Abstract
Nonrandom recurrent chromosomal abnormalities are ubiquitous in multiple myeloma (MM) and include, among others, translocations of the immunoglobulin heavy chain locus (IgH). IgH translocations in MM result in the up-regulation of oncogenes, and include more commonly t(11;14)(q13;q32), t(4;14)(p16;q32), and t(14;16)(q32;q23). Based on the recurrent nature of these translocations and their finding since the early stages of the plasma cell (PC) disorders, we hypothesized that they would confer biologic and clinical variability. In addition, deletions of 13q14 and 17p13 have also been associated with a shortened survival. We used cytoplasmic Ig—enhanced interphase fluorescent in situ hybridization to detect deletions (13q14 and 17p13.1), and translocations involving IgH in 351 patients treated with conventional chemotherapy entered into the Eastern Cooperative Oncology Group clinical trial E9486/9487. Translocations were frequently unbalanced with loss of one of the derivative chromosomes. The presence of t(4; 14)(p16;q32) (n = 42; 26 vs 45 months, P < .001), t(14;16)(q32;q23) (n = 15; 16 vs 41 months, P = .003), – 17p13 (n = 37; 23 vs 44 months, P = .005), and – 13q14 (n = 176; 35 vs 51 months, P = .028) were associated with shorter survival. A stratification of patients into 3 distinct categories allowed for prognostication: poor prognosis group (t(4;14)(p16;q32), t(14; 16)(q32;q23), and – 17p13), intermediate prognosis (– 13q14), and good prognosis group (all others), with median survivals of 24.7, 42.3, and 50.5 months, respectively (P < .001). This molecular cytogenetic classification identifies patients into poor, intermediate, and good risk categories. More importantly it provides further compelling evidence that MM is composed of subgroups of patients categorized according to their underlying genomic aberrations.
Introduction
Genetic and cytogenetic abnormalities define subgroups of hematologic neoplasms, and accordingly have been associated with unique biologic, clinical, and prognostic features.1 Recent studies with interphase fluorescent in situ hybridization (FISH) indicate that all multiple myeloma (MM) cells harbor chromosome abnormalities.2,3 Interphase FISH—detected chromosomal abnormalities studies have also been associated with dissimilar outcomes in some reports.4, 5, 6, 7, 8, 9
We and others have proposed that specific cytogenetic abnormalities can identify groups of MM patients with unique clinical and biologic features.9, 10, 11, 12 Abnormalities of chromosome 13 (Δ13), monosomy representing 85% of them, have an adverse prognosis in MM when detected by metaphase analysis and interphase FISH.4, 5, 6,8,13,14 Likewise, deletions of 17p13.1, the genomic locus of the p53 tumor suppressor gene, have been associated with an adverse patient outcome.15 Translocations involving immunoglobulin heavy chain locus (IgH) (14q32) are seen in 50% to 60% of MM patients, and involve an array of nonrandom recurrent chromosomal partners, but their prognostic significance has not been tested.16,17 The 3 most common IgH translocations in MM are t(4;14)(p16.3;q32), t(11;14)(q13;q32), and t(14;16)(q32;q23).10,11 In this paper we evaluate and integrate the clinical and biologic relevance of the most common cytogenetic abnormalities. To do so we used interphase FISH in a large cohort of MM patients who have had long duration of follow-up.
Patients and methods
Patient characteristics
Patients enrolled in the Eastern Cooperative Oncology Group (ECOG) E9486 and its associated correlative laboratory clinical trial E9487 (N = 561) had newly diagnosed MM and have been described in detail elsewhere.18 They were randomized to receive treatment with conventional chemotherapy variations.18 The median overall survival for the whole group was 40.5 months. Patients have extensive follow-up information with the minimum follow-up of survivors being 96 months (range, 96-138 months), resulting in negligible censoring. A total of 351 patients were included in this study for our analysis (Table 1), as previously described by us,19 and appeared to be no different from the larger cohort of patients when all relevant biologic and prognostic factors are considered (data not shown). Pertinent clinical and prognostic features are available for the majority of the patients including, among others, the plasma cell labeling index (PCLI), β2-microglobulin, C-reactive protein, and serum level of soluble interleukin-6 receptor (sIL-6R). These patients did not have conventional karyotype analysis requested at the time of study entry, and it is thus not available for comparison.
Baseline clinical and laboratory descriptive features of patients by abnormality
. | Cytogenetic abnormality . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable . | All (n = 351) . | t(4;14)(p16;q32) (n = 42) . | t(14;16)(q32;q23) (n = 15) . | t(11;14)(q13;q32) (n = 53) . | ▵13 (n = 176) . | Del 17p13.1 (n = 37) . | |||||
| Median age, y (range) | 63 (35-84) | 59 (35-74) | 58 (41-75) | 62 (35-80) | 62 (35-82) | 64 (40-78) | |||||
| Sex, male/female, % | 62/38 | 55/45 | 47/53 | 68/32 | 57/43 | 54/46 | |||||
| ECOG PS, % | |||||||||||
| 0 to 1 | 86 | 90 | 100 | 91 | 88 | 84 | |||||
| 2 to 4 | 14 | 10 | 0 | 9 | 12 | 16 | |||||
| Plasmacytoma, % | |||||||||||
| Yes | 10 | 12 | 13 | 6 | 11 | 24 | |||||
| Lytic bone lesions, % | |||||||||||
| Yes | 61 | 62 | 54 | 62 | 60 | 70 | |||||
| Hypercalcemia, %, | |||||||||||
| Ca2+ less than 12 mg/dL | 24 | 21 | 27 | 23 | 26 | 43 | |||||
| Serum M component, % | |||||||||||
| Present, 1 or more g/dL | 83 | 90 | 73 | 72 | 78 | 78 | |||||
| Absent | 17 | 10 | 27 | 28 | 22 | 22 | |||||
| Urine M component, % | |||||||||||
| Present, detectable | 72 | 69 | 67 | 68 | 76 | 84 | |||||
| Absent | 25 | 24 | 33 | 30 | 20 | 14 | |||||
| Unknown | 3 | 7 | 0 | 2 | 5 | 3 | |||||
| Light chain type, % | |||||||||||
| κ | 63 | 48 | 40 | 60 | 59 | 54 | |||||
| λ | 33 | 50 | 60 | 34 | 36 | 46 | |||||
| Unknown | 3 | 2 | 0 | 6 | 5 | 0 | |||||
| Hemoglobin, g/dL* | 10.7 (5.1-15.8) | 9.8 (5.1-13.9) | 10.3 (7.7-13.0) | 10.5 (6.2-15.4) | 10.6 (5.1-15.5) | 10.4 (6.9-14.0) | |||||
| Peripheral blood PCs, % | 0 (0-93) | 0 (0-8) | 0 (0-12) | 0 (0-10) | 0 (0-93) | 0 (0-56) | |||||
| Bone marrow PCs, % | 43 (2-99) | 43 (4-98) | 75 (12-99) | 50 (8-86) | 42 (2-99) | 50 (11-97) | |||||
| Creatinine, mg/dL† | 1.2 (0.4-4.9) | 1.2 (0.6-4.8) | 1.1 (0.4-4.9) | 1.3 (0.5-4.7) | 1.2 (0.5-4.8) | 1.2 (0.4-4.8) | |||||
| β2-microglobulin, mg/dL‡ | 3.7 (0.6-64.0) | 3.9 (0.6-21.3) | 5.4 (1.0-64.0) | 4.0 (0.9-18.4) | 3.8 (0.6-30.3) | 4.2 (1.0-23.9) | |||||
| PCLI, % of PC | 0.4 (0-15.4) | 0.6 (0-13.2) | 1.0 (0-10) | 0.4 (0-5.9) | 0.6 (0-13.2) | 1.1 (0-10.9) | |||||
| sIL-6R, ng/mL | 187 (50-1067) | 230 (60-500) | 313 (133-733) | 156 (65-800) | 204 (50-870) | 235 (115-573) | |||||
. | Cytogenetic abnormality . | . | . | . | . | . | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Variable . | All (n = 351) . | t(4;14)(p16;q32) (n = 42) . | t(14;16)(q32;q23) (n = 15) . | t(11;14)(q13;q32) (n = 53) . | ▵13 (n = 176) . | Del 17p13.1 (n = 37) . | |||||
| Median age, y (range) | 63 (35-84) | 59 (35-74) | 58 (41-75) | 62 (35-80) | 62 (35-82) | 64 (40-78) | |||||
| Sex, male/female, % | 62/38 | 55/45 | 47/53 | 68/32 | 57/43 | 54/46 | |||||
| ECOG PS, % | |||||||||||
| 0 to 1 | 86 | 90 | 100 | 91 | 88 | 84 | |||||
| 2 to 4 | 14 | 10 | 0 | 9 | 12 | 16 | |||||
| Plasmacytoma, % | |||||||||||
| Yes | 10 | 12 | 13 | 6 | 11 | 24 | |||||
| Lytic bone lesions, % | |||||||||||
| Yes | 61 | 62 | 54 | 62 | 60 | 70 | |||||
| Hypercalcemia, %, | |||||||||||
| Ca2+ less than 12 mg/dL | 24 | 21 | 27 | 23 | 26 | 43 | |||||
| Serum M component, % | |||||||||||
| Present, 1 or more g/dL | 83 | 90 | 73 | 72 | 78 | 78 | |||||
| Absent | 17 | 10 | 27 | 28 | 22 | 22 | |||||
| Urine M component, % | |||||||||||
| Present, detectable | 72 | 69 | 67 | 68 | 76 | 84 | |||||
| Absent | 25 | 24 | 33 | 30 | 20 | 14 | |||||
| Unknown | 3 | 7 | 0 | 2 | 5 | 3 | |||||
| Light chain type, % | |||||||||||
| κ | 63 | 48 | 40 | 60 | 59 | 54 | |||||
| λ | 33 | 50 | 60 | 34 | 36 | 46 | |||||
| Unknown | 3 | 2 | 0 | 6 | 5 | 0 | |||||
| Hemoglobin, g/dL* | 10.7 (5.1-15.8) | 9.8 (5.1-13.9) | 10.3 (7.7-13.0) | 10.5 (6.2-15.4) | 10.6 (5.1-15.5) | 10.4 (6.9-14.0) | |||||
| Peripheral blood PCs, % | 0 (0-93) | 0 (0-8) | 0 (0-12) | 0 (0-10) | 0 (0-93) | 0 (0-56) | |||||
| Bone marrow PCs, % | 43 (2-99) | 43 (4-98) | 75 (12-99) | 50 (8-86) | 42 (2-99) | 50 (11-97) | |||||
| Creatinine, mg/dL† | 1.2 (0.4-4.9) | 1.2 (0.6-4.8) | 1.1 (0.4-4.9) | 1.3 (0.5-4.7) | 1.2 (0.5-4.8) | 1.2 (0.4-4.8) | |||||
| β2-microglobulin, mg/dL‡ | 3.7 (0.6-64.0) | 3.9 (0.6-21.3) | 5.4 (1.0-64.0) | 4.0 (0.9-18.4) | 3.8 (0.6-30.3) | 4.2 (1.0-23.9) | |||||
| PCLI, % of PC | 0.4 (0-15.4) | 0.6 (0-13.2) | 1.0 (0-10) | 0.4 (0-5.9) | 0.6 (0-13.2) | 1.1 (0-10.9) | |||||
| sIL-6R, ng/mL | 187 (50-1067) | 230 (60-500) | 313 (133-733) | 156 (65-800) | 204 (50-870) | 235 (115-573) | |||||
Numbers in parentheses denote range.
Convert hemoglobin to SI units: multiply g/dL × 10 = g/L.
Convert creatinine to SI units: multiply mg/dL × 76.25 = μM.
Convert β2-microglobulin to SI units:
Bone marrow samples
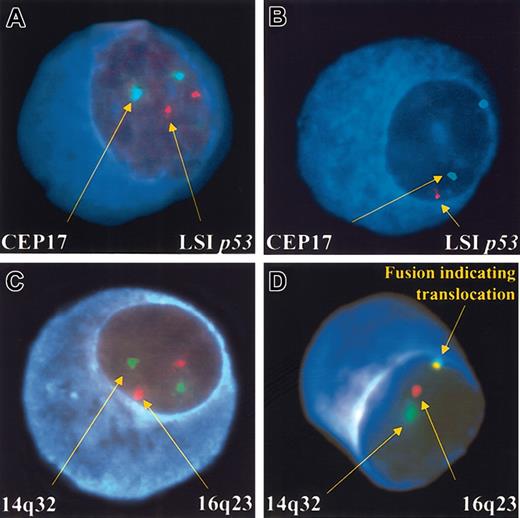
Institutional review board (IRB) approval, informed consent, and bone marrow research samples were obtained (in that order), and cytospin slides were stored for future use (at – 70°C). Aspirate samples were enriched for mononuclear cells using the Ficol method. To improve on the specificity of the scoring process we combined interphase FISH with immune-fluorescent detection of the cytoplasmic-immunoglobulin light-chain20 (Figure 1).
PCs with both the normal and abnormal pattern of hybridization. The depicted PCs show normal (A) and abnormal (B) patterns of hybridization. All panels show the blue fluorescence of the cytoplasm allowing the clone-specific interphase FISH scoring. (A) A cell with the normal configuration of 2 pairs of signals for the probes localizing to the centromere 17 (CEP17; green) and the 17p13.1 (LSI p53) (red) probe. (B) A cell with deletion of 17p13.1. There are 2 green signals arising from the centromeric probe but only 1 red signal from the p53 locus probe. (C) A normal configuration of probes used to detect the t(14;16)(q32;q23). The locus-specific 14q32 probes are labeled in green, and the 16q23 probes are labeled in red. (D) A cell with fusion of probes for 14q32 (green) and 16q23 (red). The 2 signals in proximity generate a fusion. If a significant number of cells scored showed this pattern, a patient is said to have a translocation.
PCs with both the normal and abnormal pattern of hybridization. The depicted PCs show normal (A) and abnormal (B) patterns of hybridization. All panels show the blue fluorescence of the cytoplasm allowing the clone-specific interphase FISH scoring. (A) A cell with the normal configuration of 2 pairs of signals for the probes localizing to the centromere 17 (CEP17; green) and the 17p13.1 (LSI p53) (red) probe. (B) A cell with deletion of 17p13.1. There are 2 green signals arising from the centromeric probe but only 1 red signal from the p53 locus probe. (C) A normal configuration of probes used to detect the t(14;16)(q32;q23). The locus-specific 14q32 probes are labeled in green, and the 16q23 probes are labeled in red. (D) A cell with fusion of probes for 14q32 (green) and 16q23 (red). The 2 signals in proximity generate a fusion. If a significant number of cells scored showed this pattern, a patient is said to have a translocation.
Probes
We used previously reported sets of probes to detect Δ13, t(11;14)(q31; q32), and t(4;14)(p16.3;q32).19,21,22 This same cohort of patients has been separately reported regarding the t(11;14)(q13;q32) and Δ13.7,8 To detect t(14;16)(q32;q23) we used the same 14q32 chromosome probes previously described by us,21 in combination with 2 bacterial artificial chromosome (BAC) clones (356D21 and 484H2; Research Genetics, Huntsville, AL) that localize to 16q23, and BAC clones 10205 and 10206 described by Chesi et al.23 To test for 17p13.1, we used a locus specific probe (LSI) p53 probe from Vysis (Downers Grove, IL). We used standard hybridization, validation, and scoring procedures as described previously by us.19,21 We scored 100 cells for each one of the abnormalities and recorded the percentage of cells with abnormal patterns (with special attention to the number of fusions detected for the translocations).
Statistical analysis
Descriptive and survival analysis. To test for association between abnormalities, or between abnormalities and other patient categoric treatment characteristics or response to treatment, the Fisher exact test was used.24 To test for difference in PCLI and β2-microglobulin between patients with and without an abnormality, a Wilcoxon rank sum test was used.25 The distribution for overall and progression-free survival was estimated using the method of Kaplan and Meier.26 The log-rank test was used to test for differences in survival between groups.27 We decided to score 100 cells per patient to evaluate positivity and evidence of clonal heterogeneity.
Multiple regression model. A Bayesian analysis was used to evaluate the contribution of genetic abnormalities to survival.28 This analysis allowed us to include all of the studied patients in the model, even the patients with missing data. The following clinical prognostic factors (cut-off points), dichotomized according to previously reported studies, were included in the model: PCLI (< 1% vs ≥ 1%), bone marrow PC percentage (< 30% vs ≥ 30%), serum creatinine (< 152.5 vs ≥ 152.5 μM), albumin (≤ 30 vs > 30 g/L), hemoglobin (≤ 100 vs > 100 g/L), β2-microglobulin (< 2.7 vs ≥ 2.7 mg/dL), soluble IL-6 receptor (< 270 vs ≥ 270 ng/mL), C-reactive protein (< 2 vs ≥ 2 mg/dL), serum monoclonal protein (< 10 vs ≥ 10 g/L), and stage (I-II vs III).
A Weibull distribution was used to model time to death.29 The regression coefficients and the shape parameter were given “noninformative” normal and gamma priors, respectively. Prior distributions for the covariates were assumed to be binomial (P, 1) with P distributed as uniform (0, 1). The BUGS program (Bayesian inference using the Gibbs sampling algorithm)28 was used to estimate the coefficients and obtain 95% credible confidence intervals (CIs). We performed an initial 500 burn-in of iterations followed by an additional 10 000 iterations. Parameter estimates are the mean and standard deviation based on the Gibbs samples; credible intervals are computed as the lower and upper percentiles from the last 10 000 iterations. The 5 genetic abnormalities as well as the clinical prognostic factors that appeared to be statistically significant (the 95% credible interval did not contain the null value) were included in the final model.
The appropriateness of the Weibull distribution, the adequacy of the fitted multivariate model time, and the validity of the Bayesian model (“missing at random” assumption) were checked. The results obtained using the Bayesian approach were compared with the results obtained using the Cox regression model in the subset of 275 patients that had complete data.
Hierarchic groups. Based on hazard ratios from the results of univariate and multivariate analyses, 3 hierarchic groups were created, in which 1 patient was allocated to 1 group only. These groups were based on whether patients had genetic abnormalities associated with poor, intermediate, or good prognosis.
Results
Prevalence of the abnormalities
The prevalence of chromosomal abnormalities among the 351 patients tested are as follows: t(4;14)(p16;q32) (42/332 patients, 12.7%), t(14;16)(q32;q23) (15/323 patients, 4.6%), t(11;14)(q13; q32) (53/336 patients, 15.8%), deletions 17p13.1 (37/345 patients, 10.7%), and Δ13 (176/325 patients, 54.2%). The prevalence of the abnormalities was not significantly different according to the stage of the disease or age, except for Δ13, which appeared to be more common among stage III patients (P = .014). A strong association was noted between Δ13 and the t(4;14)(p16.3;q32) as we have previously reported (38/42 patients; P < .001),22 but this association was not present in patients with t(14;16)(q32;q23) (8/13 patients) or t(11;14)(q13;q32) (24/51 patients). A significant positive correlation between deletion 17p13.1 and t(14;16)(q32;q23) was observed (5/15 patients, 33%; P = .018), and was suggested for t(4;14)(p16;q32) (8/42 patients, 20%). In contrast 17p13.1 deletions had a lower incidence in patients with t(11;14)(q13;q32) (P = .027).
Translocation patterns and relations
In this cohort of patients all translocations were mutually exclusive; that is, there were no patients with 2 coexistent translocations. However, many patients had combinations of a translocation and deletion(s) of 17p13.1 and/or Δ13. The median percentage of cells with an abnormality was more than 80% for all abnormalities. IgH translocations were usually seen in more than 95% of cells. Deletions 17p13.1 were seen in less than 50% of cells in 11 (30%) of 37 cases. Using the specific sets of probes we found that of 348 evaluable patients, 139 (40%) had 1 chromosomal abnormality detected, 77 (22%) had 2, and 10 (3%) had 3. Using these probes, a total of 122 patients had no abnormalities detected.
Prognostic features including the PCLI and β2-microglobulin
Patients had unique biologic and prognostic features according to their baseline prognostic features (Table 1). The PCLI was significantly higher among patients with Δ13 (P = .03), t(14; 16)(q32;q23) (P = .02), or deletion 17p13.1 (P = .01). Serum levels of β2-microglobulin appeared to be significantly higher in patients with deletion 17p13.1 (P = .03). Deletions of 17p13.1 were significantly associated with hypercalcemia (P = .009) and soft-tissue plasmacytomas (P = .0053). The use of λ light chain was favored in patients with t(14;16)(q23;q32) (P = .05). Serum levels of sIL6-R were higher in patients with Δ13 (P = .003), t(4;14)(p16.3;q32) (P = .025), t(14;16)(q32;q23) (P = .009), and deletions of 17p13.1 (P = .006). Patients with t(4;14)(p16.3;q32) were significantly more likely to have a serum monoclonal protein higher than 30 g/L (P = .019). Light-chain only disease was slightly more common among those patients with t(11;14)(q13; q32) (28% among patients with t(11;14)(q13;q32) and 16% in those without t(11;14)(q13;q32); Fisher exact, P = .04). The IgA isotype was slightly more common among patients with the t(4;14)(p16.3;q32) but was not significant (P > .2). No trend was observed among patients with t(14;16)(q32;q23).
Balanced versus unbalanced translocations
A predominant pattern of one fusion signal was seen in only 56 (51%) of 110 patients with evidence of a translocation by the fusion strategy. When one considers only cases in which the predominant pattern was seen in more than 90% of the clonal cells the total was 34 patients (33% of all IgH translocations). This is remarkably different than previous assumptions of balanced translocations in MM (Table 2). In a recent study by Keats et al that used reverse transcriptase—polymerase chain reaction (RT-PCR)—based strategies, they found that up to one third of patients with the t(4;14)(p16.3;q32) have unbalanced IgH translocations.30
Prevalence of unbalanced translocations
. | Patients with predominantly one signal . | All patients with each translocation . | Patients with one signal, % . | Patients/number of fusions . | Percent of cells with only one fusion more than 90% . | Patients/number of fusions . | Percent of cells with only one fusion more than 75% but less than 90% . | Patients/number of fusions . | Percent of cells with only one fusion less than 75% . |
|---|---|---|---|---|---|---|---|---|---|
| t(4;14)(p16.3;q32) | 14 | 35 | 40 | 3/14 | 21 | 6/14 | 43 | 5/14 | 36 |
| t(11;14)(q13;q32) | 35 | 53 | 66 | 25/35 | 71 | 4/35 | 11 | 6/35 | 17 |
| t(14;16)(q32;q23) | 7 | 15 | 47 | 6/7 | 86 | 1/7 | 14 | 0/7 | 0 |
| Total patients with each translocation | 56 | 34 | 11 | 11 |
. | Patients with predominantly one signal . | All patients with each translocation . | Patients with one signal, % . | Patients/number of fusions . | Percent of cells with only one fusion more than 90% . | Patients/number of fusions . | Percent of cells with only one fusion more than 75% but less than 90% . | Patients/number of fusions . | Percent of cells with only one fusion less than 75% . |
|---|---|---|---|---|---|---|---|---|---|
| t(4;14)(p16.3;q32) | 14 | 35 | 40 | 3/14 | 21 | 6/14 | 43 | 5/14 | 36 |
| t(11;14)(q13;q32) | 35 | 53 | 66 | 25/35 | 71 | 4/35 | 11 | 6/35 | 17 |
| t(14;16)(q32;q23) | 7 | 15 | 47 | 6/7 | 86 | 1/7 | 14 | 0/7 | 0 |
| Total patients with each translocation | 56 | 34 | 11 | 11 |
Response to treatment
Among patients evaluable for response, those with Δ13 had a lower likelihood of an objective response than those without the abnormality (Table 3). Otherwise there were no major differences noted.
Overall Survival (OS), progression-free survival (PFS), and objective response (OR) to treatment by abnormality
Abnormality . | N . | Median OS with abnormality, mo (95% Cl) . | Median OS without abnormality, mo (95% Cl) . | P . | Median PFS with abnormality, mo (95% Cl) . | Median PFS without abnormality, mo (95% Cl) . | P . | OR* with abnormality, n (%) . | OR* without abnormality, n (%) . | P . | 5-y OS rate with abnormality, % . | 5-y OS rate without abnormality, % . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t(4;14)(p16;q32) | 332 | 26 (21-33) | 45 (39-50) | <.001 | 17 (13-21) | 31 (28-34) | <.001 | 26 (62) | 197 (69) | .38 | 10 | 32 |
| t(14;16)(q32;q23) | 323 | 16 (13-22) | 41 (37-48) | .003 | 9 (6-13) | 30 (27-32) | .003 | 8 (53) | 204 (68) | .27 | 13 | 29 |
| t(11;14)(q13;q32) | 336 | 50 (37-60) | 39 (36-44) | .332 | 33 (28-45) | 27 (25-31) | .590 | 39 (78) | 187 (67) | .14 | 38 | 28 |
| Deletion 17p13 | 345 | 23 (20-36) | 44 (39-49) | .005 | 17 (11-24) | 30 (27-33) | .051 | 25 (68) | 208 (69) | .85 | 16 | 31 |
| Δ13 | 325 | 35 (29-41) | 51 (41-57) | .028 | 25 (21-29) | 33 (30-37) | .030 | 109 (63) | 108 (74) | .04 | 22 | 38 |
Abnormality . | N . | Median OS with abnormality, mo (95% Cl) . | Median OS without abnormality, mo (95% Cl) . | P . | Median PFS with abnormality, mo (95% Cl) . | Median PFS without abnormality, mo (95% Cl) . | P . | OR* with abnormality, n (%) . | OR* without abnormality, n (%) . | P . | 5-y OS rate with abnormality, % . | 5-y OS rate without abnormality, % . |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| t(4;14)(p16;q32) | 332 | 26 (21-33) | 45 (39-50) | <.001 | 17 (13-21) | 31 (28-34) | <.001 | 26 (62) | 197 (69) | .38 | 10 | 32 |
| t(14;16)(q32;q23) | 323 | 16 (13-22) | 41 (37-48) | .003 | 9 (6-13) | 30 (27-32) | .003 | 8 (53) | 204 (68) | .27 | 13 | 29 |
| t(11;14)(q13;q32) | 336 | 50 (37-60) | 39 (36-44) | .332 | 33 (28-45) | 27 (25-31) | .590 | 39 (78) | 187 (67) | .14 | 38 | 28 |
| Deletion 17p13 | 345 | 23 (20-36) | 44 (39-49) | .005 | 17 (11-24) | 30 (27-33) | .051 | 25 (68) | 208 (69) | .85 | 16 | 31 |
| Δ13 | 325 | 35 (29-41) | 51 (41-57) | .028 | 25 (21-29) | 33 (30-37) | .030 | 109 (63) | 108 (74) | .04 | 22 | 38 |
The number of patients evaluable for response may be slightly smaller than N given in column 2.
Survival analysis
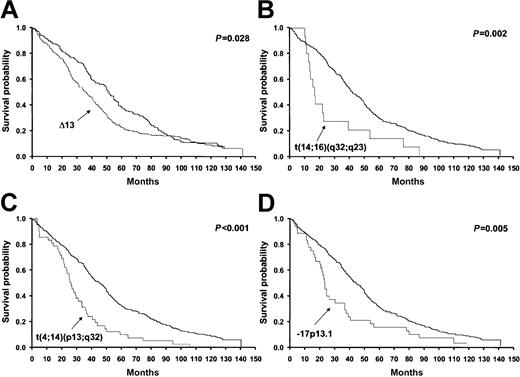
On the univariate analysis, patients with t(4;14)(p16.3;q32), t(14; 16)(q32;q32), deletions of 17p13.1, and Δ13 had a significantly worse overall survival (Figure 2 and Table 3). Progression-free survival was significantly worse in patients with t(4;14)(p16.3; q32), t(14;16)(q32;q32), and Δ13, and was of marginal significance in patients with deletions of 17p13.1 (Table 3).
Overall survival of patients stratified by the presence or absence of each one of the specific cytogenetic abnormalities showing statistical significance. The significance values are expressed next to each curve and correspond to the log-rank test. The x-axis values represent time since diagnosis, which is expressed in months. The results for the t(11;14)(q13;q32) are not shown, as they were not statistically significant. The thicker lines represent patients without the specific abnormality.
Overall survival of patients stratified by the presence or absence of each one of the specific cytogenetic abnormalities showing statistical significance. The significance values are expressed next to each curve and correspond to the log-rank test. The x-axis values represent time since diagnosis, which is expressed in months. The results for the t(11;14)(q13;q32) are not shown, as they were not statistically significant. The thicker lines represent patients without the specific abnormality.
Multiple regression model
When adjusting for the clinical factors that were statistically significant, as well as for other genetic abnormalities, t(4;14)(p16.3; q32) and t(14;16)(q32;q23) had the highest hazard ratios (1.78 and 1.67, respectively), t(4;14)(p16.3;q32) being statistically significant and t(14;16)(q32;q23) marginally significant. Deletion of 17p13.1 was marginally significant with an intermediate hazard ratio (1.34) with respect to the other 4 genetic abnormalities (Table 4). Δ13 were also statistically significant, with a hazard ratio equal to 1.28. These results were similar to those obtained using the Cox regression model for only the 275 cases with complete data. The hazard ratios (95% CI) of the genetic abnormalities in the Cox regression model, which also included creatinine, PCLI, and bone marrow PC percentage as covariates, were 1.69 (1.15-2.49) for t(4;14)(p16.3;q32), 1.42 (0.75-2.66) for t(14;16)(q32;q23), 1.47 (0.97-2.20) for – 17p13.1, 1.35 (1.04-1.74) for Δ13, and 0.94 (0.66-1.34) for t(11;14)(q13;q32).
Multivariate Bayesian analysis for survival using the Weibull proportional hazards model (n = 351)
Variable . | Coefficient . | Standard error . | Hazard ratio (95% CI) . |
|---|---|---|---|
| Constant | -10.760 | NA | NA |
| t(4;14)(p16;q32)* | 0.574 | 0.179 | 1.78 (1.23-2.50) |
| t(14;16)(q32;q23)* | 0.513 | 0.286 | 1.67 (0.92-2.83) |
| Deletion 17p13* | 0.291 | 0.188 | 1.34 (0.92-1.93) |
| ▵13* | 0.244 | 0.119 | 1.28 (1.01-1.61) |
| t(11;14)(q13;q32)* | -0.213 | 0.163 | 0.81 (0.58-1.11) |
| Creatinine, 2 or more vs less than 2 mg/dL† | 0.580 | 0.165 | 1.79 (1.29-2.44) |
| PC labeling index, 1% or higher vs less than 1% | 0.441 | 0.123 | 1.55 (1.22-1.97) |
| C-reactive protein, 2 or more vs less than 2 mg/dL | 0.435 | 0.175 | 1.54 (1.09-2.16) |
| Bone marrow involvement, 30% or higher vs less than 30% | 0.354 | 0.116 | 1.43 (1.14-1.79) |
Variable . | Coefficient . | Standard error . | Hazard ratio (95% CI) . |
|---|---|---|---|
| Constant | -10.760 | NA | NA |
| t(4;14)(p16;q32)* | 0.574 | 0.179 | 1.78 (1.23-2.50) |
| t(14;16)(q32;q23)* | 0.513 | 0.286 | 1.67 (0.92-2.83) |
| Deletion 17p13* | 0.291 | 0.188 | 1.34 (0.92-1.93) |
| ▵13* | 0.244 | 0.119 | 1.28 (1.01-1.61) |
| t(11;14)(q13;q32)* | -0.213 | 0.163 | 0.81 (0.58-1.11) |
| Creatinine, 2 or more vs less than 2 mg/dL† | 0.580 | 0.165 | 1.79 (1.29-2.44) |
| PC labeling index, 1% or higher vs less than 1% | 0.441 | 0.123 | 1.55 (1.22-1.97) |
| C-reactive protein, 2 or more vs less than 2 mg/dL | 0.435 | 0.175 | 1.54 (1.09-2.16) |
| Bone marrow involvement, 30% or higher vs less than 30% | 0.354 | 0.116 | 1.43 (1.14-1.79) |
NA indicates not applicable.
Present versus absent.
Convert creatinine to SI units: multiply mg/dL × 76.25 = μM.
Prognostic groups
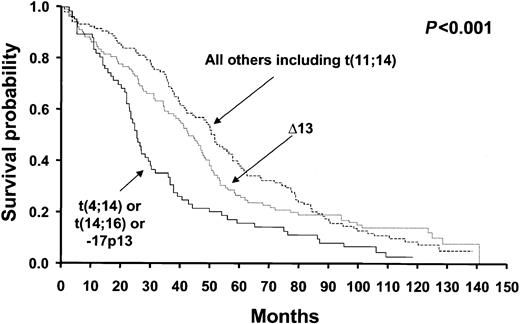
The 3 hierarchic groups identified 3 distinct prognostic groups (log-rank, P < .001). The groups are as follows: poor prognosis group—patients with t(4;14)(p16.3;q32) and/or t(14;16)(q32;q23) and/or deletion 17p13.1 (n = 66); intermediate prognosis— patients with Δ13, but not t(4;14)(p16.3;q32), t(14;16)(q32;q23), or deletion 17p13.1 (n = 103); and good prognosis—patients with only t(11;14)(q13;q32) or none of the abnormalities tested (n = 106). Their median survival times were 24.7 months, 42.3 months, and 50.5 months, respectively (Figure 3).
Overall survival of patients stratified by the hierarchic classification model proposed by our study. The survival curves show clear separation of patients into the good, intermediate, and poor prognosis category, a difference that was statistically significant. Groups were formed by the stratification according to the presence or absence of specific genetic abnormalities. The poor prognosis group includes patients with – 17p13.1, t(4;14)(p13;q32), and/or t(14;16)(q32;q23); the intermediate prognosis group includes those patients with Δ13 who did not have the aforementioned abnormalities; and the good prognosis group includes remaining patients, including those with the t(11;14)(q13;q32) and none of the aforementioned abnormalities.
Overall survival of patients stratified by the hierarchic classification model proposed by our study. The survival curves show clear separation of patients into the good, intermediate, and poor prognosis category, a difference that was statistically significant. Groups were formed by the stratification according to the presence or absence of specific genetic abnormalities. The poor prognosis group includes patients with – 17p13.1, t(4;14)(p13;q32), and/or t(14;16)(q32;q23); the intermediate prognosis group includes those patients with Δ13 who did not have the aforementioned abnormalities; and the good prognosis group includes remaining patients, including those with the t(11;14)(q13;q32) and none of the aforementioned abnormalities.
Discussion
Summary
In this study we find subgroups of MM patients classified according to their underlying cytogenetic abnormalities and show that these abnormalities alone can establish prognostic categories. Our study provides conclusive clinical evidence that MM is not a single uniform disorder, but rather a group of disorders, which can be defined by their underlying cytogenetic anomalies supported by this biologic variability.11 Patients with the t(4;14)(p16.3;q32), t(14;16)(q32;q23), and deletion of 17p13.1 have a significantly worse prognosis that others. We suspect this observation is likely due to the up-regulation of specific oncogenes involved in these translocations and to loss of the tumor suppressor gene p53. We previously demonstrated that chromosomal abnormalities define unique presenting factors for MM and may be associated with specific features such as the oligosecretory variant, λ light chain usage, or ploidy status.7,8
Biology of IgH translocations in general
Between 50% to 60% of MM patients harbor IgH translocations,31, 32, 33 and these translocations have been detected since the very early stages of the PC neoplasms (ie, monoclonal gammopathies of undetermined significance [MGUS]).34,35 This is consistent with IgH translocations being primary events, as is seen in the mouse plasmacytoma model in which they result in c-myc up-regulation.36 Of interest, all IgH translocations in MM appear to be up-regulating proliferation genes.12 We conclude that while translocations may be an early and important step,34,35 they are not sufficient in humans for malignant transformation and more likely result in the initial clone-immortalizing event. It is important to note that the translocations that impart a poor prognosis in the active MM stage have no known effect on prognosis when they are detected in MGUS. In fact we have found that patients with MGUS and the t(4;14)(p16.3;q32) or t(14;16)(q32;q23) may remain without progression to MM for prolonged periods of time.35
Genomic convergence and translocations
Despite ongoing genomic instability, IgH translocations are highly conserved, as they are not lost with advancement through the different stages of the PC neoplasms, and in fact are clonally selected as they are seen in the majority of the clonal cells.10,37 Roschke et al, exploring genomic instability in human colorectal and ovarian cancer cell lines, have previously explored the model of “signature karyotypes,” and our working hypothesis is consistent with it.38 Here we show that in virtually all abnormal MM cases IgH translocations involve a large percentage of the PCs. We observe similar patterns with Δ13 but not with – 17p13. This finding is in great contrast with aneuploid clones that more commonly affect only a fraction of cells.39,40 While the situation is less clear in MGUS, it is also suggested that, in many cases, translocations will involve the majority of the clonal cells.34,35
t(4;14)(p16.3;q32)
We have first reported the negative prognostic implications of both the t(4;14)(p16.3;q32) and the t(14;16)(q32;q23) in MM patients treated with chemotherapy.41 Similar prognostic effects have been shown by Moreau et al for patients with t(4;14)(p16.3;q32) treated with high-dose chemotherapy.9 The mechanisms resulting in the negative prognostic associations with the t(4;14)(p16.3;q32) are not known.
In contrast to other B-cell malignancies, IgH translocations in MM can deregulate 2 or more oncogenes.12 This is because in MM, IgH translocations occur into switch regions causing segregation of the Eμ and 3′α enhancers. An example of this is the t(4;14)(p16.3; q32). While other genes may be up-regulated by the t(4;14)(p16.3; q32),42 FRGR3 and MMSET are best characterized. However, FGFR3 is not up-regulated in all cases of a t(4;14)(p16.3;q32), and it has been found that der14 chromosome can be lost in primary samples or cell lines (Michael Kuehl, oral communication, June 2002).30 This implies that MMSET deregulation is needed for clone survival. The probes used in this study bracket all reported human MM breakpoints (for all translocations) and should in theory always result in 2 detectable fusion signals. However, in this study we have found that IgH translocations in MM will frequently be unbalanced (Table 2).
t(14;16)(q32;q23)
Unlike the study by Avet-Loiseau et al,10 we have found the t(14;16)(q23;q32) as recurrent in MM. We observed the t(14; 16)(q32;q23) in 5% of patients and had a clear association with an adverse outcome, with a shorter survival and features of aggressiveness. We have also detected the t(14;16)(q32;q23) in MGUS without transformation to MM,35 but nevertheless when the abnormality is seen in the active stages of the disease it still confers an aggressive phenotype. While c-maf up-regulation is believed to be a culprit, a recently described gene, WWOX, is also disrupted by breakpoints at 16q23.43 A possible tumor suppressor role is being sought for this gene, which spans several hundred kilobases at 16q32 and is located at the fragile site Fra16D.43 C-maf, has been shown to have transforming activity in chicken fibroblasts.44 The t(14;16)(q32;q23) is observed in 25% of human MM cell lines but is seen in only 5% of primary MM samples. In one of these human MM cell lines, KMS-11, the 16q23 breakpoint places c-maf at more than 700 kb of the IgH enhancer in the translocated allele, without evidence of intervening deletion or inversion (R.F., unpublished observations, September 2001). This highlights the possibility that oncogenes other than those described to date may be up-regulated by any IgH translocation, other than those currently believed important for pathogenesis.
Chromosome 13 abnormalities
Our study confirms that Δ13 have a negative impact on prognosis. Others and we have found that Δ13 detected by FISH is an independent prognostic variable on a multivariate analysis.4,6 The genes associated with the negative prognostic implications of Δ13 have yet to be defined. Detailed molecular analysis has revealed that in the majority of cases Δ13 are indicative of monosomy.19,45 While a minimally deleted region has been postulated as being in 13q14, other sites may be involved as well.19,45 The role of Δ13 in the pathogenesis of MM is still being elucidated. In the setting of widespread genomic instability, chromosome 13 is almost never seen as trisomic, suggesting clonal intolerance to the gain.46
Deletion of 17p13.1 (p53)
Deletions at the p53 locus also confer an adverse prognosis, even when they are observed in only a small proportion of patients.15 While most other abnormalities (ie, IgH translocations and Δ13) showed little heterogeneity, p53 deletions could be seen in smaller percentages of cells and suggest early clonal evolution. Furthermore, patients with this specific abnormality were more likely to have other features of aggressiveness, such as plasmacytomas and hypercalcemia. All this information suggests that even when p53 deletions may be detected at the time of diagnosis, p53 deletions are likely markers of an advanced clone.
Therapeutic implications
As targeted therapy evolves, different treatment interventions will have variable success, depending on the underlying genetic nature of the clone.47 For instance, the development of effective MMSET or FGFR3 small molecule inhibitors may allow for patients with t(4;14)(p16.3;q32) to become a better prognostic category. The use of inhibitors of the cyclin D1/CDK pathways, such as flavopiridol, seems especially suited for patients with t(11;14)(q13;q32). It is also worth noting that it appears that specific treatments may be better tailored for patients with specific chromosomal abnormalities. A comparison of our results (in patients treated with conventional chemotherapy) with those of Moreau et al9 (in patients treated with high-dose therapy) suggests that high-dose chemotherapy provides little, or no, survival advantage for patients with Δ13 or t(4;14)(p16.3;q32).8 In contrast it appears that high-dose chemotherapy provides a significant survival increment for patients with t(11;14)(q13;q32).7,9 While these observations are speculative for now, as they are based on a retrospective comparison, they are highly provocative and in need of confirmation in prospective clinical trials.
Statistical aspects
A Bayesian approach was used in this study to assess the impact of genetic abnormalities on survival, adjusting for known clinical prognostic factors. The advantage of the Bayesian analysis is that it allows all subjects to be included in the model, even those that have missing data in their covariates. In our study sample 22% of subjects had missing data, either in the genetic abnormalities or in the clinical variables. Imputation of missing data is done in the Gibbs sampling framework by treating missing values as additional unknown quantities and randomly selecting values from their conditional distributions. Conditional distributions are a function of the observed individual data and the current sampled values of the other missing data for a particular individual. There was no indication of nonrandom “missingness” in our data, one of the assumptions of the Bayesian analysis. Also, the assumptions of Weibull distribution, proportional hazards, and adequacy of the multivariate model were assessed with satisfactory results. When the results of the Bayesian approach (which included all of the 351 studied patients) were compared with those of the Cox proportional hazards regression (which included only the 275 patients with complete data), the hazard ratios were similar. Differences were observed mostly for the genetic abnormalities with the smallest prevalence: t(14;16)(q32;q23) and – 17p13.
It is important to cautiously interpret the hierarchic group survival analysis, as patients in the poor prognosis group could have more abnormalities than patients in the intermediate or good prognosis groups. For instance, patients in the poor prognosis group could possibly have Δ13 deletion in addition to one or more of the poor prognosis abnormalities. To make sure this was not the only reason that patients in the poor prognosis group did poorly, we switched the order of the hierarchic grouping. In the new grouping, patients with Δ13 were in one group; patients with any of the 3 ((t(4;14)(p16.3;q32), t(14;16)(q32;q23), deletion 17p13.1) poor prognosis abnormalities, but not Δ13 were in another group; and patients with only t(11;14)(q13;q32) or none of the 5 tested abnormalities were in the third group. While the median survival times differed slightly from the originally hierarchic grouping, the trend in median survival times was the same. Patients with the poor prognosis abnormalities (t(4;14)(p16.3;q32), t(14;16)(q32;q23), and deletion 17p13.1) did worse than patients with Δ13, and those patients did worse than patients with only t(11;14)(q13;q32) or none of the 5 tested abnormalities. We thus conclude that patients with t(4;14)(p16.3;q32), t(14;16)(q32;q23), or deletion 17p13.1 seem to make up a poor prognosis group.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-10-3017.
R.F. is a Lilly Clinical Investigator of the Damon Runyon—Cancer Research Fund, and a Leukemia and Lymphoma Society Translational Research Awardee. Supported in part by Public Health Service grant R01 CA83724-01 (R.F.) and P01 CA62242 (R.A.K., P.R.G.) from the National Cancer Institute. P.R.G. is supported by the ECOG grant CA21115-25C from the National Cancer Institute.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal