Abstract
B-cell chronic lymphoid leukemia (BCLL) is a highly heterogeneous human malignancy, presumably reflecting specific molecular alterations in gene expression and protein activity that are thought to underlie the variable disease outcome. Most B-CLL cell samples undergo apoptotic death in response to DNA damage. However, a clinically distinct aggressive subset of B-CLL is completely resistant in vitro to irradiation-induced apoptosis. We therefore addressed 2 series of microarray analyses on 4 sensitive and 3 resistant B-CLL cell samples and compared their gene expression patterns before and after apoptotic stimuli. Data analysis pointed out 16 genes whose expression varied at least 2-fold specifically in resistant cells. We validated these selected genes by real-time quantitative reverse transcription–polymerase chain reaction (RT-PCR) on 7 microarray samples and confirmed their altered expression level on 15 additional B-CLL cell samples not included in the microarray analysis. In this manner, in 11 sensitive and 11 resistant B-CLL cell samples tested, 13 genes were found to be specific for all resistant samples: nuclear orphan receptor TR3, major histocompatibility complex (MHC) class II glycoprotein HLA-DQA1, mtmr6, c-myc, c-rel, c-IAP1, mat2A, and fmod were up-regulated, whereas MIP1a/GOS19-1 homolog, stat1, blk, hsp27, and ech1 were down-regulated. In some cases, the expression profile may be dependent on the status of p53. Some of these genes encode general apoptotic factors but also exhibit lymphoid cell specificities that could potentially be linked to the development of lymphoid malignancies (MIP1α, blk, TR3, mtmr6). Taken together, our data define new molecular markers specific to resistant B-CLL subsets that might be of clinical relevance.
Introduction
B-cell chronic lymphoid leukemia (B-CLL) results in an accumulation of mature CD5+/CD23+ B cells in peripheral blood, not due to excessive proliferation (at the early stage of disease, cells are mainly arrested in G0/G1 of the cell cycle), but rather to decreased cell mortality resulting from an uncharacterized defect in apoptotic cell death.1 The most revealing alteration of gene expression that may explain the prolonged survival of malignant B-cells is the overexpression of the bcl-2 gene resulting from its hypomethylation.2 Although this overexpression as well as deregulated expression of some other bcl-2 family members1 is consistent with prolonged cell survival, it does not clearly correlate with disease aggressiveness, clinical outcome, or cell sensitivity to apoptosis induced in vitro. Another gene involved in the apoptotic process through its transducing role in the cellular response to DNA damage is ATM, which is deleted or mutated in an aggressive form of B-CLL.3, 4, 5 These alterations can lead to dysfunction of p53, whose direct inactivation by mutation is a relatively rare molecular event in B-CLL.6 Deregulation of p53 can also result from an altered function of the proteasome system, which we observed in all B-CLL samples tested.7 Together, these observations make deregulated control of the apoptotic death pathway more easily conceivable. Because the ubiquitin-proteasome system, an essential cellular biochemical safeguard,8 is known to be involved in apoptotic death control,9 an alteration of its function may at least partly explain the commonly observed defect of B-CLL cells to die in this manner. Recent studies based on microarray approaches have revealed a common B-CLL gene expression profile related to memory B cells as well as a subset of genes specifically expressed in B-CLL cells.10, 11, 12 The expression profile proper to CLL may be specific for a subset harboring somatic mutations of immunoglobulin (Ig) genes10,11 and may correlate with patient survival or clinical stage or both.12
Because of high sample heterogeneity that may underlie the individual gene expression “signature” of each B-CLL case, the aim of this study was to analyze the expression profile of 2 subsets of B-CLL cells according to, as principal criteria, their sensitivity to undergo or not in vitro DNA damage–induced apoptosis. Further, we sought whether the use of a minimal number of samples for microarray analysis may be useful to identify genes specifically expressed in one of 2 subsets and which can further be confirmed by another most reliable and reproducible method on large series of B-CLL samples. Thus, in the first step, we used an oligonucleotide DNA microarray approach (Hu-FL GeneChips; Affymetrix, Santa Cruz, CA) to explore mRNA expression levels of about 7070 genes in 3 resistant B-CLL cell samples before and 3 hours after exposure of cells to apoptotic stimuli through ionizing irradiation, and compared them to the expression levels in 4 cell samples that activate apoptosis in response to the same stimulus. Although this new technology is a powerful method to rapidly define global mRNA expression levels,13, 14, 15, 16, 17 the data it provides can sometimes be unreliable or confusing due to conceptual or technical difficulties. For this reason, in a second step, we validated the mRNA expression levels of the genes selected in the DNA microarray analysis by a reliable real-time quantitative polymerase chain reaction (PCR) method (LightCycler; Roche Diagnostics, Basel, Switzerland) on the same samples analyzed by microarray approach. In this manner, we confirmed the microarray data for 14 (with one redundant gene) of the 16 selected genes whose expression appeared to be specifically deregulated at least 2-fold before or after apoptosis signaling, in B-CLL samples resistant to irradiation-induced apoptosis. These selected genes were then confirmed by PCR on another 15 B-CLL (7 sensitive and 8 resistant) samples not included in the microarray analysis. In the 22 studied cases, mutational status of p53 has been also defined. Although one of the genes selected by microarray analysis could not be confirmed by PCR, due to unresolved technical problems possibly related to its short nucleotide sequence, our data on the whole suggest that rigorous analysis and selection of microarray data can be a useful and reliable method to define differential gene expression profiles between resistant and sensitive B-CLL cells. In the 11 resistant B-CLL samples (including 3 microarray samples and 8 additional cases studied by reverse transcription–PCR [RT-PCR] alone) we observed, independently of p53 mutation status, a constitutive down-regulation of 4 genes, MIP1α, a human homolog of murine G0S19-1, stat1, hsp27, and blk, and upregulation of 3 genes, nuclear orphan receptor TR3, HLA-DQA1, and mtmr6 (myotubularin-related protein 6, D13S824E 13q12 locus). After irradiation in the same B-CLL subset, 3 genes were up-regulated, c-myc (2 microarray probes, c-myc-P64 and c-myc-ORF114, were validated and confirmed), mat2A, and c-rel, and 2 down-regulated, ech1 and hypothetical protein A4. Only 2 genes were specifically modified according to p53 mutation status: c-IAP1 (a human B homolog of MIH), in 6 resistant samples expressing wild-type p53 (p53wt) and fmod (fibromodulin) in 5 resistant samples expressing mutated p53 (p53mt). Together, these modifications in the gene expression profile characterize a resistant subset of B-CLL and may be implicated in the defect of apoptotic cell death specifically expressed in this subset.
Patients, materials, and methods
Patient selection
This study was approved by the Paris-Cochin Ethics Commitee (Comité Consultatif de Protection des Personnes dans la Recherche Biomédicale, Paris-Cochin). The cell samples were selected according to their defined sensitivity to in vitro irradiation-induced apoptosis and their previously established p53 status.7 None of the included resistant samples expressing p53wt showed loss of heterozygosity (LOH) for the ATM gene as defined by analyzing 4 microsatellite markers (D11S1817, D11S1819, D112179, and D11S1294).18 All samples expressing p53wt disclosed an accumulation of the nuclear p53 following irradiation. At the time of the study, the 22 patients included were all in stage A (according to the Binet staging system), had lymphocytosis ranging from 5 × 109/L to 20 × 109/L, and hadreceived no treatment for at least 3 months prior to blood sampling. The common phenotype of all included patients was CD5+/CD23+ (except one with CD23+/–), weak or negative FMC7, weak expression of surface IgM, and a negative Coombs test. The 11 patients with malignant B cells sensitive to in vitro radiation-induced apoptosis had never been treated, whereas the 8 of 11 patients with resistant disease were previously treated with fludarabine, cyclophosphamide, chlorambucil, or theophylline/chlorambucil and were revealed to be resistant to treatment. Three patients from this subset died during the course of the study.
Isolation of B-CLL lymphocytes and total RNA
Peripheral blood lymphocytes from 22 leukemic patients were collected, with their informed consent, in heparin-coated tubes as previously described.7,9 The mean percentages of apoptotic cells for resistant samples were 7.2% ± 2.3% in absence of irradiation and 6.7% ± 3.7% after 24-hour postirradiation culture. Sensitive samples had 15% ± 4.3% of spontaneous apoptotic cells and 87.1% ± 7.2% following irradiation. Thus, clinical resistance to treatment appeared to be consistent with the resistance of B-CLL cells to in vitro irradiation-induced apoptosis.
Total RNA from resistant or sensitive malignant B lymphocytes was extracted immediately after lymphocyte isolation or 3 hours after irradiation culture (10 Gy using a 137Cs source) using a fresh RNA-now kit (Biogentex, Seabrook, TX) according to the supplier's instructions. The quality of the total RNA preparation was verified by both agarose gel and PCR and, when necessary, further purification to remove chromatin DNA was performed with the Rneasy cleanup system (Quiagen, Valencia, CA).
Samples and oligonucleotide DNA microarray
Two series of microarrays were prepared. The first series of 5 hybridizations (commercial conditioning of the human Hu-FL GeneChip by Affymetrix, Santa Cruz, CA) comprised a pool of 3 sensitive RNA samples mixed in equal quantities and RNA from patient cells resistant to irradiation-induced apoptosis, all expressing p53wt. RNAs were extracted and purified before and 3 hours after cell irradiation. The fifth hybridization was performed with RNA derived from one patient whose cells were resistant to irradiation-induced apoptosis and expressed p53mt, before irradiation only. The second series of microarrays was performed using 2 different samples: one sensitive and one resistant sample both expressing p53wt, before and after irradiation and the resistant p53mt cell sample, which has been previously used in the first microarray, but with total RNA being extracted at 3 hours after irradiation. The detailed protocol for the microarray hybridizations, sample preparation, and the human Hu-FL GeneChip array design is available from Affymetrix. Briefly, 10 μg purified total RNA was transcribed into a first cDNA using Superscript RT (Invitrogen; Cergy Pontoise, France), in the presence of T7-(T)24-oligo-dT primer, doxyribonucleoside triphosphates (dNTPs), and T7 RNA polymerase promoter (Gibco Invitrogen Life Technology). The second cDNA synthesis of all mRNAs was performed using Escherichia coli DNA polymerase I. The double-stranded (ds)–cDNA was cleaned and an in vitro transcription reaction was then performed to generate biotinylated cRNA which, after fragmentation, was used in a hybridization assay on a Hu-FL GeneChip microarray targeting 7070 human genes or expressed sequence tags (ESTs) as selected from the National Center for Biotechnology Information (NCBI) GenBank database. Before hybridization, the efficiency of cDNA synthesis was estimated on a test chip by calculating the ratios for 5′ and middle intensities relative to 3′ for the control genes actin and glyceraldehydes-3-phosphate dehydrogenase (GAPDH). Biotinylated RNA used as a probe in the microarray hybridization was revealed by streptavidinphycoerythrin conjugation and amplification with a second antibody. Fluorescence intensity was measured for each microarray and the results were normalized by global scaling to the average fluorescence intensity of the entire microarray, rather than by normalization based on the expression levels of actin or GAPDH, because these were found to be variable (among different microarrays) as well. The final results were imported into Microsoft Excel (Microsoft France, Cortaboeuf, France).
Western blotting
Stabilization of p53 following irradiation was examined by Western blot analysis as described elsewhere.7 DO-7 monoclonal p53 antibody (Abcam, Cambridge, United Kingdom) recognizing N-terminal domain of both wild-type and mutated form of p53, was used to monitor p53 protein level.
Data analysis
Comparative analysis was performed for each gene or EST and for each microarray separately. The first comparison value was “pairs in average,” meaning the number of probe pairs used in the computation of “average difference” and “log average.” “Log average ratio” is an intensity ratio (pm/MM, perfect match signal/mismatch signal) for each probe pair used, calculated by dividing the intensity of the pm by the intensity of the MM. Thus, an average of the intensity ratios was calculated for each gene from the pairs in average and multiplied by 10. Average difference means an intensity difference (pm/MM) for each probe pair used, which was calculated by subtracting the intensity of the mismatch from the intensity of the perfect match. “Absolute call” can be expressed as present (P), absent (A), or marginal (M). “Increase” means the number of probe pairs, for each gene, for which (pm) – (MM) is significantly greater in the experimental data than in the baseline data; to be considered significant 2 conditions must be met:
(pm – MM)experimental – (pm – MM)baseline ≥ CT (change threshold)
[(pm – MM)experimental – (pm – MM)baseline]/(pm – MM)baseline ≥ PCT (percent change)
Similarly, “decrease” means the number of probe pairs for each gene for which pm-MM is significantly greater in the baseline data than in the experimental data. “Difference call” can result in 5 possible outcomes indicating that the level of expression of an RNA is decreased (D), increased (I), marginally increased (MI), marginally decreased (MD), or with no detectable change (NC). “Fold change” represents the ratio of the average differences between the experimental and baseline files and reflects the magnitude but not the direction of the change. Finally, the “sort score,” which is based on both fold change and average difference change, was calculated for each gene or EST. Taking into account all of these parameters, the data for each gene on each of the 2 microarrays were analyzed. Then, to identify genes whose expression appeared to be modified before and after apoptotic stimuli in the 2 B-CLL cell populations, a comparison rather than a clustering method was performed between the data for the same gene on the 2 microarrays, taking into account the following parameters: average difference, absolute call, fold change (FC, ≥ 2 or ≤ –2) and sort score (SC, > 0.5 or < –0.5). Thus, expression levels were compared between the samples resistant and sensitive to irradiation-induced apoptosis before and after apoptotic stimuli on each microarray, and genes common to these 2 microarrays were selected (Figure 1).
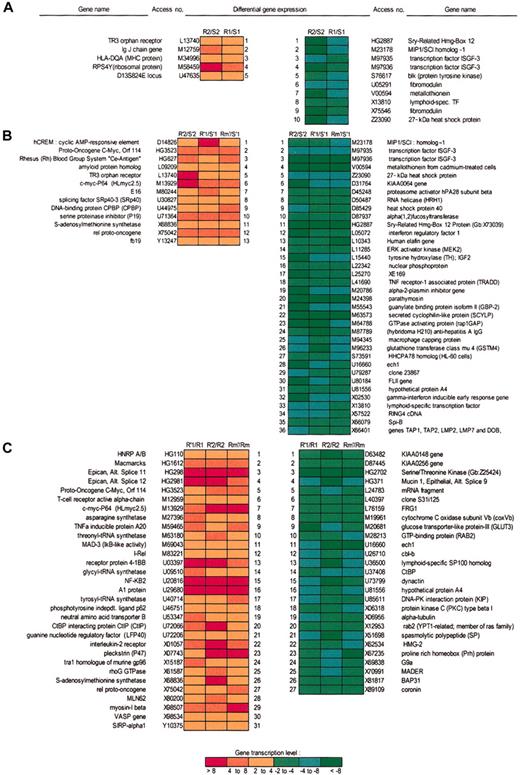
Microarray data analysis showing relative gene expression levels of the B-CLL subset resistant to irradiation-induced apoptosis. (A) Comparative data analyses and ratios between resistant (R) and sensitive (S) B-CLL samples both expressing p53wt from first (R1/S1) and from second (R2/S2) microarray analysis revealed that 5 transcripts were up-regulated (red boxes) and 10 transcripts were down-regulated (green boxes) in a constitutive manner in resistant B-CLL samples. (B) Comparisons between all resistant (R′) versus sensitive (S′) samples in 2 microarrays at 3 hours after irradiation only, expressing both p53wt (R′1/S′1, R′2/S′2), and p53mt (Rm′/S′1), showed that 36 transcripts were down-regulated and 13 transcripts were up-regulated in all resistant B-CLL samples, regardless of p53 status. (C) Comparisons between resistant samples only, before and after irradiation (R′1/R1, R′2/R2 Rm′/Rm1), showed modified expression of 58 transcripts (31 up- and 27 down-regulated). The green (down-regulation) to red (up-regulation) color scale indicates the relative values (fold change, FC) of gene expression levels.
Microarray data analysis showing relative gene expression levels of the B-CLL subset resistant to irradiation-induced apoptosis. (A) Comparative data analyses and ratios between resistant (R) and sensitive (S) B-CLL samples both expressing p53wt from first (R1/S1) and from second (R2/S2) microarray analysis revealed that 5 transcripts were up-regulated (red boxes) and 10 transcripts were down-regulated (green boxes) in a constitutive manner in resistant B-CLL samples. (B) Comparisons between all resistant (R′) versus sensitive (S′) samples in 2 microarrays at 3 hours after irradiation only, expressing both p53wt (R′1/S′1, R′2/S′2), and p53mt (Rm′/S′1), showed that 36 transcripts were down-regulated and 13 transcripts were up-regulated in all resistant B-CLL samples, regardless of p53 status. (C) Comparisons between resistant samples only, before and after irradiation (R′1/R1, R′2/R2 Rm′/Rm1), showed modified expression of 58 transcripts (31 up- and 27 down-regulated). The green (down-regulation) to red (up-regulation) color scale indicates the relative values (fold change, FC) of gene expression levels.
Real-time PCR
The genes selected in the comparative microarray data analysis, whose expression appeared to be specifically associated with resistance to irradiation-induced apoptosis (16 selected genes; Figures 3 and 4) were confirmed by a real-time reverse transcription assay (LightCycler, Roche Diagnostics) according to the manufacturer's instructions. The PCR primers shown in Table 1 were designed using WebPrimer online software (www.alces.med.umn.edu) and are based on published sequences from the NCBI database. Primers were synthesized by Genset (Paris, France). All probes were provided by Roche Diagnostics and all reactions were performed in duplicate in a LightCycler. Briefly, the PCR conditions were as follows: 100 ng RNA was mixed with 0.5 μM of each primer and 6 mM MgCl2. SybrGreen mix and RT-PCR enzyme mix (Roche Diagnostics) were added successively. Cytokine RNA dilutions with established copy number were used as control. For all genes except one, temperature cycles were as follows: 10 minutes at 55°C (reverse transcription) and 3 seconds at 95°C (initial denaturation), 45 amplification cycles for 0 seconds at 95°C (denaturation), 10 seconds at 55°C (annealing), and 13 seconds at 72°C (elongation). For the D90070 gene, the initial denaturing was increased to 60 seconds and the amplification cycles to 5 seconds. The amplified RT-PCR products were checked for specificity by gradual denaturing from 65°C to 95°C over 5 minutes. This allows an assessment of the melting curves for each amplified PCR product so as to discriminate between primer dimers and specific products. Each RNA sample was quantified in duplicate for the selected gene and for GAPDH as internal control. The median values of each calculated ratio were less than 0.2.
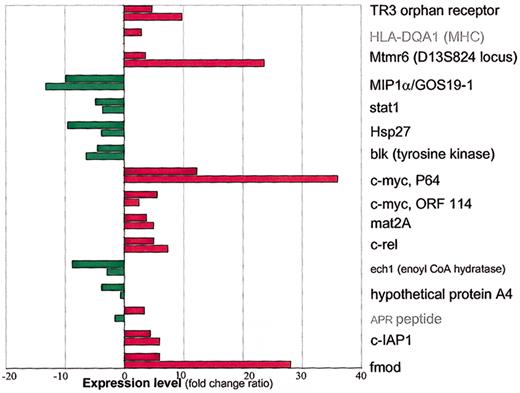
Quantitative real-time RT-PCR (LightCycler) validation of 16 genes selected by microarray analysis on resistant B-CLL samples. The relative RT-PCR and microarray expression levels of the same resistant B-CLL samples are shown; fold change (FC) ratios for microarray analysis and the ratios for RT-PCR were calculated by taking into account the expression levels of GAPDH. The microarray data are shown as shaded columns and RT-PCR data as light columns. Down-regulation is shown in green and up-regulation in red. Note that one gene (APR peptide, access number D90070) was found to be up-regulated by microarray analysis and down-regulated by RT-PCR. Note that APR transcript (access number D90070) was not validated and that the RT-PCR result for one selected gene (HLA-DQA1 MHC protein, access number M34996) is not given.
Quantitative real-time RT-PCR (LightCycler) validation of 16 genes selected by microarray analysis on resistant B-CLL samples. The relative RT-PCR and microarray expression levels of the same resistant B-CLL samples are shown; fold change (FC) ratios for microarray analysis and the ratios for RT-PCR were calculated by taking into account the expression levels of GAPDH. The microarray data are shown as shaded columns and RT-PCR data as light columns. Down-regulation is shown in green and up-regulation in red. Note that one gene (APR peptide, access number D90070) was found to be up-regulated by microarray analysis and down-regulated by RT-PCR. Note that APR transcript (access number D90070) was not validated and that the RT-PCR result for one selected gene (HLA-DQA1 MHC protein, access number M34996) is not given.
Validation of microarray selected genes by quantitative real-time RT-PCR; confirmation of validated genes on 22 B-CLL samples. Expression patterns of 16 selected genes by microarray analyses specifically regulated in the resistant B-CLL subset were verified by RT-PCR on 22 B-CLL samples: 7 samples included in microarray analyses, 7 new sensitive, 4 resistant expressing p53wt as well as 4 new resistant samples expressing p53mt. Microarray data on the expression levels of 14 genes were validated by RT-PCR and all but one gene were confirmed on all included B-CLL cell samples. The names of genes are indicated in red or green characters according to their up- or down-regulation. Data are expressed as means of duplicate experiments for each of 22 samples. The median + SD, median + SE, median, median – SE and median – SD are shown. (A) Relative constitutive expression levels (in absence of irradiation) in sensitive (S) and resistant (R) subset revealed 3 up-regulated transcripts (TR3 orphan receptor, HLA-DQA, and mtmr6), and 4 down-regulated transcripts (stat1, blk, MIP1α, and hsp27). (B) Relative expression levels before and after 3 hours of irradiation in sensitive (S and S′, respectively) and resistant (R and R′, respectively) samples revealed 4 up-regulated (mat2A, c-myc-P64, c-rel, and c-myc ORF114), and 2 down-regulated transcripts (ech1 and hypothetical protein [hp] A4); only ech1 was confirmed in all resistant samples. (C) RT-PCR ratios in resistant samples expressing p53mt before (Rmt) and after irradiation (R′mt) and p53wt (R and R′) pointed out c-IAP1 as the more efficiently up-regulated transcript following irradiation in resistant samples expressing p53wt. Following irradiation, APR peptide appeared as up-regulated in p53mt-expressing samples and down-regulated in resistant samples expressing p53wt. Note that this transcript was neither validated nor confirmed as modified in resistant samples according to p53 mutation status. Fibromodulin (fmod, X75546) was validated as being induced by irradiation in resistant samples expressing p53mt (R′mt) as compared to sensitive (S′) and resistant (R′) samples, both expressing p53wt.
Validation of microarray selected genes by quantitative real-time RT-PCR; confirmation of validated genes on 22 B-CLL samples. Expression patterns of 16 selected genes by microarray analyses specifically regulated in the resistant B-CLL subset were verified by RT-PCR on 22 B-CLL samples: 7 samples included in microarray analyses, 7 new sensitive, 4 resistant expressing p53wt as well as 4 new resistant samples expressing p53mt. Microarray data on the expression levels of 14 genes were validated by RT-PCR and all but one gene were confirmed on all included B-CLL cell samples. The names of genes are indicated in red or green characters according to their up- or down-regulation. Data are expressed as means of duplicate experiments for each of 22 samples. The median + SD, median + SE, median, median – SE and median – SD are shown. (A) Relative constitutive expression levels (in absence of irradiation) in sensitive (S) and resistant (R) subset revealed 3 up-regulated transcripts (TR3 orphan receptor, HLA-DQA, and mtmr6), and 4 down-regulated transcripts (stat1, blk, MIP1α, and hsp27). (B) Relative expression levels before and after 3 hours of irradiation in sensitive (S and S′, respectively) and resistant (R and R′, respectively) samples revealed 4 up-regulated (mat2A, c-myc-P64, c-rel, and c-myc ORF114), and 2 down-regulated transcripts (ech1 and hypothetical protein [hp] A4); only ech1 was confirmed in all resistant samples. (C) RT-PCR ratios in resistant samples expressing p53mt before (Rmt) and after irradiation (R′mt) and p53wt (R and R′) pointed out c-IAP1 as the more efficiently up-regulated transcript following irradiation in resistant samples expressing p53wt. Following irradiation, APR peptide appeared as up-regulated in p53mt-expressing samples and down-regulated in resistant samples expressing p53wt. Note that this transcript was neither validated nor confirmed as modified in resistant samples according to p53 mutation status. Fibromodulin (fmod, X75546) was validated as being induced by irradiation in resistant samples expressing p53mt (R′mt) as compared to sensitive (S′) and resistant (R′) samples, both expressing p53wt.
Primers used for real-time RT-PCR
Gene . | Primer 1 (forward, 5′-3′) . | Primer 2 (reverse, 5′-3′) . |
|---|---|---|
| GAPDH | TCATCCATGACAACTTTGGTATCG | TGGCAGGTTTTTCTAGACGGC |
| L13740 | ATGGTGAAGGAAGTTGTCCGAAC | CCGGAGAGCAGGTCGTAGAAC |
| M34996 | AATTTGATGGAGATGAGGAGTTCTACG | AGCAGCGGTAGAGTTGTAGCGTT |
| U47635 | GGAACATGTACCATCAGTTTGATCG | TCGAAGAGCATCATTTACGGGTAG |
| M23178 | CCTGCTGCTTCAGCTACACCTC | TGGCTGCTCGTCTCAAAGTAGTC |
| M97935 | GTTACTTTCCCTGACATCATTCGC | GAATTCTACAGAGCCCACTATCCGA |
| Z23090 | CAGGACGAGCATGGCTACATCT | GGGATGGTGATCTCGTTGGAC |
| S76617 | CTTGGCTCGAATCATCGACAG | CATCAGGAGGACTCCAAACGAC |
| M13929 | CCAACAGGAACTATGACCTCGACTAC | CTCGAATTTCTTCCAGATATCCT |
| HT4899 | CTCCCTATCTACACTAACATCCCACG | TCCTTGTCCTGTGAGTATAAATC |
| X68836 | AACTGGCAGAACTACGCCGTAA | TGTATCCTCATCAAGGTATTTCG |
| X75042 | CGAACCCAATTTATGACAACCG | TTTTGTTTCTTTGCTTTATTGCCG |
| U16660 | GTAGAGTGCTTCAACAAGATTTCGAGA | CGTTGAAGGTCTCCTGGTATCG |
| U81556 | CCACTTGCCCCAGAGAAGCTA | AACCCGACAGCAAATAATGACTAGG |
| D90070 | TGGAAGTCGAGTGTGCTACTCAACT | AGATTCAGAAGTTTCTGCCGGAA |
| U37547 | TTCCAAGGTGTGAGTTCTTGATAC | AGAGCCATTCTGTTCTTCCGAAT |
| X75546 | CCCTATGACCCTTACCCGTATGA | CTGTGGCATTGTCAAAGACGC |
Gene . | Primer 1 (forward, 5′-3′) . | Primer 2 (reverse, 5′-3′) . |
|---|---|---|
| GAPDH | TCATCCATGACAACTTTGGTATCG | TGGCAGGTTTTTCTAGACGGC |
| L13740 | ATGGTGAAGGAAGTTGTCCGAAC | CCGGAGAGCAGGTCGTAGAAC |
| M34996 | AATTTGATGGAGATGAGGAGTTCTACG | AGCAGCGGTAGAGTTGTAGCGTT |
| U47635 | GGAACATGTACCATCAGTTTGATCG | TCGAAGAGCATCATTTACGGGTAG |
| M23178 | CCTGCTGCTTCAGCTACACCTC | TGGCTGCTCGTCTCAAAGTAGTC |
| M97935 | GTTACTTTCCCTGACATCATTCGC | GAATTCTACAGAGCCCACTATCCGA |
| Z23090 | CAGGACGAGCATGGCTACATCT | GGGATGGTGATCTCGTTGGAC |
| S76617 | CTTGGCTCGAATCATCGACAG | CATCAGGAGGACTCCAAACGAC |
| M13929 | CCAACAGGAACTATGACCTCGACTAC | CTCGAATTTCTTCCAGATATCCT |
| HT4899 | CTCCCTATCTACACTAACATCCCACG | TCCTTGTCCTGTGAGTATAAATC |
| X68836 | AACTGGCAGAACTACGCCGTAA | TGTATCCTCATCAAGGTATTTCG |
| X75042 | CGAACCCAATTTATGACAACCG | TTTTGTTTCTTTGCTTTATTGCCG |
| U16660 | GTAGAGTGCTTCAACAAGATTTCGAGA | CGTTGAAGGTCTCCTGGTATCG |
| U81556 | CCACTTGCCCCAGAGAAGCTA | AACCCGACAGCAAATAATGACTAGG |
| D90070 | TGGAAGTCGAGTGTGCTACTCAACT | AGATTCAGAAGTTTCTGCCGGAA |
| U37547 | TTCCAAGGTGTGAGTTCTTGATAC | AGAGCCATTCTGTTCTTCCGAAT |
| X75546 | CCCTATGACCCTTACCCGTATGA | CTGTGGCATTGTCAAAGACGC |
The expression levels of the genes selected from the microarray data analyses were validated and confirmed by real-time quantitative RT-PCR (LightCycler). Primers were designed with online software (WebPrimer, http://alces.med.umn.edu) and used in RT-PCR as described.
To validate the accuracy of the genes selected on the microarray, the same RNA samples extracted from 7 patients with CLL were used for both microarray hybridization and quantitative RT-PCR; 3 sensitive samples included as a single pooled sensitive sample in the first microarray were verified individually by RT-PCR. Fifteen new B-CLL cell RNA samples not included in the microarray hybridization were analyzed to further confirm the microarray results. Thus, 7 new sensitive and 8 new resistant samples were tested by this method.
Results
Comparative data analysis of specific modifications of gene expression levels in B-CLL cells resistant to irradiation-induced apoptosis
Based on the values of the 2 previously defined parameters, average difference (as > 0) and absolute call (as P or M), the comparison between resistant and sensitive samples before and after irradiation indicates that 472 genes before and 255 genes after irradiation appeared to be specifically expressed in resistant cells only, in both series of microarrays. However, a significantly higher number of genes were differentially expressed before rather than after irradiation, suggesting that irradiation may modify the expression of some genes common to both cell populations. To investigate these genes, cross-comparisons were performed using 2 other parameters: fold change equal to or more than 2 and sort score equal to or more than 0.5. Three levels of comparative analysis and transcript selection were carried out: R1/S1 and R2/S2 for resistant versus sensitive samples before and R′1/S′1, R′2/S′2 after irradiation in a first and second series of microarray data, respectively; R′m1/S′1 is a comparison of the expression profiles between resistant samples expressing p53mt and sensitive samples following irradiation; in a third cross-comparison, R′1/R1, R′2/R2, and Rm′1/Rm1, the data for resistant samples after irradiation were compared to those before irradiation for both microarrays (Figure 1). Both sensitive and resistant B-CLL samples expressing p53wt disclosed an accumulation of nuclear p53 level following irradiation (Figure 2). Thus, among 301 genes before and 449 genes after irradiation in a first series and 666 before and 924 after irradiation in a second series of microarray data—the comparison being made between resistant versus sensitive cell samples both expressing p53wt—the modified expression levels of 106 genes before and 160 genes after irradiation appeared to be specific to resistant cells. Ten identical transcripts were found to be down-regulated and 5 were up-regulated (R1/S1 and R2/S2). When the resistant samples expressing p53mt (Rm1 before and R′m1 after irradiation) were included in the comparison, then the expression levels of 35 and 81 genes varied before and after irradiation, respectively. Thirteen transcripts were up-regulated in resistant compared to sensitive samples after irradiation. Five transcripts found in a comparison of 14 up-regulated transcripts before and 49 after irradiation appeared to be specific for the resistant samples expressing p53wt. Similarly, 5 identical transcripts of 36 (ie, 32 transcripts specific to irradiated cells) were down-regulated by irradiation. Cross-comparison of these 5 transcripts with 10 transcripts found to be down-regulated in resistant versus sensitive samples (both expressing p53wt) revealed 5 transcripts that were specifically down-regulated before and after irradiation in all resistant samples tested, independently of p53 status. Additional comparisons among all resistant versus all sensitive samples after irradiation revealed 81 transcripts that were altered in resistant samples (13 up-regulated, 36 down-regulated, and 31 with unclear results between 2 respective samples, which were excluded). Resistant samples after irradiation compared with each other before irradiation revealed 93 transcripts whose expression levels were affected by irradiation. Finally, when sensitive samples before and after irradiation were included in a cross-comparison with the previous one, then 32 transcripts were found to have altered expression levels in all resistant B-CLL samples before and after irradiation. Cross-comparisons of these 3 groups of transcripts (81, 93, and 28) revealed 27 down-regulated and 31 up-regulated transcripts from which we finally selected 4 transcripts whose levels were up-regulated and 2 whose levels were down-regulated by irradiation specifically in resistant B-CLL samples, independently of p53 status. Finally, the number of genes selected for RT-PCR validation and confirmation was restricted to 16 (all shown in Figure 3) because all transcripts with values of absolute call (P and/or M) less than 0 before or after irradiation were excluded (overall comparison data are available on the Blood website; see the Supplemental Data Set link at the top of the online article).
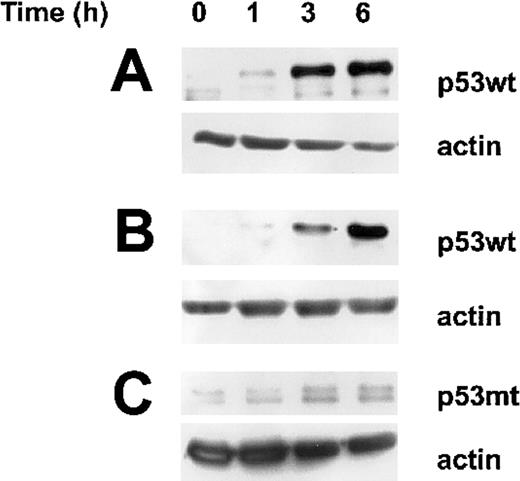
Accumulation of p53wt following irradiation in both sensitive and resistant B-CLL cells. Western blot analysis of the nuclear protein levels of wild-type (wt) and mutated (mt) p53 at indicated time after irradiation was compared to untreated cells (time 0). According to cell susceptibility to undergo or not apoptosis following irradiation, an equal amount of total proteins derived from 1.5 × 106 B-CLL cells sensitive (A) or resistant (B) both expressing p53wt and resistant cells expressing p53mt (C) was deposited in each lane and evidenced by actin protein level as control. The results are representative analyses of 11 sensitive and 6 resistant samples expressing p53wt as well as 5 resistant B-CLL samples expressing p53mt.
Accumulation of p53wt following irradiation in both sensitive and resistant B-CLL cells. Western blot analysis of the nuclear protein levels of wild-type (wt) and mutated (mt) p53 at indicated time after irradiation was compared to untreated cells (time 0). According to cell susceptibility to undergo or not apoptosis following irradiation, an equal amount of total proteins derived from 1.5 × 106 B-CLL cells sensitive (A) or resistant (B) both expressing p53wt and resistant cells expressing p53mt (C) was deposited in each lane and evidenced by actin protein level as control. The results are representative analyses of 11 sensitive and 6 resistant samples expressing p53wt as well as 5 resistant B-CLL samples expressing p53mt.
Microarray analysis of genes whose expression was potentially modified in a p53-dependent manner
Comparative data analyses were also performed to identify the potential p53 targets whose expression levels may be modified in a constitutive as well as irradiation-inducible manner (Figure 1). The included B-CLL samples expressing p53wt did not show LOH at ATM gene locus. In addition, the stabilization of p53wt following irradiation does not suggest a dysfunction of the factors involved in its upstream regulation (Figure 2). The first comparison crossed microarray expression levels in resistant samples expressing p53mt with those in sensitive samples expressing p53wt. This type of comparison indicated that 181 transcripts before and 279 after irradiation were up-regulated or down-regulated, respectively, in B-CLL samples expressing p53mt. When these genes were compared with those identified by comparing resistant versus sensitive B-CLL samples both expressing p53wt (2 different B-CLL samples for each comparison), then only one transcript was affected in a constitutive manner in the resistant sample expressing p53mt (sry-related hmg-box 12 protein, HG2887/X73039). Similarly, after irradiation the expression level of only fmod (fibromodulin, X75546) was deregulated in resistant samples expressing p53mt. When the comparison was made between resistant samples only, before and after irradiation, then the expression levels of 2 transcripts were affected in a p53-dependent manner following irradiation: APR peptide and c-IAP1, a human B homolog of MIH (D90070 and U37547). Thus, the expression levels of only 4 genes appeared as potentially modified in a p53-dependent manner.
Real-time quantitative RT-PCR validation of genes selected in the microarray analysis
The same RNA samples used in the microarray hybridizations and selected by comparative data analysis were further tested by quantitative, reproducible real-time RT-PCR. The efficacy of RT-PCR, represented by the slope of CP [CP = f[log(1/dilution)] with a value between –3.3 (100% efficacy) and –4 (80% efficacy), was calculated for each RT-PCR by introducing samples with known copy number (LightCycler; Roche Biochemicals). The specificity of the amplified products was tested during each experiment by plotting a melting slope obtained from progressive final denaturation (65°C-95°C in 5 minutes). The ratio of the fluorescence signal derivative/temperature derivative allowed control of the melting temperature for each amplified PCR product. Each product was confirmed to be a single peak and product size was checked on an agarose gel. Quantification of mRNA copy numbers was normalized to GAPDH copy number. All but one of the genes selected in the microarray analysis were validated by quantitative RT-PCR. Thus, genes whose expression was modified in resistant cells in the microarray analysis were retested for altered expression by RT-PCR. As expected, the fold change observed in the microarray analysis was often different from the ratios of increase or decrease calculated by PCR, but the direction of change (increase or decrease) was always the same. As in the case of the microarray hybridizations, RNA from sensitive cell samples (S1) was first tested as a pool of equal amounts of total RNA from 3 different sensitive B-CLL samples. Each B-CLL cell sample was then tested separately for the accuracy of the microarray data. Although the direction of change was the same in both the pooled and individual RNA samples, the latter showed significant differences by PCR. The gene expression ratios based on PCR/microarray data presented in Figure 3 show that 14 of the 16 genes selected by comparative microarray analysis were validated by quantitative PCR. The uncertainty for one gene (access number DO90070 corresponding to the EST of a small putative peptide) may arise from technical difficulties relating to the RT-PCR conditions and specific to this gene (short GC-rich sequence), rather than an inaccurate microarray hybridization result. Figures 3 and 4 show the real-time RT-PCR results that validated and confirmed the selected genes.
Additional B-CLL samples included in quantitative RT-PCR confirmation of the selected and validated genes
Fifteen new B-CLL samples (7 sensitive and 8 resistant) were included to extend and confirm the 14 genes previously selected by comparative microarray analysis and validated by real-time RTPCR. In this manner we were able to confirm 13 genes in all B-CLL samples tested. These confirmed genes are shown in Figures 2 and 3. TR3 nuclear orphan receptor, HLA-DQA1 (major histocompatibility complex [MHC] class II glycoprotein) and mtmr6 (myotubularinrelated protein 6; access numbers L13740, M34996 and U47635, respectively), were constitutively up-regulated, whereas MIP1α, stat1, blk, and hsp27 (access numbers M23178, M97935, S76617, and Z23090, respectively) were constitutively down-regulated in all resistant B-CLL samples tested. Three hours after irradiation, the expression levels of c-myc (present in 2 array probes: c-myc-P64 and c-myc-ORF114), mat2A, and c-rel (access numbers M13929, HT4899, X68836, and X75 042) were up-regulated, whereas that of ech1 (U16660) was down-regulated.
Discussion
The fate of B-CLL cells exposed to DNA damaging agents in vivo during clinical treatment or in vitro in studies of their sensitivity to DNA damage-induced apoptosis, should depend, at least in part, on both constitutive and inducible gene expression. Exposure to such agents can result in cell survival, if checkpoint controls and DNA repair are effectively activated following cell stress, or in cell death when these first events are defective and when apoptotic mechanisms are functional. The main obstacle to the molecular characterization of B-CLL is the marked heterogeneity of this pathology. Recent microarray data suggest that the prognosis of B-CLL may be related to an altered expression profile of the genes involved in lymphocyte trafficking.12 The previously defined worst prognostic group of CLL patients lacking IgV mutations is thought to correlate with a specific gene expression pattern in this CLL subtype.10,11
One of the common features of B-CLL cells is their prolonged survival in peripheral blood, which is obviously due to not yet defined defect in apoptotic cell death.1 Although this defect may be specific to cell death mechanisms, it may also result from an altered function of the biochemical safeguards essential for cell life (such as we reported for the ubiquitin-proteasome system7,9 ), thus underscoring the complex processes at work. All of these data point toward the existence of complex alterations, not only in gene expression but also protein activities, that occur in B-CLL cells simultaneously.
In this work, to understand why a subset of B-CLL cells does not activate apoptotic death pathway in response to DNA damage, we used a DNA microarray approach already shown to be useful in defining global gene expression patterns in cancer cells.19, 20, 21, 22, 23, 24 Two large microarray studies on B-CLL expression profiling10,11 have underlined significant differences according to IgVH mutational status. These differences are obviously due to sample heterogeneity of each series of patients and may suggest that in addition to the common B-CLL gene pattern, the individual gene expression signature cannot be avoided even by using large patient series. In this regard, our initial concept was to point out the genes specifically expressed in the subsets resistant to irradiation-induced apoptosis, using a minimal number of CLL samples for microarray analysis. The genes issued from this analysis were then validated and confirmed on large series of CLL by a reliable and quantitative real-time RT-PCR. Essential criteria of sample inclusions was their susceptibility to undergo or not apoptotic death pathway induced by irradiation The time point of RNA sampling at 3 hours after irradiation was arbitrarily chosen on the basis of the recovery of residual DNA damage in B-CLL.25 Further at this time point, the irradiation-induced apoptosis is still in a phase of initiation but not of execution. Effectively, other studies previously showed that apoptosis induced by γ irradiation in human lymphocytes occurs 24 hours after irradiation9 and that the process is reversible in the initiation phase up to 6 hours after irradiation.26 We also showed that resistant, as compared to sensitive, B-CLL cells display an accelerated kinetics of recovery of DNA damage (single- and double-strand breaks) and that repair was achieved within 2 hours after irradiation in both CLL subsets.25 Thus, the purpose of this study was to identify the specific mRNA expression pattern before and at the time of apoptotic signaling (3 hours after irradiation) rather than to establish the kinetics of gene activation in response to irradiation, which per se is of interest and deserves further studies.
The comparative gene expression patterns between sensitive and resistant B-CLL cells (both expressing p53wt) on a single microarray revealed that the expression levels of 301 genes before and 449 genes after irradiation were modified equal to or more than 2-fold in resistant B-CLL cells. In a second series of microarrays using RNA from B-CLL samples not included in the first series, this number was 666 before and 924 after irradiation, respectively. This significant difference is obviously due to the heterogeneity in gene expression among B-CLL samples (displaying the same radiation sensitivity). Accordingly, the number of genes whose expression levels varied at least 2-fold before and after irradiation was found to be similar in resistant B-CLL samples expressing p53mt, when compared to each other and to 2 different sensitive samples (S1 and S2; modified expression levels of 460 transcripts before and 443 after irradiation). This clearly indicates a heterogeneous expression before and after irradiation in each B-CLL cell sample (sensitive and resistant). For this reason we further compared the genes whose expression levels were modified at least 2-fold in the 2 microarray analyses. Comparative data analysis led to the identification of 14 genes, with one redundant c-myc sequence, and 2 ESTs. The expression levels of these genes were specifically modified before or after irradiation in resistant cells only.
Interestingly, among the up- and down-regulations were genes coding for transcriptional factors whose direct or indirect roles in apoptotic death control have already been described. For instance, mitochondrial translocation of nuclear orphan receptor TR3 and cytochrome c release, but not its DNA binding and transcriptional functions, are associated with apoptosis in the human prostate cancer cell line LNCaP.27 However, the fact that the expression of this transcription factor was found to be constitutively up-regulated in resistant B-CLL cells suggests a transcriptional rather than a proapoptotic role. Similarly, the overexpression of c-myc after irradiation in resistant cells, if truly followed by overexpression at the protein level, relates more to its transcriptional functions than to its role in inducing apoptosis.28 In parallel, bcl-2, which is constitutively overexpressed in all B-CLL samples,29 might inhibit the apoptotic function of overexpressed c-myc, thereby explaining the bypass of Myc-associated apoptosis induction observed in murine lymphomas.30 The observed overexpression of the c-rel oncogene, shown to protect B cells from apoptosis activation,31 may also impair c-Myc apoptotic functions32 in B-CLL cells expressing p53wt. Although activation of nuclear factor κB (NF-κB) is often associated with tumor cell resistance to apoptosis in general, an overexpression of c-rel (a member of the NF-κB transcription factor family) has never been reported to be associated with an apoptosis-resistant subset of B-CLL. Methionine adenosyltransferase mat2A, which catalyzes S-adenosyl-methionine formation, was up-regulated in resistant cells after irradiation, suggesting a methylation defect (as in the case of the bcl-2 gene in B-CLL cells), as observed in human hepatocarcinoma cells.33 Some genes, including hsp27, involved in differentiation of human leukemic cells34 ; a gene coding for a signal transducer and activator of transcription 1 (STAT1α, β), a member of the family of STAT factors35,36 ; macrophage inhibitor protein 1α (MIP1α), a homolog of the inhibitor of lymphocyte proliferation G0S19,37 which is believed to be an antioncogene; and a gene encoding the B-cell specific src-containing tyrosine kinase blk38 were found to be constitutively down-regulated. These genes are involved in various cell processes such as differentiation, proliferation, and apoptosis. Our data suggest that the defect linked to differentiation and proliferation in this subset of B-CLL may relate to their resistance to efficient apoptosis activation. This defect sustains the specific subset of B-CLL and may also help explain the observed B-CLL heterogeneity by the additional potential oncogenic properties of blk,39 an oncogenic fusion of TEC as the family member of TR3, was described in myxoid chondrosarcomas40 and mtmr6 (locus D13S824E), which is found here to be constitutively overexpressed. This latter locus is reported to be associated in reciprocal translocation 13q12 and 8p11 in myeloid leukemias.19,41
Concerning apoptotic death susceptibility in response to DNA damage, deregulation of mtmr6 and blk may be of particular interest because both participate in phosphatidylinositol 3-kinase signaling and thus potentially in repair of DNA damage. This may also reveal a new signaling pathway controlling DNA repair or apoptosis that appears to be deregulated in a resistant subset that disclosed normal p53wt stabilization following irradiation. The limited number of potential p53-responsive genes associated with apoptotic death susceptibility should come as no surprise. Indeed, the microarray approach used to define p53-responsive genes revealed that DNA damage-induced genes are cell-type specific but also that the majority of responsive genes show no significant p53-dependent regulation.19,23 Moreover, a potentially large class of p53-independent genes inducible by irradiation stress has already been suggested for leukemic cell lines.19 In our study, 460 genes before and 443 genes after irradiation appeared to be specifically expressed in resistant B-CLL cell samples expressing mutated p53. Unexpectedly, irradiation deregulated the expression of 1028 genes in resistant B-CLL cells expressing p53mt but only 186 genes in resistant cells expressing p53wt. When genes whose expression increased after irradiation in resistant p53mt samples were compared with down-regulated transcripts in resistant samples expressing p53wt and with sensitive samples, then only 4 genes were expressed in a p53-dependent manner. Furthermore, RT-PCR confirmed that only 2 genes appeared to be specifically upregulated in a p53-dependent manner: MIHB, a homolog of the c-IAP1 inhibitor of apoptosis, in p53wt-resistant B-CLL cells, and fibromodulin in cells expressing p53mt. Although a direct role of MIHB/c-IAP1 in apoptotic death control has previously been reported,42 that of fibromodulin43 has never been suggested elsewhere and may not be linked to the apoptotic response at all. Its up-regulation (7-fold induction in the microarray analysis) after irradiation in resistant B-CLL cells expressing p53mt simply suggests that its expression may be negatively regulated by p53, whose regulation is altered in all p53wt B-CLL samples.7 Fibromodulin, a member of the multifunctional family of leucine-rich proteoglycans,44 interacts with fibrillar collagens to allow correct matrix assembly, as well as with growth factors such as transforming growth factor β (TGF-β), which is then negatively regulated, thus expanding its role to the promotion of cell growth and differentiation.45 Decorin, a structural and functional homolog of fibromodulin,42 was suggested to exert antioncogenic activity by acting as a signal of suppression of growth and differentiation in breast carcinoma cells.46 The overexpression of fibromodulin may be a hallmark of malignant B-CLL cells, as previously observed in CLL versus memory B cells.10 Our data further indicate that its higher expression is associated with a specific subset of resistant B-CLL cells expressing p53mt.
The present work highlights the utility of microarray analysis followed by quantitative RT-PCR confirmation in defining both constitutive and inducible gene expression profiles. Such an approach should contribute toward a better understanding of the heterogeneous response of B-CLL cells to DNA damage-induced apoptosis. The constitutive and inducible defects found here are mainly linked to proliferation/oncogenesis and appear specific for the resistant B-CLL subset, suggesting potentially new and unsuspected roles in apoptotic death control for certain genes such as nuclear orphan receptor TR3, mtmr6, and HLA-DQA1. The latter gene has already been indirectly implicated in inhibition of cell proliferation,47 suggesting that this resistant subset may also harbor specific alterations in addition to the general proliferation defects observed in B-CLL cells. Microarray analysis has already led to the identification of global gene expression patterns that may be a specific “signature” of B-CLL cells.10,11 It may also be used for differential gene profiling to discriminate between B-CLL cells sensitive or resistant to a given treatment, serving to complement the specific profile observed in the B-CLL subset with IgV mutations and thus providing a new prognostic marker of CLL.
Prepublished online as Blood First Edition Paper, February 13, 2003; DOI 10.1182/blood-2002-06-1743.
Supported by Fondation de France, Association pour la Recherche sur le Cancer (ARC), Electricité de France (EDF), Institut Curie, CEA, and CEC RADINSTAB FIGH-1999-00003. L.V. received a fellowship from Académie Nationale de Médecine.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We are grateful to all the volunteer leukemic blood donors.

![Figure 4. Validation of microarray selected genes by quantitative real-time RT-PCR; confirmation of validated genes on 22 B-CLL samples. Expression patterns of 16 selected genes by microarray analyses specifically regulated in the resistant B-CLL subset were verified by RT-PCR on 22 B-CLL samples: 7 samples included in microarray analyses, 7 new sensitive, 4 resistant expressing p53wt as well as 4 new resistant samples expressing p53mt. Microarray data on the expression levels of 14 genes were validated by RT-PCR and all but one gene were confirmed on all included B-CLL cell samples. The names of genes are indicated in red or green characters according to their up- or down-regulation. Data are expressed as means of duplicate experiments for each of 22 samples. The median + SD, median + SE, median, median – SE and median – SD are shown. (A) Relative constitutive expression levels (in absence of irradiation) in sensitive (S) and resistant (R) subset revealed 3 up-regulated transcripts (TR3 orphan receptor, HLA-DQA, and mtmr6), and 4 down-regulated transcripts (stat1, blk, MIP1α, and hsp27). (B) Relative expression levels before and after 3 hours of irradiation in sensitive (S and S′, respectively) and resistant (R and R′, respectively) samples revealed 4 up-regulated (mat2A, c-myc-P64, c-rel, and c-myc ORF114), and 2 down-regulated transcripts (ech1 and hypothetical protein [hp] A4); only ech1 was confirmed in all resistant samples. (C) RT-PCR ratios in resistant samples expressing p53mt before (Rmt) and after irradiation (R′mt) and p53wt (R and R′) pointed out c-IAP1 as the more efficiently up-regulated transcript following irradiation in resistant samples expressing p53wt. Following irradiation, APR peptide appeared as up-regulated in p53mt-expressing samples and down-regulated in resistant samples expressing p53wt. Note that this transcript was neither validated nor confirmed as modified in resistant samples according to p53 mutation status. Fibromodulin (fmod, X75546) was validated as being induced by irradiation in resistant samples expressing p53mt (R′mt) as compared to sensitive (S′) and resistant (R′) samples, both expressing p53wt.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/11/10.1182_blood-2002-06-1743/5/m_h81134381004.jpeg?Expires=1770855206&Signature=fkHvWYsac5ooeFSBk79kCwB4UhkztYSfX5H~Je-96fgc1dxILoqGc-byy3FKiaweyoL7nbrsbTXBdMSQLn0RksgK71froIYXtuDehoLRNs0q~vKrfMkXxfgIkgTF07bXoauBTw5N0vjcO17SXH8eyLXPvubus~u~cZ87CIw218-4g0yKjvj-ZCn5tvZ3u2rBwdRwtvOB8DB3tGJHl02E1Lhd2oLxrsP9nBQEACSmGZF4vl1lz9vFxdLEuAvp016x1V-gnmBY2-aMAiFyfC9Qmoqfbnos3cq8fPVpwOi6rfPmOpit3s7capXabfI1p1uLOej9WqSafkJiXX5MpEI9rg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal