Abstract
CD4, the primary receptor for entry of HIV, is known to be expressed on T cells and monocytes/macrophages; healthy natural killer (NK) lymphocytes; in vitro human herpesvirus 6 (HHV6)–infected CD8+, NK, and γδ T lymphocytes; CD34+ progenitor cells; and a subset of eosinophils and basophils. We here report the unconventional expression of CD4 at the surface of peripheral blood neutrophils derived from 4 of 51 (7.8%) HIV-1–infected and 3 of 25 (12%) uninfected donors, with similar frequency within the 2 groups. The percentage of CD4+ neutrophils ranged from 39% to 97% of the total neutrophil population. Both surface and cytoplasmic forms of CD4 were present in neutrophils. Quantitative RNA polymerase chain reaction (PCR) revealed that neutrophils contain levels of CD4 mRNA comparable to those of peripheral blood mononuclear cells derived from the same donor. The conformation of CD4 expressed at the surface of neutrophils was similar to that of CD4 expressed on T lymphocytes as determined by the binding of monoclonal antibodies specific for conformational epitopes and the binding of recombinant HIV-1 gp120. Thus, our data provide evidence that neutrophils express endogenous CD4 and bind HIV. Owing to their abundance in peripheral blood, CD4+ neutrophils may influence significantly the biodistribution of HIV delivering it to sites of inflammation or to additional tissue reservoirs.
Introduction
CD4 was originally identified as a T-lymphocyte marker and specifically associated to the subset of helper T lymphocytes. In addition to its function as a ligand of major histocompatibility complex (MHC) class II molecules,1 CD4 was defined as the primary receptor of HIV viruses.2 Binding of HIV envelope to CD4 is the first event that allows its binding to other coreceptors, namely the chemokine receptors CCR5 and CXCR4, followed by membrane fusion and entry.3,4 CD4 is also expressed on monocytes/macrophages.5 In addition, further studies have reported the expression of CD4 on cells other than helper T lymphocytes and monocytes, including eosinophils6 ; CD34+ progenitor cells7,8 ; in vitro human herpesvirus 6 (HHV-6)–infected CD8+,9 natural killer (NK),10 and γδ T lymphocytes11 ; mast cells/basophils12 ; and normal NK lymphocytes.13 To our knowledge the presence of CD4 on peripheral blood neutrophils has not yet been documented.
Traditionally, flow cytometry-based phenotyping is performed either on separated peripheral blood mononuclear cells or, in routine diagnostic investigation, on whole blood lymphocytes selected by morphologic gating; therefore, information regarding neutrophils is not gathered nor recorded. In our clinic we have set up whole blood-based flow cytometry protocols that analyze all leukocyte populations.
By this mean we identified an HIV-infected patient (pt 1) whose neutrophils were unexpectedly stained by a CD4-specific monoclonal antibody (mAb). We, therefore, (1) attempted a preliminary evaluation of the frequency of individuals whose neutrophils display CD4, (2) confirmed the specificity by staining with mAb directed toward different conformational epitopes of CD4, (3) tested whether CD4+ neutrophils could bind soluble HIV-1 envelope protein gp120, and (4) tested the endogenous origin of CD4 in neutrophils.
Patients, materials, and methods
Patients
Fifty-one individuals infected with HIV were tested among patients with chronic HIV infection regularly followed at our Clinic of Infectious Diseases. Three of the 4 HIV-infected patients who displayed CD4+ neutrophils had significant levels of HIV replication (> 10 000 HIV RNA copies/mL) in the presence of antiretroviral treatment. One of these 3 patients (pt 1) was resistant to several classes of antiretroviral drugs; the fourth patient had plasma viremia below the detection level (< 80 copies/mL). All 4 patients had CD4 cell counts more than 500. Control individuals were healthy laboratory workers.
Cellular purifications
Whole blood was drawn by venipuncture in EDTA (ethylenediaminetetraacetic acid)–containing Vacutainer (BD Biosciences Labware, Plymouth, United Kingdom) tubes and immediately processed for flow cytometry staining and cellular purification. Peripheral blood mononuclear cells (PBMCs) were obtained (floating ring) by gradient density centrifugation (Lymphoprep Nycomed, Oslo, Norway); purified neutrophils were prepared by recovering the white blood cell layer over the red blood cells (RBCs) after the gradient centrifugation and allowing a 30-minute sedimentation on a 4% dextran solution at room temperature followed by lysis of contaminating RBCs with NaCl solution (1.2% + 0.2%). Extensive washing of both PBMCs and neutrophils was performed with 1 × phosphate-buffered saline (PBS). Purity of the neutrophil preparation was controlled by flow cytometry (CD3–, CD14dim+, and CD66b+) and found to be 98.0% or more. Purified populations were used either for flow cytometry or immunofluorescence or were dry-pelletted (1 × 106 cells/tube) and frozen at –80°C for CD4 mRNA quantification. For the HIV-infected patient 1, besides PBMCs and neutrophils, CD14+ monocytes and CD8-depleted lympho-cytes (CD4-enriched) were also prepared by sequential magnetic bead (Dynabeads, Oxoid S.p.A., Garbagnate M.se, Italy) purification following the manufacturer's directions. Each cell population was dry-pelletted (1 × 106 cells) and frozen at –80°C for HIV-1 DNA load quantitation.
Flow cytometry
Whole blood (100 μL/tube) cells were stained with fluorochrome directly conjugated mAbs: phycoerythrin (PE)–anti-CD8 (OKT8, mouse (m) immunoglobulin G2a (IgG2a; Ortho Diagnostic Systems, Raritan, NJ), tricolor (TC)–anti-CD4 (clone S3.5, mIgG2) (Caltag Laboratories, Burlingame, CA), fluorescein isothiocyanate (FITC)–anti-CD66b (clone G10F5, mIgM) (Pharmingen BD), as well as with fluorochrome- and isotype-matched control mAbs. After a 30-minute incubation at 4°C, cells were fixed, and RBCs were lysed with the Cal-Lyse lysing solution (Caltag). Staining of purified PBMCs and neutrophils was performed with the unconjugated OKT4, 13B8-2, and C9F11 mouse mAbs followed by FITC-conjugated rabbit antimouse IgG (Dako S.p.A.) and fixed with 1% formaldehyde. Only the second-step reagent was added to control tubes. Stained samples were acquired with a fluorescence-activated cell sorter scan (FACScan) cytometer and analyzed by Cell Quest and Lysis software (BD Biosciences).
Immunofluorescence
PBMCs and neutrophils were fixed for 15 minutes at 4°C with 4% paraformaldehyde in 125 mM PBS and washed with the same buffer. The cells were then treated for 30 minutes with a solution containing 0.3% Triton X-100, 15% filtered goat serum, 0.45 M NaCl, and 10 mM phosphate buffer, pH 7.4. After washing, the preparations were exposed (90 minutes at 37°C) to the primary Ab (nonconjugated anti-CD4, clone SK3, mIgG1; BD) diluted in the above Triton X-100 and goat serum-containing solution. After an additional thorough wash, the cells were treated with the appropriate rhodamine-labeled goat antimouse Ab (1:100 dilution in the Triton X-100–goat serum solution, 60 minutes, 37°C), washed again, and mounted in glycerol to be examined with a laser scanning confocal microscope (MRC 1024; Bio-Rad House).
Quantitation of CD4 mRNA
Total RNA from PBMCs and neutrophil pellets was extracted, quantified, and reverse transcribed for generating cDNA using MultiScribe Reverse Transcriptase from the TaqMan Reverse Transcription Reagents (PE Biosystems). A cDNA amount corresponding to 200 ng total RNA was amplified. Quantitative real-time polymerase chain reaction (PCR) was performed using the SYBR Green PCR Master mix (PE Biosystems) and 300 nM gene-specific forward and reverse primers designated using the PrimerExpress software (Applied Biosystems, Foster City, CA): CD4, for 5′-CTG GCC CTT GAA GCG AAA A-3′, reverse 5′-CCA CCA GGT TCA CTTCCTGATG-3′; β-actin, for 5′-CCC AAG GCC AAC CGC GAG AAG AT-3′, reverse 5′-GTC CCG GCC AGC CAG GTC CAG-3′. The reaction conditions were 10 minutes at 95°C (one cycle) and 15 seconds at 95°C and 1 minute at 60°C (40 cycles). Gene-specific products were continuously measured by means of an ABI PRISM 5700 detection system (Perkin Elmer, Norwalk, CT). Samples were normalized using the housekeeping gene β-actin. Three replicates for each experimental point were performed, and differences were assessed with the 2-tailed Student t test.
Cell-associated viral load
Proviral HIV-1 DNA load was measured in purified PBMCs, neutrophils, CD14+ monocytes, and CD8-depleted lymphocytes of patient 1 using an in-house Taqman “Real-Time” PCR assay specifically developed to accurately quantitate the major clades belonging to the HIV-1 M group (Scarlatti et al, unpublished observations, June 20, 1999). Primers and probe were derived from highly conserved sequences placed in the LTR-gag junction. The cellular content of each sample was evaluated by a second in-house Taqman real-time PCR assay developed with a highly conserved fragment (from nucleotide +3175 to nucleotide +3255) of the single-copy human CCR5 gene. Three replicates for each experimental point were performed in both PCR assays.
Reverse-transcriptase (RT) activity
The virion-associated RT activity was measured in culture supernatants by a homemade radioactive assay, as previously described.14
p24 enzyme-linked immunosorbent assay (ELISA)
Content of the HIV-1 p24 core antigen was evaluated by an in-house ELISA, as previously reported.15
Results
Dual-color staining with CD4 and CD8 mAbs of whole blood derived from an HIV-infected patient (pt 1) revealed an unexpected high proportion (51%) of CD4+ neutrophils in their morphologic gate (Figure 1).
Detection of CD4 on whole blood leukocyte populations derived from an HIV-infected individual (pt 1). The first panel displays the morphologic parameters forward scatter (FSC) and side scatter (SSC) of whole blood leukocyte populations. Three morphologic gates are drawn that encompass lymphocytes (R1), monocytes (R2), and granulocytes (R3). The 3 panels on the right show the fluorescence intensities of each gated population on dual staining with CD4 and CD8 mAbs. The numbers indicate the percentage of positive cells in each quadrant; the quadrant markers are set on the fluorescence intensities of the isotype-matched control mAb within each gate.
Detection of CD4 on whole blood leukocyte populations derived from an HIV-infected individual (pt 1). The first panel displays the morphologic parameters forward scatter (FSC) and side scatter (SSC) of whole blood leukocyte populations. Three morphologic gates are drawn that encompass lymphocytes (R1), monocytes (R2), and granulocytes (R3). The 3 panels on the right show the fluorescence intensities of each gated population on dual staining with CD4 and CD8 mAbs. The numbers indicate the percentage of positive cells in each quadrant; the quadrant markers are set on the fluorescence intensities of the isotype-matched control mAb within each gate.
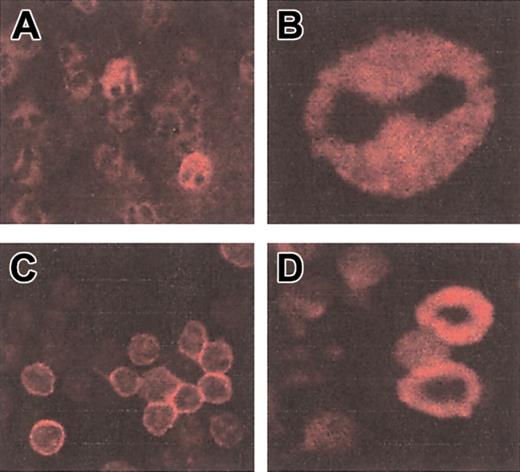
Immunofluorescence staining with a CD4 mAb on purified neutrophils and PBMCs from patient 1 is shown in Figure 2. Neutrophils were identifiable by the typical drumstick morphology of their nucleus. Staining was performed on permeabilized cells and revealed both membrane and intracellular CD4 molecules in neutrophils. The intensity of fluorescence was higher in PBMCs compared with neutrophils.
Staining of neutrophils and lymphocytes with a CD4-specific mAb. Immunofluorescence with CD4 mAb on permeabilized purified neutrophils (A-B) and PBMCs (C-D) from patient 1. Original magnifications × 63.
Staining of neutrophils and lymphocytes with a CD4-specific mAb. Immunofluorescence with CD4 mAb on permeabilized purified neutrophils (A-B) and PBMCs (C-D) from patient 1. Original magnifications × 63.
To evaluate the frequency of subjects with CD4+ neutrophils, we screened a group of HIV-infected and uninfected individuals for the presence of cells that double stain with CD4 and a mAb to the antigen CD66b. CD66b is a selective marker of neutrophils16 that, also in our experiments, was found consistently negative in lymphocytes and monocytes (not shown). This screening system was selected to exclude from our analysis cells that could contaminate the flow cytometric neutrophil gate. Four of 51 (7.8%) HIV-infected individuals and 3 of 25 (12%) uninfected control volunteers displayed a proportion of CD4+CD66b+ cells ranging from 39% to 97% of the total neutrophil population. Figure 3 shows the data obtained from 2 HIV-positive and 2 HIV-negative individuals who were either negative or positive for the expression of CD4 on CD66b+ neutrophils. The average mean fluorescence intensity (MFI) was 15.3 (± 5.5 SD), with the exception of the control individual shown in Figure 3 who displayed a much higher value (MFI, 180). Thus, the expression of CD4 on neutrophils is not restricted to HIV-infected individuals but can also be observed in uninfected individuals. The frequency of individuals displaying CD4+ neutrophils was not significantly different between the 2 groups as determined by the chi-square test (P = .868).
Detection of CD4 on CD66+ cells. Flow cytometric analyses of whole blood from 2 representative HIV-infected (top panels) and 2 uninfected (bottom panels) individuals on dual staining with CD4 and CD66b mAbs. Shown are the fluorescence intensities within morphologic gate R3 (neutrophils). The numbers indicate the percentage of positive cells in each quadrant. Ten percent of neutrophils from one of the uninfected donors were also positive on staining with another directly conjugated anti-CD4 mAb (anti–CD4-PE, clone SK3, mIgG1; BD Biosciences).
Detection of CD4 on CD66+ cells. Flow cytometric analyses of whole blood from 2 representative HIV-infected (top panels) and 2 uninfected (bottom panels) individuals on dual staining with CD4 and CD66b mAbs. Shown are the fluorescence intensities within morphologic gate R3 (neutrophils). The numbers indicate the percentage of positive cells in each quadrant. Ten percent of neutrophils from one of the uninfected donors were also positive on staining with another directly conjugated anti-CD4 mAb (anti–CD4-PE, clone SK3, mIgG1; BD Biosciences).
The possibility that the CD4+ cells detected within neutrophils were contaminating monocytes or lymphocytes was ruled out for the following reasons: (1) the proportion of CD4+ cells within the neutrophil gate was too high to be derived from cell populations other than neutrophils, (2) the MFI of CD4+ neutrophils was consistently different from that of lymphocytes and monocytes, (3) CD4 was detected at the surface of CD66b+ cells, and (4) CD4 could be directly visualized in neutrophils by immunofluorescence.
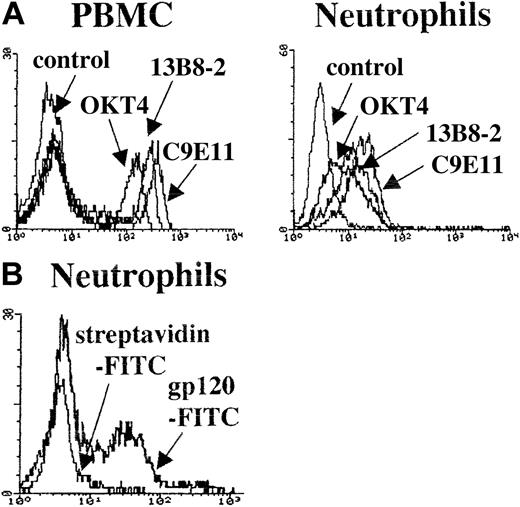
To rule out that binding of the CD4-specific mAb was due to a CD4-mimicking epitope present on an unrelated molecule, we tested 3 mAbs specific for different CD4 conformational determinants: OKT4 recognizes domain 3 and 4,17 13B8 binds the CDR3-like region of domain 1,18 and C9E11 recognizes domain 1 and 2 (S. Burastero, unpublished observations, April 15, 1999). All 3 mAbs, tested on neutrophils and PBMCs derived from the same HIV-uninfected donor who had CD4+ CD66b+ neutrophils (control 1), bound both PBMCs and neutrophils (Figure 4A). Also in this case the MFI was higher for PBMCs compared with neutrophils. The patterns of mAb binding to the 2 cell populations were very similar, with C9E11 and 13B8 being more reactive than OKT4. Thus, the molecular conformation of CD4 expressed at the surface of neutrophils appears to be identical to that of CD4 expressed on PBMCs. Furthermore, CD4 on neutrophils was cleaved by the enzyme pronase that is known to specifically cleave CD4, CD8, and CD28 on the surface of lymphocytes19 (data not shown). We then tested the ability of purified neutrophils from control 1 to bind the HIV-1 envelope surface unit, gp120. The experiment was performed with a FITC-labeled recombinant HIV-1 gp120 derived from the T-cell line adapted strain HIV-1LAI. To control for nonspecific binding we used FITC-conjugated streptavidin. As shown in Figure 4B, there was a bimodal pattern of staining with 37% of cells that stained positive (MFI = 83) and a second population that displayed nonspecific staining overlapping that of streptavidin-FITC. The high proportion of neutrophils that bound HIV-1 gp120 ruled out potential binding by lymphocytes or monocytes contaminating the neutrophil preparation.
CD4 epitope expression on neutrophils and PBMCs and neutrophil binding of HIV-1 gp 120. (A) Histogram plots depicting overlaid fluorescence intensities of purified PBMCs and neutrophils from an HIV-uninfected control after indirect staining with the anti-CD4 mAbs OKT4, 13B8-2, and C9E11. The control histogram derives from cells stained with second-step reagent only. (B) The histogram plots show the fluorescence intensities of purified neutrophils stained with FITC-directly-conjugated gp120; streptavidin-FITC was used as control.
CD4 epitope expression on neutrophils and PBMCs and neutrophil binding of HIV-1 gp 120. (A) Histogram plots depicting overlaid fluorescence intensities of purified PBMCs and neutrophils from an HIV-uninfected control after indirect staining with the anti-CD4 mAbs OKT4, 13B8-2, and C9E11. The control histogram derives from cells stained with second-step reagent only. (B) The histogram plots show the fluorescence intensities of purified neutrophils stained with FITC-directly-conjugated gp120; streptavidin-FITC was used as control.
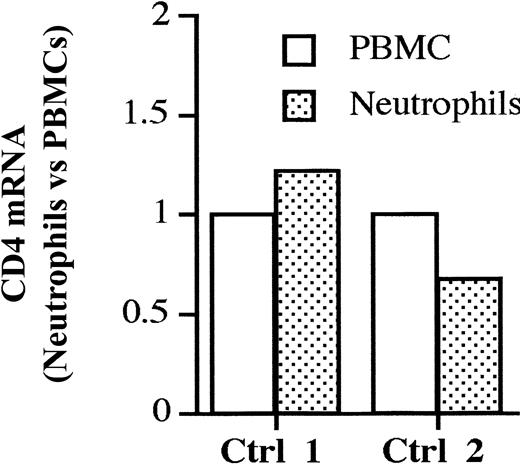
To prove that neutrophils express endogenous CD4 we tested the presence of CD4 mRNA by real-time PCR in purified neutrophils and PBMCs derived from one donor displaying CD4+ neutrophils (control 1) and one donor with CD4– neutrophils (control 2) (Figure 5). The data are expressed as the relative fold increase/decrease of CD4 mRNA in neutrophils versus PBMCs, used as internal calibrator. Neutrophils displayed levels of CD4 mRNA that were similar to the ones detected in PBMCs, consistent with the capacity of these cells to synthesize CD4. Surprisingly, also the individual with CD4– neutrophils (control 2) displayed similar amounts of CD4 mRNA, suggesting that the regulation of CD4 expression in this cell lineage may be under translational control. Similar levels of CD4 mRNA were found in other 2 purified neutrophil populations from uninfected individuals.
Comparison of CD4 mRNA content in neutrophils and PBMCs. Levels of CD4 mRNA in purified PBMCs and neutrophils from uninfected control 1 (displaying CD4+ neutrophils) and control 2 (displaying CD4–neutrophils) were determined by real-time PCR assay. The amount of CD4 mRNA in autologous PBMCs is used as an internal calibrator in each donor. Purified PBMCs and neutrophils from control 1 prepared in an independent experiment yielded similar results in terms of CD4 mRNA content (not shown).
Comparison of CD4 mRNA content in neutrophils and PBMCs. Levels of CD4 mRNA in purified PBMCs and neutrophils from uninfected control 1 (displaying CD4+ neutrophils) and control 2 (displaying CD4–neutrophils) were determined by real-time PCR assay. The amount of CD4 mRNA in autologous PBMCs is used as an internal calibrator in each donor. Purified PBMCs and neutrophils from control 1 prepared in an independent experiment yielded similar results in terms of CD4 mRNA content (not shown).
Among the different chemokine receptors expressed at the surface of neutrophils, CXCR4, one of the HIV entry coreceptors3,4 is expressed constitutively.20 Thus, CD4+ neutrophils express the molecular machinery required for entry of HIV viruses that use CXCR4 (X4 viruses).21 We tested whether neutrophils derived from patient 1 were infected in vivo by cocultivating them with allogeneic phytohemagglutinin (PHA)–activated blasts. Unfractionated PBMCs, positively selected CD14+ monocytes, and CD4+ T-cell-enriched (CD8-depleted) populations were each cocultured (1:1 ratio) with the same blasts as controls. The HIV structural p24 antigen and RT activity were measured in culture supernatants. We consistently failed to rescue replicating virus from cocultures containing unfractionated PBMCs, monocytes, and neutrophils by both RT and p24 assays. In contrast, the CD4 T-cell–enriched population yielded positive results (771 cpm/μL RT activity and 7524 pg/mL p24 Ag at day 12 of culture). We also looked at the cell-associated HIV-1 DNA load in the 4 cell populations using a quantitative real-time PCR and found a detectable amount of HIV DNA in neutrophils and monocytes (59 and 57 genome equivalent × 106 cells, respectively). In comparison, the amounts of HIV-1 DNA measured in PBMCs and the CD8-depleted population were 10-fold higher. Although the granulocyte preparation used in these experiments was more than 98.0% pure, we cannot rule out the possibility that the low levels of HIV DNA detected in this cell population was derived by contaminating CD4+ mononuclear cells. Kaneda et al22 previously reported the presence of defective HIV-1 provirus in peripheral neutrophils derived from an HIV-infected patient. Thus, the question of whether neutrophils are infected by HIV in vivo deserves further investigation.
Discussion
Taken together our data show that (1) a large fraction of peripheral blood neutrophils of 4 of 51 HIV-infected individuals and 3 of 25 uninfected control volunteers express surface CD4, (2) the molecular conformation of neutrophil-expressed CD4 is very similar to that of CD4 expressed at the surface of lymphocytes, (3) CD4+ neutrophils bind HIV-1 gp120, and (4) neutrophils express CD4 mRNA consistent with CD4 being directly synthesized by these cells. This is to our knowledge the first report on the expression of CD4 on neutrophils.
Comparable levels of CD4 mRNA in purified neutrophil populations were found in the 2 donors expressing CD4+ and CD4– neutrophils. The possibility that this expression could be due to contaminating residual lymphocytes or monocytes was ruled out because our neutrophil preparations were consistently 97% to 98% CD66b+. Thus, 2% to 3% of contaminating lympho-monocytic cells, which include also CD8+ lymphocytes, cannot account for the amount of CD4 mRNA derived from neutrophils, which was similar to that derived from PBMCs (which encompass more than 70% CD4+ lympho-monocytes in healthy donors). Rather, the presence of CD4 mRNA in neutrophils that do not express CD4 at the surface suggest the existence of mechanisms that act at the translational level. The hematopoietic CD34+ progenitor cell also expresses CD4 at the surface.7,8 Thus, the CD4 gene within the myelomonocytic lineage, which ultimately gives rise to CD4+ monocytes and CD4– neutrophils, most likely undergoes mechanisms regulating its expression or suppression.
Our report raises the question of whether CD4+ neutrophils may have a particular functional phenotype. In this regard, we have investigated other cell surface markers, finding that a particular chemokine receptor, CXCR3, was also concomitantly elevated on neutrophils in those individuals with CD4+ neutrophils (P. Biswas et al, unpublished observations, June 28, 2002). CXCR3, together with CCR5, is a major receptor for inflammatory chemokines.23,24 Thus, it is possible that neutrophils expressing unusual surface markers such as CD4 and CXCR3 may be endowed with particular migratory patterns toward inflammation sites.
The presence of CD4+ neutrophils in the peripheral blood of healthy uninfected control volunteers indicated that this unusual expression of CD4 is not a consequence of HIV infection but rather preexists infection. It will be interesting to determine whether the presence of CD4+ neutrophils may have some influence on the course of HIV infection and on disease progression. Differently from the other unconventional CD4+ cell populations described so far, neutrophils are by far the most abundant among peripheral leukocytes and may influence significantly the spread of HIV. We have provided evidence that CD4+ neutrophils bind the HIV envelope surface unit gp120. Therefore, CD4+ neutrophils may bind HIV and play an active role in its propagation, similarly to that of DC-specific intercellular adhesion molecule (ICAM)3-grabbing nonintegrin (DC-SIGN)–positive dendritic cells that have been shown to promote infection of bystander cells without being themselves infected.25 In addition, neutrophils constitutively express CXCR4 and may selectively bind X4 viruses, which are generally associated to loss of CD4 T lymphocytes and disease progression, favoring the emergence of these viruses.
Prepublished online as Blood First Edition Paper, January 16, 2003; DOI 10.1182/blood-2002-10-3056.
Supported by the Japan Foundation for AIDS Prevention.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal