antibody maturation | ++ | +/+++ |
| M29927 | OAT | Metabolism; ornithine aminotransferase | ++ | +/+++ |
| S74017 | NFE2L2 | Transcription | ++ | +/+++ |
| HT4604 | GYG | Metabolism; glycogen biogenesis | ++ | +/+++ |
| M55531 | SLC2A5 | Metabolism; fructose transporter | ++ | +/++++ |
| M60750 | H2BFL | Histone; nucleosome | ++ | +/++++ |
| L19437 | TALDO1 | Metabolism; transaldolase | ++ | ++/+++ |
| M10901 | NR3C1 | Transcription; glucocorticoid receptor | ++ | ++/+++ |
| L41887 | SFRS7 | mRNA splicing factor | ++ | ++/+++ |
| M34423 | GLB1 | Metabolism; galactosidase | ++ | ++/++++ |
| X15414 | AKR1B1 | Metabolism; aldose reductase | +++ | +/++++ |
| J04456 | LGALS1 | Signaling; inhibits CD45 phosphatase | +++ | +/++++ |
| X92493 | PIP5K1B | Signaling; kinase | +++ | +/++++ |
| U51478 | ATP1B3 | Na+, K+transporter | +++ | ++/++++ |
| X91257 | SARS | Seryl-tRNA synthetase | +++ | ++/++++ |
| D30655 | EIF4A2 | Translation initiation | +++ | ++/++++ |
| D31887 | KIAA0062a | Unknown | +++ | ++/++++ |
| X04106 | CAPN4 | Cysteine protease; calcium dependent | +++ | ++/++++ |
| D87442 | NCSTNa | Nicastrin | +++ | ++/++++ |
| L76191 | IRAK1 | Signaling; cytokine receptor kinase | +++ | +++/++++ |
| HT1428 | HBB | Hemoglobin | ++++ | −/++++ |
| U44975 | COPEB | Oncogene; transcription factor | ++++ | −/++++ |
| X55733 | EIF4B | Translation initiation | ++++ | +/++++ |
| L09604 | PLP2 | Signaling; colonic epithelium differentiation | ++++ | +/++++ |
| HT1614 | PPP1CA | Signaling; phosphatase | ++++ | +++/++++ |
| L26247 | SUI1a | Translation initiation; probable | ++++ | +++/++++ |
| Accession no. . | Symbol . | Function . | Quantitative gene expression . | |
|---|---|---|---|---|
| BPCs . | MM . | |||
| U61145 | EZH2 | Transcription; SET domain | − | −/+ |
| HT4000 | SYK | Signaling; lymphocyte kinase | − | −/++ |
| X89986 | BIK | Signaling; apoptosis inducer | − | −/++++ |
| D85181 | SC5DL | Metabolism; sterol-C5-desaturase | + | −/+ |
| M98045 | FPGS | Metabolism; folylpolyglutamate synthase | + | −/++ |
| L41559 | PCBD | Transcription; enhances TCF1 activity | + | −/++ |
| L25876 | CDKN2 | Cell cycle; CDK inhibitor; phosphatase | + | +/++ |
| U76638 | BRAD1 | Transcription; BRCA1 heterodimer | + | +/++ |
| L05072 | IRF1 | Transcription; IRF family | + | +/++ |
| D87440 | KIAA0252a | Unknown | + | +/++ |
| U02680 | PTK9 | Tyrosine kinase | + | +/++ |
| U28042 | DDX10 | Oncogene; ATP-dependent RNA helicase | + | +/++ |
| L20320 | CDK7 | Cell cycle; kinase | + | +/++ |
| X56494 | PKM2 | Metabolism; pyruvate kinase | + | +/++++ |
| M12959 | TCRA | Signaling; T-cell receptor | ++ | −/++ |
| HT3981 | INSL3 | Signaling; insulinlike peptide; IGF family | ++ | −/++ |
| U21931 | FBP1 | Metabolism; fructose bisphophatase | ++ | −/++++ |
| Z48054 | PXR1 | Metabolism; peroxisome biogenesis | ++ | +/++ |
| D84145 | WS-3a | Dynatin 6 | ++ | +/++ |
| D14661 | KIAA0105a | Transcription; WT1-associating protein | ++ | +/++ |
| X77548 | NCOA4 | Transcription; nuclear receptor coactivator | ++ | +/++ |
| M90696 | CTSS | Cysteine protease | ++ | +/++ |
| D11086 | IL2RG | Cytokine receptor | ++ | +/++ |
| U70426 | RGS16 | Signaling; GTPase activating protein | ++ | +/+++ |
| X14850 | H2AX | Histone; required for antibody maturation | ++ | +/+++ |
| M29927 | OAT | Metabolism; ornithine aminotransferase | ++ | +/+++ |
| S74017 | NFE2L2 | Transcription | ++ | +/+++ |
| HT4604 | GYG | Metabolism; glycogen biogenesis | ++ | +/+++ |
| M55531 | SLC2A5 | Metabolism; fructose transporter | ++ | +/++++ |
| M60750 | H2BFL | Histone; nucleosome | ++ | +/++++ |
| L19437 | TALDO1 | Metabolism; transaldolase | ++ | ++/+++ |
| M10901 | NR3C1 | Transcription; glucocorticoid receptor | ++ | ++/+++ |
| L41887 | SFRS7 | mRNA splicing factor | ++ | ++/+++ |
| M34423 | GLB1 | Metabolism; galactosidase | ++ | ++/++++ |
| X15414 | AKR1B1 | Metabolism; aldose reductase | +++ | +/++++ |
| J04456 | LGALS1 | Signaling; inhibits CD45 phosphatase | +++ | +/++++ |
| X92493 | PIP5K1B | Signaling; kinase | +++ | +/++++ |
| U51478 | ATP1B3 | Na+, K+transporter | +++ | ++/++++ |
| X91257 | SARS | Seryl-tRNA synthetase | +++ | ++/++++ |
| D30655 | EIF4A2 | Translation initiation | +++ | ++/++++ |
| D31887 | KIAA0062a | Unknown | +++ | ++/++++ |
| X04106 | CAPN4 | Cysteine protease; calcium dependent | +++ | ++/++++ |
| D87442 | NCSTNa | Nicastrin | +++ | ++/++++ |
| L76191 | IRAK1 | Signaling; cytokine receptor kinase | +++ | +++/++++ |
| HT1428 | HBB | Hemoglobin | ++++ | −/++++ |
| U44975 | COPEB | Oncogene; transcription factor | ++++ | −/++++ |
| X55733 | EIF4B | Translation initiation | ++++ | +/++++ |
| L09604 | PLP2 | Signaling; colonic epithelium differentiation | ++++ | +/++++ |
| HT1614 | PPP1CA | Signaling; phosphatase | ++++ | +++/++++ |
| L26247 | SUI1a | Translation initiation; probable | ++++ | +++/++++ |
Please see Table 2 note.
The range of QGE levels for genes with variable expression in MM are given as lowest level/highest level.
Hierarchical cluster analysis with vEDGs, v1LDGs, and v2LDGs reveals relationships between gene expression–based and developmental stage–based groups of MM
To identify whether the variability in gene expression seen in MM might be used to identify subgroups of disease, we performed hierarchical cluster analysis of 74 newly diagnosed MM cases, 7 tonsil BC, 7 tonsil PC, and 7 bone marrow PC samples using the vEDGs (Figure3A). The cluster analysis created 2 major branches, one containing the tonsil BCs and one containing the tonsil PCs and bone marrow PCs intermingled. A total of 22 of the 74 MM cases clustered with the tonsil BCs. We have previously shown that the 74 MM cases used in this study could be separated into 4 distinct gene expression–defined subgroups (MM1 through MM4).42 An analysis of the 22 MM cases clustering with the tonsil BCs revealed that 13 of 18 MM4, 5 of 15 MM3, 1 of 21 MM2, and 3 of 20 MM1 cases (P = .000 05) made up this group (Table 7). An identical clustering approach was applied to the v1LDGs (Figure 3B). As expected, v1LDG clustering segregated bone marrow PCs and tonsil PCs into 2 major cluster branches. The tonsil BCs were tightly clustered together on a separate subbranch of the tonsil PC branch. A total of 29 MM cases were clustered with the tonsil PCs. Here, 3 of 18 MM4, 14 of 15 MM3, 4 of 21 MM2, and 8 of 20 MM1 clustered with the tonsil PCs (P = .000 008) (Table 7). Clustering with the v2LDGs again created 2 major branches segregating the bone marrow PCs from the tonsil BCs and PCs (Figure 3C). A subbranch on the bone marrow PC branch contained all bone marrow PC samples and 20 MM cases. Here, the gene expression subgroup distribution of the MM cases was 0 of 18 MM4, 0 of 15 MM3, 14 of 21 MM2, and 6 of 20 MM1 (P = .000 001; Table 7). Whereas all MM3 cases were able to be classified, 6 MM1, 5 MM2, and 3 MM4 cases did not cluster with any of the normal cell groups in 3 cluster analyses performed. A total of 3 MM1, 2 MM2, 4 MM3, and 1 MM4 cases could be clustered in 2 groups. With the exception of sample P241, which clustered with the bone marrow PCs and tonsil BCs, all cases clustering with 2 different normal cell types were always in adjacent, temporally appropriate, groups, such as tonsil PC and bone marrow PC. No samples were found to cluster with all 3 normal cell types. Thus, these data suggest that MM4, MM3, and MM2 subtypes have similarities to tonsil BCs, tonsil PCs, and bone marrow PCs, respectively. MM1 represented the only subgroup with no strong correlation with the normal cell counterparts tested here.
A 2-dimensional hierarchical cluster analysis of experimental expression profiles and gene behavior of (A) vEDGs, (B) v1LDGs, and (C) v2LDGs.
Genes (HUGO-approved gene symbols, right side) are plotted along the vertical axis and experimental samples are plotted along the top horizontal axis by their similarity. A cluster-ordered data table was used to analyze 7 tonsil BC samples (red vertical bars), 7 tonsil PC samples (blue vertical bars), 7 bone marrow PC samples (yellow vertical bars), and 74 newly diagnosed MM cases. The nomenclature for the 74 MM samples is as indicated in Zhan et al.42 The normal cell-defined cluster for tonsil BCs (horizontal red bar), tonsil PCs (horizontal blue bar), and bone marrow PCs (horizontal yellow bar) are indicated. The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are adjacent to each other. Experimental samples are arranged the same way along the horizontal axis; those with the most similar patterns of expression across all genes are adjacent to each other. Sample groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean.
A 2-dimensional hierarchical cluster analysis of experimental expression profiles and gene behavior of (A) vEDGs, (B) v1LDGs, and (C) v2LDGs.
Genes (HUGO-approved gene symbols, right side) are plotted along the vertical axis and experimental samples are plotted along the top horizontal axis by their similarity. A cluster-ordered data table was used to analyze 7 tonsil BC samples (red vertical bars), 7 tonsil PC samples (blue vertical bars), 7 bone marrow PC samples (yellow vertical bars), and 74 newly diagnosed MM cases. The nomenclature for the 74 MM samples is as indicated in Zhan et al.42 The normal cell-defined cluster for tonsil BCs (horizontal red bar), tonsil PCs (horizontal blue bar), and bone marrow PCs (horizontal yellow bar) are indicated. The clustering is presented graphically as a colored image. Along the vertical axis, the analyzed genes are arranged as ordered by the clustering algorithm. The genes with the most similar patterns of expression are adjacent to each other. Experimental samples are arranged the same way along the horizontal axis; those with the most similar patterns of expression across all genes are adjacent to each other. Sample groupings can be further described by following the solid lines (branches) that connect the individual components with the larger groups. The color of each cell in the tabular image represents the expression level of each gene, with red representing an expression greater than the mean, green representing an expression less than the mean, and the deeper color intensity representing a greater magnitude of deviation from the mean.
MM subgroup distribution in normal cell type–defined cluster after hierarchical clustering using vEDGs, v1LDGs, and v2LDGs
| Normal cell type–defined cluster . | Gene expression– defined MM subgroups . | P . | |||
|---|---|---|---|---|---|
| MM1 (n = 20) . | MM2 (n = 21) . | MM3 (n = 15) . | MM4 (n = 18) . | ||
| vEDGs (n = 22) | 3 | 1 | 5 | 13 | .000 05 |
| v1LDGs (n = 29) | 8 | 4 | 14 | 3 | .000 008 |
| v2LDGs (n = 20) | 6 | 14 | 0 | 0 | .000 001 |
| Normal cell type–defined cluster . | Gene expression– defined MM subgroups . | P . | |||
|---|---|---|---|---|---|
| MM1 (n = 20) . | MM2 (n = 21) . | MM3 (n = 15) . | MM4 (n = 18) . | ||
| vEDGs (n = 22) | 3 | 1 | 5 | 13 | .000 05 |
| v1LDGs (n = 29) | 8 | 4 | 14 | 3 | .000 008 |
| v2LDGs (n = 20) | 6 | 14 | 0 | 0 | .000 001 |
Gene expression–defined MM subgroups are as described in Zhan et al.42
Normal cell–defined cluster indicates MM cases clustering with CD19-enriched BCs using vEDGs (n = 22), CD138-enriched tonsil PCs using v1LDGs, or CD138-enriched bone marrow PCs using v2LDGs, defined by vEDGs as described in Figure 2. The MM cases within each normal cell type–defined cluster were separated based on whether the sample belonged to MM1, 2, 3, or 4.
The cyclin B gene (CCNB1), which is expressed in proliferating cells, was identified as a vEDG (Table 4). CCNB1exhibited high average difference calls in tonsil BCs and the MM4 subgroup. CD19-enriched tonsil BCs and MM4 also exhibit elevated expression of other proliferation-associated genes includingMKI67 and PCNA (Zhan et al42 and our unpublished data, January 2002). In order to extend the relationship between tonsil BCs and MM4, we compared the expression patterns of a panel of proliferation-associated genes (CCNB1, CKS1, CKS2, SNRPC,EZH2, KNSL1, PRKDC, andPRIM1) across all normal samples and the 4 MM subgroups (Figure 4A). Kruskal-Wallis tests revealed that expression differed across the samples (P < 4.25 × 10−5) and box charts revealed that with the exception of SNRPC and PRIM1, all genes showed a progressive reduction in the median expression from tonsil BCs to tonsil PCs to bone marrow PCs. The box charts also showed strong similarity between tonsil BCs and MM4. In addition, althoughPRIM1 expression was significantly different across the entire group (P = 4.25 × 10−5), no differences existed between tonsil BCs and MM4 (WRSP = .1) or between tonsil PCs and MM3 (WRSP = .6).
Box charts of expression profiles of a panel of proliferation genes and XBP1 show similarities between tonsil BCs and MM4.
(A) The 7 tonsil BC (TBC), 11 tonsil PC (TPC), 31 bone marrow PC (BPC), 20 MM1, 21 MM2, 15 MM3, and 18 MM4 samples are distributed along the x axis and the natural log-transformed average difference call is plotted on the y axis. The top, bottom, and middle lines of each box correspond to the 75th percentile (top quartile), 25th percentile (bottom quartile), and 50th percentile (median) of the natural log-transformed average difference call for each gene, respectively. The whiskers extend from the 10th percentile (bottom decile) and top 90th percentile (top decile). The Kruskal-Wallis test for differences in expression of each gene across the groups are: EZH2,P = 7.61 × 10−11; KNSL1,P = 3.21 × 10−8; PRKDC,P = 2.86 × 10−11; SNRPC,P = 5.44 × 10−12; CCNB1,P = 2.54 × 10−8; CKS2,P = 9.49 × 10−11; CKS1,P = 5.86 × 10−9; PRIM1,P = 4.25 × 10−5. Note the similarity in expression of each gene in TBC and MM4. (B) Box chart ofXBP1. Note that all MM subgroups have lower median expression levels than BPCs and that MM4 has the lowest level of XBP1 in the MM subgroups. Lack of expression of XBP1 in MM4 may account for the relationship between MM4 and TBCs.
Box charts of expression profiles of a panel of proliferation genes and XBP1 show similarities between tonsil BCs and MM4.
(A) The 7 tonsil BC (TBC), 11 tonsil PC (TPC), 31 bone marrow PC (BPC), 20 MM1, 21 MM2, 15 MM3, and 18 MM4 samples are distributed along the x axis and the natural log-transformed average difference call is plotted on the y axis. The top, bottom, and middle lines of each box correspond to the 75th percentile (top quartile), 25th percentile (bottom quartile), and 50th percentile (median) of the natural log-transformed average difference call for each gene, respectively. The whiskers extend from the 10th percentile (bottom decile) and top 90th percentile (top decile). The Kruskal-Wallis test for differences in expression of each gene across the groups are: EZH2,P = 7.61 × 10−11; KNSL1,P = 3.21 × 10−8; PRKDC,P = 2.86 × 10−11; SNRPC,P = 5.44 × 10−12; CCNB1,P = 2.54 × 10−8; CKS2,P = 9.49 × 10−11; CKS1,P = 5.86 × 10−9; PRIM1,P = 4.25 × 10−5. Note the similarity in expression of each gene in TBC and MM4. (B) Box chart ofXBP1. Note that all MM subgroups have lower median expression levels than BPCs and that MM4 has the lowest level of XBP1 in the MM subgroups. Lack of expression of XBP1 in MM4 may account for the relationship between MM4 and TBCs.
Additional evidence for a link between MM4 and tonsil BCs is supported by the expression pattern of the transcription factor XBP1(Figure 4B). XBP1 was identified as an EDG (Table 2) but was not in the list of the 50 most significant vEDGs. A Kruskal-Wallis test revealed significant differences across the groups (P = 3.85 × 10−10) and a box chart showed a prominent up-regulation in the tonsil PCs (median of log value of average difference call = 10.9) compared with tonsil BCs (median of log value of average difference call = 9.5). However, XBP1showed a reduction in bone marrow PCs and a progressive reduction across the 4 MM subgroups from MM1 to MM4. Other EDG and LDG transcription factors linked to PC differentiation, for example,BCL6, CIITA, and IRF4, did not show significant differences in expression across the MM subgroups (data not shown).
Discussion
CD19-enriched BCs from human tonsil and CD138-enriched PCs from tonsil and bone marrow were used to compare the gene expression changes associated with late-stage BC differentiation. This global survey allowed the identification of previously defined and novel genes discriminating these cell types, which should aid in the elucidation of the genetic pathways involved in PC development. It is important to note that it is likely that many more novel genes remain to be discovered because only 6800 of the estimated 35000 human genes were investigated in this analysis. Although the CD19-enriched cells used in this study represented a heterogenous mixture of BCs, they appeared to represent an adequate cell population for identifying genes modulated as BCs progress to the tonsil PC stage of development, as genes known to change during this process showed significant variation in the comparison of these 2 groups. In addition, hierarchical clustering analysis with 3288 genes created 2 major branches containing either tonsil BC or PC samples. Furthermore, the 3 types of PC samples (tonsil PCs, bone marrow PCs, and MM PCs) could be further distinguished. Overall, the expression differences were consistent with the cells representing distinct stages of maturation in a direction of tonsil BCs to tonsil PCs to bone marrow PCs.
Consistent with the terminal differentiation of PCs, many genes involved in cell-cycle control and DNA metabolism were found to be down-regulated in tonsil PCs. The downward modulation of the DNA ligase, LIG1, repair enzymes MSHC andRPA1, the checkpoint gene CDC20, and the cyclinsCCNG2 and CCNF may have important consequences in inducing the quiescent state of PCs. The telomeric repeat binding protein, TERF2, which is one of 2 recently cloned mammalian telomere binding proteins, acts to protect telomere ends, prevent telomere end-to-end fusion, and may be important in maintaining genomic stability.47,48 It will be of interest to determine ifTERF2 is down-regulated during the terminal differentiation of all cell types, and whether the lack of this gene product in tumors results in structural chromosome rearrangements, a common feature of MM. The CDC28 protein kinase 2 gene, CKS2, which binds to the catalytic subunit of the cyclin-dependent kinases and is essential for their biologic function, was the only cell-cycle gene in the LDGs being expressed in tonsil PCs, cells capable of modest proliferation,49 and extinguished in bone marrow PCs. Thus, shutting down CKS2 expression may be critical in ending the proliferative capacity of bone marrow PCs.
Overall, the largest group of genes altered in these comparisons represented transcription factors. Surprisingly, 4 members of theets family of transcription factors, ETS1, SPI1, SP1B, and ELF1, known to be expressed in the BC lineage,50 were shut down in tonsil PCs. ETS1knock-out mice show massive increases in both splenic and peripheral blood PCs,51-53 supporting the notion that reduction of this protein is critical in PC differentiation. Given that multiple transcription factors appear to be modulated during PC differentiation, more extensive global expression profiling combined with sophisticated data mining tools may help elucidate the transcriptional networks driven by each of the various classes of transcription factors discovered in this study.
MM PCs are derived from the bone marrow and are thought to represent a transformed counterpart of normal terminally differentiated bone marrow PCs. However, the dramatic differences in survival, which can range from several months to more than 10 years, suggest that MM may represent a constellation of several subtypes of disease that may reflect differences in the cell of origin. Using microarray profiling we previously demonstrated that MM can be classified into 4 distinct gene expression–based subgroups that exhibit differences in proliferation characteristics as well as clinical parameters associated with poor outcome.42 Variability in expression of the 359 EDGs and 500 LDGs in 74 newly diagnosed MM cases provided a means of classifying this malignancy, in that 3 of the 4 gene expression–defined subgroups had distinct similarities to the 3 normal cell populations studied, such that MM4, MM3, and MM2 have tonsil BC–like, tonsil PC–like, or bone marrow PC–like expression features, respectively.
Most of the vEDGs used to classify MM4 as a tonsil BC–like subtype of disease belonged to a range of gene classes, including adhesion, transcription, signaling, and metabolism, with very few vEDGs being associated with cell proliferation. However, comparison of expression of a panel of proliferation genes across MM and normal cell types advanced the relationship between the MM4 group and tonsil BCs. In addition, the expression of XBP1, a transcription factor essential for PC differentiation, was significantly different across the 4 MM subgroups, with MM4 having the lowest level of the MM subgroups and thus being more similar to the tonsil BCs. A future question will be whether reduced XBP1 is a cause or effect in the apparent de-differentiated state of the MM4 subtype. It is of note that other transcription factors important in regulating PC development, for example, IRF4, BCL6, CIITA, STAT6, andPAX5, did not show the down-regulation seen withXBP1.
Using the same type of analysis described in this study, we identified a panel of genes distinguishing 7 CD19-enriched peripheral blood BCs (kind gift from B. Klein) from the 7 CD19-enriched tonsil BCs. Hierarchical clustering with the most variable of these genes showed no link (P = .39) between the 4 MM subgroups and CD19-enriched peripheral blood BCs (our unpublished data, June 2002), suggesting that similarities between MM and normal BC development stages may be limited. It is important to note that MM1 was the only gene expression–defined subgroup lacking strong similarities to any of the normal cell types analyzed in our current study. It is possible that MM1 may be related to mucosal-derived PCs or peripheral blood PCs, which have recently been shown to represent a distinct type of PC.18
Although the distribution of MM2, MM3, and MM4 subgroups in the normal cell–defined clusters was significant, there were outliers. For example, the tonsil BC cluster consisted mainly of MM4 cases, but 5 MM3, 1 MM2, and 3 MM1 cases were also found in this cluster. In addition, 10 of 60 cases that could be clustered with normal cell types clustered with 2 different normal cell types depending on whether the genes used in the cluster analysis were VEDG, V1LDG, or V2LDG. Furthermore, 3 MM4, 5 MM2, and 6 MM1 cases could not be clustered with any of the normal cell types. These data demonstrate a lack of complete correlation between the class systems and that the possible plasticity seen suggests that outlier cases may have intermediate characteristics and may represent distinct clinical entities. It will be important to determine if our unsupervised gene expression–based or developmental stage–based classification system alone or in combination will represent a robust clinical stratification of MM. We anticipate that in the next year or two the clinical response data on this cohort of newly diagnosed MM cases treated with high-dose therapy and stem cell support will mature and allow us to answer this question.
We thank Eric Siegel for helpful discussions, Karin Tarte and Bernard Klein for helpful discussions and use of the peripheral blood BC samples, Dr David Parham and colleagues for providing tonsil samples, members of the Lambert Laboratory, Jena Derrick, Kelly McCastlain, John Smith, Yan Xiao, and Hongwei Xu, for technical assistance, and the clinical faculty of the MIRT for providing clinical MM samples.
Prepublished online as Blood First Edition Paper, September 26, 2002; DOI 10.1182/blood-2002-06-1737.
Supported through private funding and by grants CA55819 (B.B. and J.S.) and CA97513 (J.S.) from the National Cancer Institute, Bethesda, MD.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
John Shaughnessy Jr, Donna and Donald Lambert Laboratory of Myeloma Genetics, University of Arkansas for Medical Sciences, 4301 W Markham St, Slot 776, Little Rock, AR 72205; e-mail: shaughnessyjohn@uams.edu.

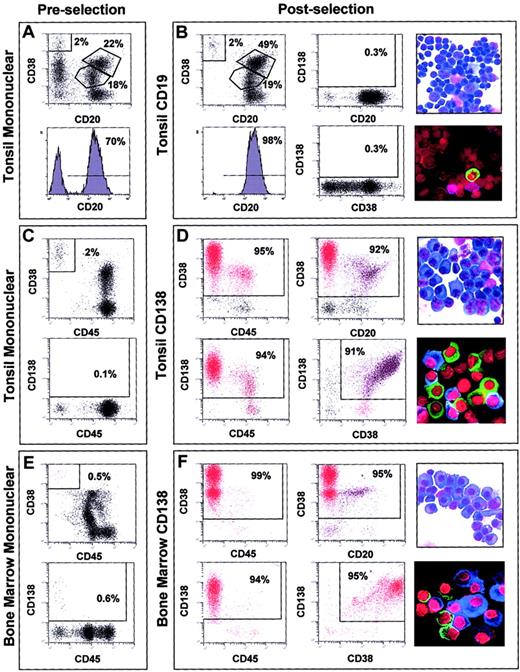
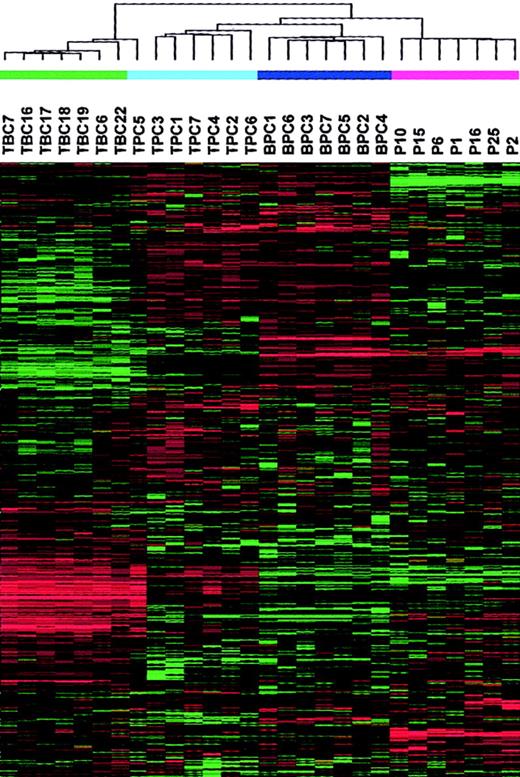
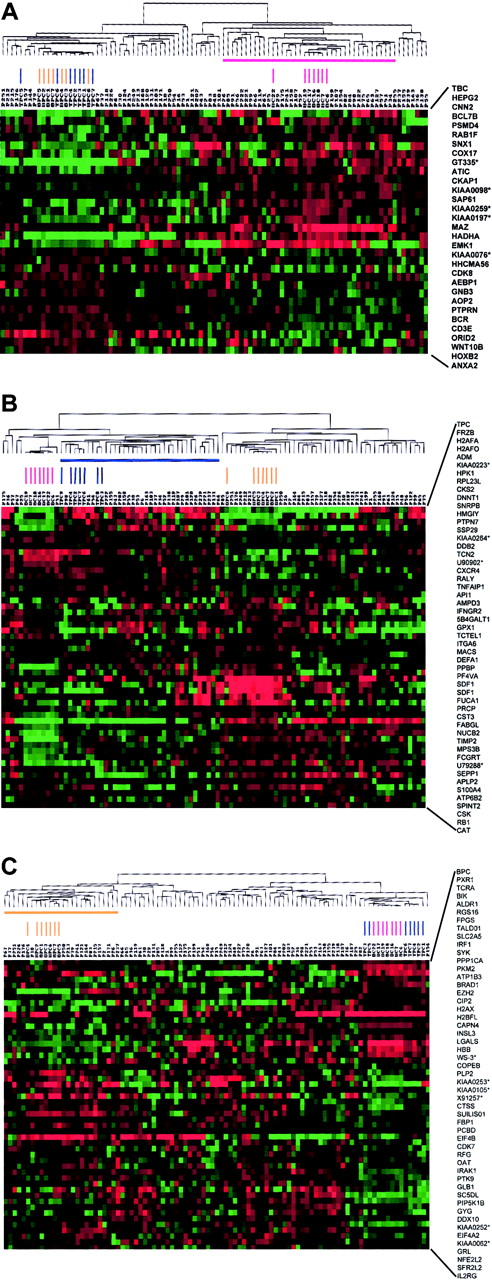
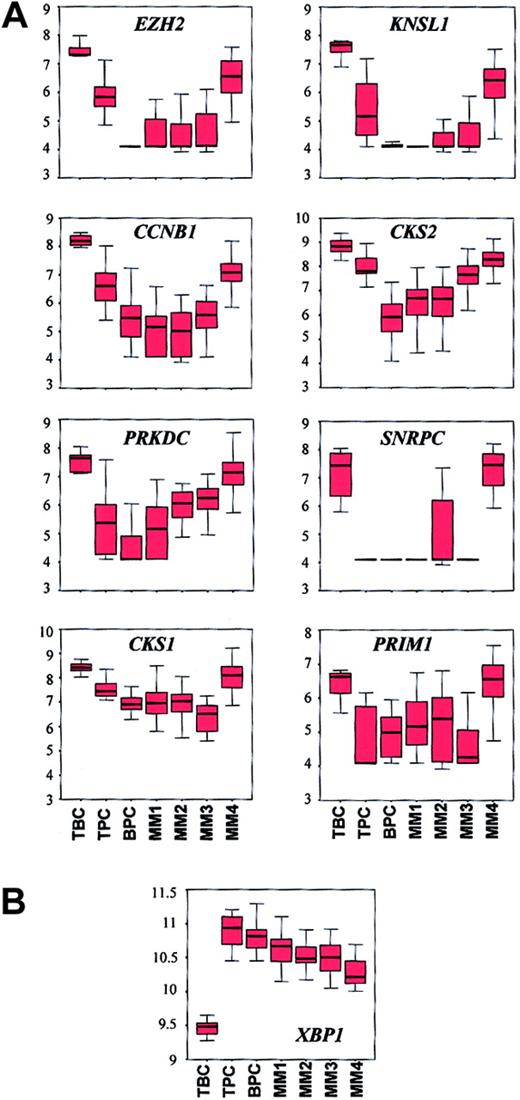
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal