Abstract
Megakaryocytic and erythroid lineages derive from a common bipotential progenitor and share many transcription factors, most prominently factors of the GATA zinc-finger family. Little is known about transcription factors unique to the megakaryocytic lineage that might program divergence from the erythroid pathway. To identify such factors, we used the K562 system in which megakaryocyte lineage commitment is dependent on sustained extracellular regulatory kinase (ERK) activation and is inhibited by stromal cell contact. During megakaryocytic induction in this system, the myeloid transcription factor RUNX1 underwent up-regulation, dependent on ERK signaling and inhibitable by stromal cell contact. Immunostaining of healthy human bone marrow confirmed a strong expression of RUNX1 and its cofactor, core-binding factor β (CBFβ), in megakaryocytes and a minimal expression in erythroblasts. In primary human hematopoietic progenitor cultures, RUNX1 and CBFβ up-regulation preceded megakaryocytic differentiation, and down-regulation of these factors preceded erythroid differentiation. Functional studies showed cooperation among RUNX1, CBFβ, and GATA-1 in the activation of a megakaryocytic promoter. By contrast, the RUNX1-ETO leukemic fusion protein potently repressed GATA-1–mediated transactivation. These functional interactions correlated with physical interactions observed between GATA-1 and RUNX1 factors. Enforced RUNX1 expression in K562 cells enhanced the induction of the megakaryocytic integrin proteins αIIb and α2. These results suggest that RUNX1 may participate in the programming of megakaryocytic lineage commitment through functional and physical interactions with GATA transcription factors. By contrast, RUNX1-ETO inhibition of GATA function may constitute a potential mechanism for the blockade of erythroid and megakaryocytic differentiation seen in leukemias with t(8;21).
Introduction
Despite divergent phenotypes, megakaryocytic and erythroid lineages originate from a common bipotent progenitor, known variously as the blast-forming unit erythroid/megakaryocyte (BFU-E/MK) or the megakaryocyte erythroid progenitor (MEP).1, 2, 3 As further evidence of a developmental link, human erythroblasts at relatively late stages of development retain the potential for megakaryocytic transdifferentiation.4 The molecular basis for this developmental relationship appears to reside in the extensive sharing of lineage-restricted transcription factors. Many transcription factors initially identified as critical in erythroid development have been found through gene knock-out experiments to be important in megakaryocytic development.5, 6, 7
GATA-1 is the prototypic erythro-megakaryocytic transcription factor, cooperating with its cofactor FOG-1 to serve essential roles in erythroid and megakaryocytic differentiation.8 Enforced GATA-1 expression in myeloid cell lines promotes erythroid, megakaryocytic, or combined differentiation, depending on the cell type.9, 10, 11 Knock-out of either the GATA-1 or the FOG-1 gene results in midgestation embryonic lethality because of severe anemia associated with abnormal or absent megakaryopoiesis.7,12 Lineage-selective knock-down in mice of GATA-1 expression in megakaryocytes causes increased megakaryocyte proliferation coupled with impaired maturation.6,13 Knock-in mice with compound GATA-1 and GATA-2 mutations, causing the loss of FOG-1 binding, display a complete absence of megakaryopoiesis, a phenocopy of FOG-1 null mice.8 Human hereditary mutations in the amino terminal zinc-finger (N-finger) of GATA-1, which also disrupt FOG-1 binding, underlie a syndrome of X-linked thrombocytopenia with or without associated anemia.14,15 It has been suggested that distinct domains of FOG-1 differentially influence GATA-1–dependent megakaryocytic versus erythroid differentiation,16 implying a role for FOG-1 in the divergence of erythroid and megakaryocytic lineages. However, despite the demonstrated importance of GATA and FOG factors in erythroid and megakaryocyte differentiation, no mechanisms have been discerned for lineage-specific function. It is likely that additional transcription factors exist that function as specificity determinants for GATA factors.
RUNX1 (previously known as AML1) is the evolutionarily conserved DNA-binding subunit of a transcription complex known as core-binding factor (CBF).17 CBFβ, the non-DNA binding component of CBF, binds RUNX1 and enhances its DNA-binding ability.18 The RUNX1 gene is the frequent target of chromosomal translocations in leukemia—for example, t(8;21) in acute myeloid leukemia—resulting in a dominant-negative RUNX1-ETO fusion protein.19,20 The CBF complex is essential for definitive hematopoiesis, and knock-out of RUNX1 or CBFβ in mice is lethal at midgestation because of central nervous system hemorrhage and necrosis.17 Human germline mutations of RUNX1 were recently associated with autosomal dominant familial platelet disorder with predisposition to acute myelogenous leukemia.21,22 RUNX1 haploinsufficiency in humans impairs some aspect of megakaryopoiesis, but the mechanistic details of this effect are unknown.
This report provides evidence for RUNX1 and CBFβ involvement in divergence of the megakaryocytic from the erythroid lineage. RUNX1 manifested a unique expression pattern in human bone marrow, with high levels in megakaryocytes and minimal levels in erythroid cells. This pattern contrasted with GATA-1, in which similar expression was seen in both lineages. Furthermore, a novel functional interaction among RUNX1, CBFβ, and GATA-1 occurred in the activation of the αIIb integrin promoter; a corresponding physical interaction of RUNX1 and GATA-1 was also demonstrable. Notably, the amino terminal transcription activation domain of GATA-1, which undergoes deletion in acute megakaryoblastic leukemias of Down syndrome (M7-DS), was required for physical and functional interactions with RUNX1.23 The leukemic fusion protein RUNX1-ETO also physically interacted with GATA-1, but strongly inhibited its activation of the αIIb promoter. Thus RUNX1, through its interaction with GATA-1, may participate in programming the divergence of the megakaryocytic from the erythroid lineages. Conversely, one of the leukemogenic mechanisms of RUNX1-ETO may consist of inhibition of the GATA factor function. Furthermore, one of the leukemogenic mechanisms of the GATA-1 mutations in M7-DS may be loss of interaction with RUNX1.
Materials and methods
Plasmids and constructs
pEF-GATA-1 FL and related deletion mutants were provided by Dr Jane Visvader (Walter and Eliza Hall Institute of Medical Research, Melbourne, Australia).24 pCMV-RUNX1 FL (AML1b), pCMV-RUNX1 1-381, and pCMV-RUNX1-ETO plasmids were provided by Dr Scott Hiebert (Vanderbilt University School of Medicine, Nashville, TN).25 pCMV-CBFβ was provided by Dr Alan Friedman (Johns Hopkins Medical Institutions, Baltimore, MD).26 pCMV-RUNX1 41-190 was generated by subcloning of an XbaI-BamHI fragment from pET3C-RUNX1 41-190 into the corresponding sites of pCMV5; pET3C-RUNX1 41-190 was provided by Dr John Bushweller (University of Virginia, Charlottesville). The FLAG-tagged GATA-1 mutant FLAG-Δ1-85 was generated by subcloning an EcoRI fragment from pXM-GATA-127 into the EcoRI site of pCMV-Tag 2C. For mammalian expression of glutathione-S-transferase (GST) fusions, full-length GATA-1 and RUNX1 coding sequences were inserted in-frame as BglII-NotI fragments into the BamHI-NotI sites of pEBG, a vector provided by Dr Janet Cross (University of Virginia, Charlottesville). αIIb-598-Luciferase, αIIb-348-Luciferase, and αIIb-98-Luciferase, produced in our laboratory, have been previously described.28 Consensus and variant RUNX-binding sites within αIIb-598 were identified using 2 web-based programs, TFSEARCH (www.cbrc.jp/research/db/TFSEARCH.html) and TESS (www.cbil.upenn.edu/tess), both of which use the TRANSFAC database. Identified sites (—432, —413, —273, and —69) were individually mutated to the non-RUNX–binding sequence TGTTAG29 using overlap polymerase chain reaction (PCR) followed by cloning into pGL3-Basic (Promega, Madison, WI) and subsequent sequence verification. For retroviral transduction, full-length RUNX1 (RX1-FL) was inserted into the EcoRI site of MSCV-IRES-GFP30 to yield MIG-RX1-FL.
Cell culture
The bone marrow stromal cell line HESS-5 and the human hematopoietic cell line K562 were grown as previously described.31 Megakaryocytic induction used 10 to 25 nM phorbol ester (TPA [12-O-tetradecanoylphorbol-13-acetate]) for 48 hours. Inhibition of MKK1/2 made use of 20 μM U0126 (Promega) versus dimethyl sulfoxide (DMSO) solvent control added 30 minutes before megakaryocytic induction. Purified human peripheral blood, granulocyte–colony-stimulating factor (G-CSF)–mobilized CD34+ cells (Poietic Technologies, Gaithersburg, MD) were cultured at 105 cells/mL in serum-free defined medium (StemPro; Gibco, Grand Island, NY) under erythroid (3 U/mL erythropoietin [EPO], 25 ng/mL stem cell factor [SCF]) or megakaryocytic (40 ng/mL thrombopoietin [TPO], 25 ng/mL SCF, and 25 nM ingenol dibenzoate) conditions. C3H10T1/2 cells were maintained in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum at 37°C, 5% CO2. HEK293T cells were maintained in DMEM with 10% fetal bovine serum at 37°C, 5% CO2. 293GPG retroviral packaging cells32 were grown in DMEM supplemented with 10% fetal bovine serum, 1 μg/mL tetracycline, 2 μg/mL puromycin, and 0.3 mg/mL G418.
Immunohistochemistry
Healthy human marrow clot sections obtained to rule out metastatic tumor were fixed in standard neutral-buffered formalin. Immunoperoxidase staining was performed using avidin-biotin complex (ABC; Vector, Burlingame, CA) on paraffin-embedded samples. Counterstaining was performed with Harris hematoxylin. Antibodies consisted of rabbit polyclonal anti-RUNX1, anti–GATA-1, and mouse monoclonal anti-CBFβ (all from Geneka, Montreal, Canada).
Transient transfection and reporter assays
K562 cells (2 × 106) were transfected with a fixed quantity of DNA (6.5 μg) using DOTAP transfection reagent (Roche, Indianapolis, IN) according to the manufacturer's instructions. All transfections included 1.5 μg Luciferase reporter and 0.5 μg pCMV-βGal. In addition, 1.5 μg each indicated expression plasmid alone or in various combinations was included. Parent vector was used to maintain a constant DNA amount. For dosage dependency of RUNX1-ETO inhibition of GATA-1, a constant amount of pEF-GATA-1 (1.5 μg) was cotransfected with variable amounts of pCMV-RUNX1-ETO. Total DNA was kept constant with parent vector. C3H10T1/2 cells were transfected using calcium phosphate precipitation with 20 μg DNA per 10-cm plate—4.5 μg each expression vector, 4.5 μg Luciferase reporter, and 2 μg pCMV-βGal. Cells were lysed 40 hours after transfection with cell culture lysis buffer (Promega) and were assayed for Luciferase activities by a Luciferase assay kit (Promega). Transfection efficiencies were normalized with β-gal assays (Tropix/Applied Biosystems, Bedford, MA).
Immunoblot
K562 whole cell lysates (105 viable cell equivalents in 10 μL per lane) were subjected to standard immunoblot analysis with rabbit polyclonal anti-RUNX1 (Geneka), rabbit anti–phospho-ERK (Promega), and mouse antitubulin (Sigma, St Louis, MO). Primary human progenitor cells were subjected to whole cell lysis at different time points after megakaryocytic or erythroid induction, and 105 viable cell equivalents in 10 μL per lane were immunoblotted with the following: rabbit polyclonal anti-RUNX1, mouse monoclonal anti-CBFβ (Geneka), rat monoclonal anti–GATA-1 (N1; Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-gpIIb (kindly provided by Dr Dan Rosson, Lankenau Medical Research Center, Wynnewood, PA), and rabbit anti-HbA (AXL 241) (Accurate Chemical & Scientific, Westbury, NY). For transiently transfected K562 cells from Luciferase assays, whole cell lysates from duplicate transfections were subjected to immunoblotting for GATA-1, RUNX1, CBFβ, and tubulin as above, except that RUNX1-ETO protein was detected with rabbit antibody to the runt homology domain (rhd; Oncogen Research Products, Boston, MA). Immunoblotting for the α2 integrin protein in K562 cells transduced with control vector or the RUNX1 expression construct used rabbit anti-integrin α2 (H-293; Santa Cruz Biotechnology).
Coimmunoprecipitation
293T cells were cotransfected with 6 μg indicated expression constructs per 10-cm plate using FuGENE6 (Roche). For initial experiments, the carboxy terminal truncated mutant RUNX1 1-381 was used because it is readily extractable from the nucleus as opposed to the inextractable full-length RUNX1.33 Initially, CBFβ coexpression was used as a means to stabilize RUNX1.34 Cells harvested 48 hours after transfection were extracted for 20 minutes on ice with 1 × 106 cells/100 μL extraction buffer (50 mM Tris HCl, pH 7.6, 150 mM NaCl, 0.5% NP-40, 1 mM MgCl2,50 μM ZnSO4,1× EDTA [ethylenediaminetetraacetic acid]–free protease inhibitor cocktail [Roche], 10 mM NaF, 1 mM Na3VO4). Extracts were precleared by centrifugation twice at 14 000 rpm at 4°C for 15 minutes. Immunoprecipitation was performed for 2 hours at 4°C with either monoclonal rat anti–GATA-1 (N6) or rabbit anti-RUNX1 (rhd) followed by complex recovery with protein G-agarose beads, which were washed 4 to 6 times with ice-cold extraction buffer. Immune complexes were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblotting. Antibodies used for immunoblotting were as described above, except that rabbit anti–GATA-1 (Geneka) was used to probe rat anti–GATA-1 immunoprecipitates. For GST pull-down, transfection of 293T cells, extraction, and preclearance were performed as for the coimmunoprecipitations. Extracts were diluted 2- to 3-fold in extraction buffer (depending on initial immunoblot determinations), and 25 μL packed volume glutathione agarose beads was added to 300 μL extract. After 90-minute incubation at 4°C, beads were washed 5 to 6 times in ice-cold extraction buffer. Proteins were then eluted in SDS-PAGE loading buffer and analyzed by immunoblot as for the coimmunoprecipitations. Immunoblotting for RUNX1 made use of rabbit antibody to the runt homology domain (rhd; Oncogen Research Products). Immunoblotting for GATA-1 used the N1 rat monoclonal antibody (Santa Cruz Biotechnology). Immunoblotting for FLAG epitope used the mouse monoclonal M2 antibody (Sigma).
Retroviral transduction
Retroviral supernatants were prepared by transfecting 293GPG packaging cells with 6 μg plasmid/10-cm plate using FuGENE 6 (Roche, Indianapolis, IN). After 16 hours, cells were washed and grown in DMEM containing only 15% fetal bovine serum; supernatants were collected daily between 48 and 120 hours after transfection. K562 cells were transduced by spinoculation, in which 5 × 105 cells were suspended in 2 mL viral supernatant containing 2 mM (5 μg/mL) polybrene (hexadimethrine bromide) and spun at 2000 rpm for 90 minutes at room temperature. GFP+ cells were isolated
Results
In vitro and in vivo RUNX1 expression in megakaryocytic but not erythroid differentiation
Previously, a proteomic approach was applied to identify cellular targets of the ERK/MAPK signaling pathway in K562 cells induced to undergo megakaryocytic differentiation.35 We have used this cellular system to identify candidate transcription factors involved in megakaryocytic differentiation. In this system, prolonged treatment of K562 cells with TPA leads to the up-regulation of multiple megakaryocyte-specific genes and the down-regulation of erythroid genes. Activation of this differentiation program is dependent on sustained ERK/MAPK activation and is specifically blocked by bone marrow stromal cell contact.31,36 We have identified the hematopoietic transcription factor RUNX1 as a cellular target in this system. In the absence of stromal contact, RUNX1 protein underwent significant up-regulation during 48 hours of megakaryocytic induction (Figure 1A). The presence of a stromal monolayer clearly inhibited this up-regulation of RUNX1, correlating with stromal blockade of megakaryocytic differentiation. Furthermore, the up-regulation of RUNX1 during megakaryocytic induction also correlated with ERK/MAPK phosphorylation (Figure 1B). Inhibition of MKK1/2 with U0126 blocked RUNX1 up-regulation and diminished RUNX1 basal levels.
Expression of RUNX1 correlates with megakaryocytic induction of K562 cells. (A) Immunoblottting of whole cell lysates with rabbit anti-RUNX1. K562 cells were grown for 48 hours with or without TPA and with or without a bone marrow stromal monolayer. Immunoblotting with mouse antitubulin was performed to control for lane loading. (B) Immunoblotting of whole cell lysates with rabbit anti-RUNX1, rabbit anti–phospho-ERK, and mouse antitubulin. K562 cells were grown for 48 hours with or without TPA and with or without the MKK1/2 inhibitor U0126.
Expression of RUNX1 correlates with megakaryocytic induction of K562 cells. (A) Immunoblottting of whole cell lysates with rabbit anti-RUNX1. K562 cells were grown for 48 hours with or without TPA and with or without a bone marrow stromal monolayer. Immunoblotting with mouse antitubulin was performed to control for lane loading. (B) Immunoblotting of whole cell lysates with rabbit anti-RUNX1, rabbit anti–phospho-ERK, and mouse antitubulin. K562 cells were grown for 48 hours with or without TPA and with or without the MKK1/2 inhibitor U0126.
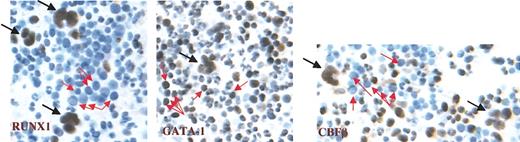
To determine the in vivo pattern of RUNX1 expression in human hematopoietic progenitor cells, bone marrow biopsy samples were subjected to immunohistochemical staining (Figure 2). Strikingly, RUNX1 expression was restricted to megakaryocytes (black arrows) and was weak to absent in erythroblasts (red arrows). Occasional immature myeloid precursors also expressed RUNX1. By contrast, GATA-1 showed equivalent expression in megakaryocytes (black arrows) and erythroblasts (red arrows). CBFβ, the cofactor for RUNX1, manifested an expression pattern similar to that of RUNX1, present in megakaryocytes and weak or absent in erythroblasts; unlike RUNX1, however, CBFβ also displayed expression in more mature myeloid cells.
Expression profile of RUNX1, GATA-1, and CBFβ in healthy human bone marrow. Left panel, RUNX1; middle panel, GATA-1; right panel, CBFβ. Black arrows indicate megakaryocytes, and red arrows indicate erythroblasts. Immunoperoxidase staining was performed using avidin-biotin complex on paraffin-embedded marrow clot section. Slides were counterstained with Harris hematoxylin (original magnification, × 400).
Expression profile of RUNX1, GATA-1, and CBFβ in healthy human bone marrow. Left panel, RUNX1; middle panel, GATA-1; right panel, CBFβ. Black arrows indicate megakaryocytes, and red arrows indicate erythroblasts. Immunoperoxidase staining was performed using avidin-biotin complex on paraffin-embedded marrow clot section. Slides were counterstained with Harris hematoxylin (original magnification, × 400).
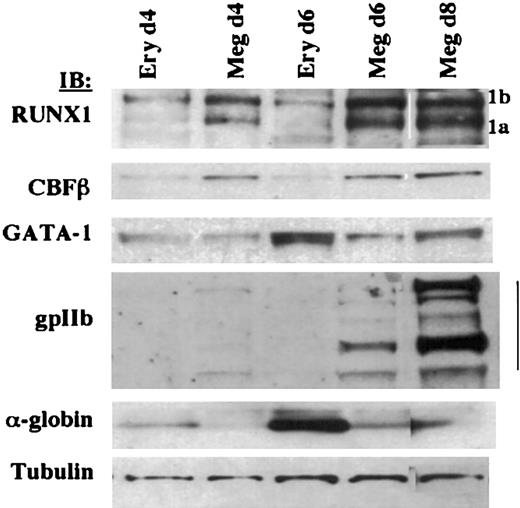
To characterize the kinetics of RUNX1 expression during megakaryocytic and erythroid differentiation, primary human CD34+ hematopoietic progenitor cells cultured under megakaryocytic and erythroid conditions were analyzed. RUNX1 and CBFβ exhibited differential expression in megakaryocytic versus erythroid culture conditions as early as 4 days (Figure 3) before the appearance of lineage-specific markers (gpIIb and α-globin). GATA-1, by contrast, maintained similar expression in day 4 erythroid and megakaryocytic cultures and underwent up-regulation in day 6 erythroid cultures, concurrent with the up-regulation of α-globin.
Expression profile of RUNX1, GATA-1, and CBFβ in primary CD34+human hematopoietic progenitors undergoing megakaryocytic and erythroid differentiation. Immunoblotting was performed on whole cell lysates with rabbit anti-RUNX1, mouse anti-CBFβ, rat anti–GATA-1 (N1), rabbit anti-gpIIb, rabbit anti–α-globin, and mouse antitubulin. Human CD34+ cells were grown in megakaryocytic or erythroid differentiation medium, and cells were harvested at the indicated durations. 1b and 1a refer to the AML1b and AML1a isoforms.
Expression profile of RUNX1, GATA-1, and CBFβ in primary CD34+human hematopoietic progenitors undergoing megakaryocytic and erythroid differentiation. Immunoblotting was performed on whole cell lysates with rabbit anti-RUNX1, mouse anti-CBFβ, rat anti–GATA-1 (N1), rabbit anti-gpIIb, rabbit anti–α-globin, and mouse antitubulin. Human CD34+ cells were grown in megakaryocytic or erythroid differentiation medium, and cells were harvested at the indicated durations. 1b and 1a refer to the AML1b and AML1a isoforms.
These data confirm the differential expression of RUNX1 and CBFβ in the megakaryocytic versus erythroid lineages. Results with CBFβ corroborate the recent findings of Kundu et al,37 who showed with a Cbfb-GFP knock-in mouse model that Cbfb promoter activity is maintained during megakaryocytic differentiation but is extinguished during erythroid differentiation. The expression pattern we observed for RUNX1 and CBFβ stands in contrast to the GATA factors, FOG1, NF-E2, and SCL/tal, all of which show expression in erythroid and megakaryocytic lineages.3,38,39 Furthermore, our data show that this pattern of RUNX1 and CBFβ differential expression occurs before the appearance of lineage-specific markers (αIIb integrin and α-globin). Thus, the pattern and developmental timing of RUNX1/CBFβ expression in primary human marrow cells are compatible with a role in programming the divergence of the megakaryocytic and erythroid lineages.
Functional cooperation of RUNX1 and GATA-1
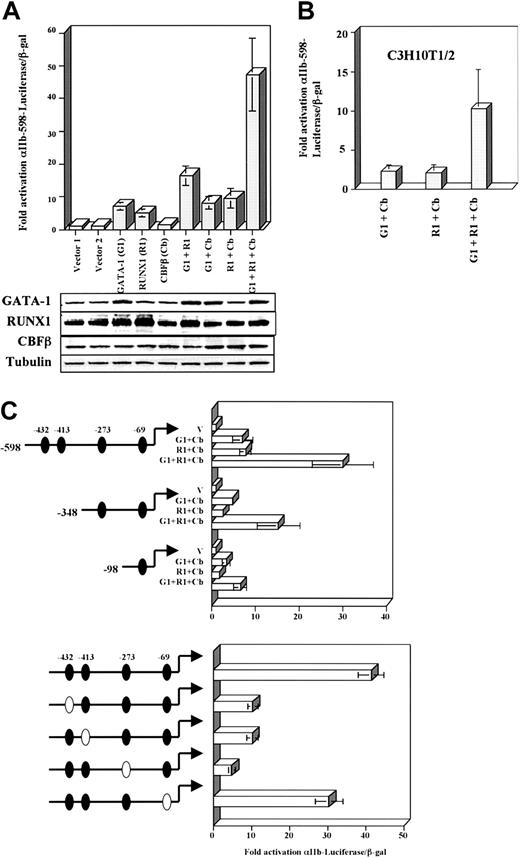
To address potential functional contributions to megakaryocytic lineage commitment, we examined the ability of RUNX1 to activate the αIIb integrin promoter. The —598 to +32 core αIIb promoter region has been demonstrated to dictate megakaryocyte-specific gene expression in transgenic embryonic stem cells.40 A standard transient cotransfection approach initially addressed the ability of RUNX1, alone or with other factors, to activate an αIIb-598–Luciferase reporter. RUNX1 alone and GATA-1 alone each transactivated αIIb-598-Luc to a similar degree (Figure 4A); CBFβ alone had minimal activity. Modest functional cooperation occurred with GATA-1 plus RUNX1, and marked synergy resulted with GATA-1 plus RUNX1 plus CBFβ. Immunoblots of duplicate parallel transfections demonstrated only moderate up-regulation of proteins encoded by the transfected genes, highlighting the potency of the synergy and ruling out construct expression artifacts. To rule out any effects possibly caused by cellular context, a similar analysis was carried out in C3H10T1/2 fibroblasts, which do not express appreciable amounts of endogenous GATA-1, RUNX1, or CBFβ (data not shown). As in K562 cells, GATA-1 showed marked synergy with RUNX1 plus CBFβ in the activation of the αIIb promoter in C3H10T1/2 fibroblasts (Figure 4B).
Cooperation of GATA-1, RUNX1, and CBFβ in transcriptional activation of the αIIb promoter. (A) Transactivation of αIIb-598-Luciferase reporter by GATA-1, RUNX1, and CBFβ. K562 cells were transiently cotransfected with the indicated vectors. Results were expressed as fold activation relative to empty expression vector control and were normalized for transfection efficiency with β-gal assays. Results represent the mean ± SEM of 3 independent experiments. To confirm transgene expression, whole cell lysates from duplicate transfections were subjected to immunoblot with the indicated antibodies. (B) Transactivation of αIIb-598-Luciferase reporter by GATA-1, RUNX1, and CBFβ in transiently transfected C3H10T1/2 fibroblasts. Results are expressed as fold activation relative to empty expression vector control and are normalized for transfection efficiency with β-gal assays. Results represent the mean ± SEM of 3 independent experiments. (C) Cis-acting sequences involved in transcriptional activation by GATA-1 plus RUNX1/CBFβ. Shown in the upper panel are αIIb 5′ promoter truncations: αIIb-348-Luciferase and αIIb-98-Luciferase were compared with αIIb-598-Luciferase in cotransfection assays, as above. Positions of RUNX sites, as determined by the TFSEARCH and TESS programs, are indicated by ovals. Shown in the lower panel are αIIB-598 promoter mutants in which individual RUNX sites have been mutated to the non-RUNX–binding sequence TGTTAG29 (hollow ovals). These Luciferase reporters were cotransfected into K562 cells with expression vectors for GATA-1 and RUNX1/CBFβ. Results are expressed as fold activation relative to empty expression vector control, normalized with β-gal assays. Results represent the mean ± SEM of 3 independent experiments.
Cooperation of GATA-1, RUNX1, and CBFβ in transcriptional activation of the αIIb promoter. (A) Transactivation of αIIb-598-Luciferase reporter by GATA-1, RUNX1, and CBFβ. K562 cells were transiently cotransfected with the indicated vectors. Results were expressed as fold activation relative to empty expression vector control and were normalized for transfection efficiency with β-gal assays. Results represent the mean ± SEM of 3 independent experiments. To confirm transgene expression, whole cell lysates from duplicate transfections were subjected to immunoblot with the indicated antibodies. (B) Transactivation of αIIb-598-Luciferase reporter by GATA-1, RUNX1, and CBFβ in transiently transfected C3H10T1/2 fibroblasts. Results are expressed as fold activation relative to empty expression vector control and are normalized for transfection efficiency with β-gal assays. Results represent the mean ± SEM of 3 independent experiments. (C) Cis-acting sequences involved in transcriptional activation by GATA-1 plus RUNX1/CBFβ. Shown in the upper panel are αIIb 5′ promoter truncations: αIIb-348-Luciferase and αIIb-98-Luciferase were compared with αIIb-598-Luciferase in cotransfection assays, as above. Positions of RUNX sites, as determined by the TFSEARCH and TESS programs, are indicated by ovals. Shown in the lower panel are αIIB-598 promoter mutants in which individual RUNX sites have been mutated to the non-RUNX–binding sequence TGTTAG29 (hollow ovals). These Luciferase reporters were cotransfected into K562 cells with expression vectors for GATA-1 and RUNX1/CBFβ. Results are expressed as fold activation relative to empty expression vector control, normalized with β-gal assays. Results represent the mean ± SEM of 3 independent experiments.
To determine whether cis-acting promoter sequences contributed to the synergy, truncations of the αIIb promoter were initially analyzed for responsiveness to GATA-1, RUNX1, and CBFβ. Notably, functionally equivalent GATA-binding sites are present in all 3 promoter truncations.28 Elimination of the 5′ 250 bp (—598 to —348) caused a significant loss in responsiveness to RUNX1/ CBFβ alone or with GATA-1 (Figure 4C), suggesting a role for the RUNX consensus site at —413 (ACCACA). However, residual functional interaction between RUNX1/CBFβ and GATA-1 existed with αIIb —348 and, to a lesser degree, with αIIb —98, suggesting roles for the variant RUNX sites at —273 and —69 or for DNA-independent effects. To ascertain the importance of specific binding sites, we mutated each of the 4 RUNX-binding sites predicted from database searches (TFSEARCH and TESS). As shown in Figure 4C (lower panel), individual mutations in 3 of the RUNX-binding sites (—432, —413, and —273) markedly diminished responsiveness of the αIIb promoter to GATA-1 plus RUNX1/CBFβ. Surprisingly, the most drastic loss of responsiveness occurred with mutation of the variant RUNX site at —273 (TGGGGT). Therefore, multiple RUNX-binding sites, including consensus and variant sites, are required for optimal responsiveness of the αIIb promoter to GATA-1 plus RUNX1/CBFβ.
Leukemic fusion RUNX1-ETO inhibits GATA-1
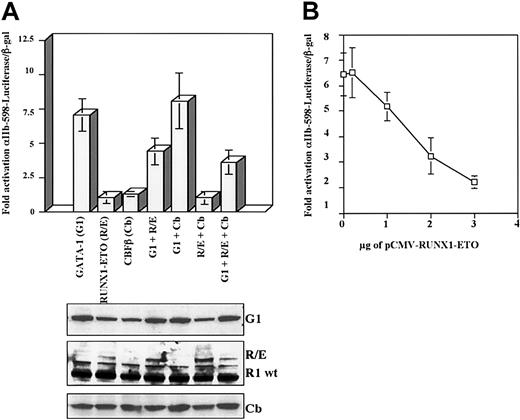
Because wild-type RUNX1 augmented GATA-1–mediated promoter activation, we examined whether the dominant-negative fusion RUNX1-ETO might antagonize GATA-1 function. As predicted, GATA-1 transactivation of the αIIb promoter was clearly inhibited by coexpression of the RUNX1-ETO fusion protein (Figure 5). The inhibition of GATA-1 function by RUNX1-ETO did not appear to be significantly enhanced by CBFβ, which is known to enhance DNA binding by RUNX1-ETO.18 The inhibitory effects of RUNX1-ETO seemed to be potent, as suggested by the corresponding immunoblot in which levels of transfected RUNX1-ETO were significantly less than the levels of endogenous RUNX1. A dose-response titration (Figure 5B) confirmed direct dosage dependency of RUNX1-ETO in its inhibition of GATA-1.
Inhibition of GATA-1–mediated transcriptional activation by RUNX1-ETO. (A) RUNX1-ETO inhibition of GATA-1 function in transactivation of the αIIb-598 promoter. Cotransfections were performed exactly as in Figure 4A, except that RUNX1-ETO was expressed instead of RUNX1. Results represent the mean ± SEM of 3 independent experiments. Whole cell lysates from duplicate transfections were immunoblotted with the indicated antibodies. (B) Dosage dependency of RUNX1-ETO mediated inhibition of GATA-1. A constant amount of pEF-GATA-1 (1.5 μg) was cotransfected with the indicated amounts of pCMV-RUNX1-ETO. Activation of the αIIb-598-Luc reporter was determined as in Figure 4A. Results represent the mean ± SEM of 3 independent experiments.
Inhibition of GATA-1–mediated transcriptional activation by RUNX1-ETO. (A) RUNX1-ETO inhibition of GATA-1 function in transactivation of the αIIb-598 promoter. Cotransfections were performed exactly as in Figure 4A, except that RUNX1-ETO was expressed instead of RUNX1. Results represent the mean ± SEM of 3 independent experiments. Whole cell lysates from duplicate transfections were immunoblotted with the indicated antibodies. (B) Dosage dependency of RUNX1-ETO mediated inhibition of GATA-1. A constant amount of pEF-GATA-1 (1.5 μg) was cotransfected with the indicated amounts of pCMV-RUNX1-ETO. Activation of the αIIb-598-Luc reporter was determined as in Figure 4A. Results represent the mean ± SEM of 3 independent experiments.
Physical association of RUNX1 and GATA-1
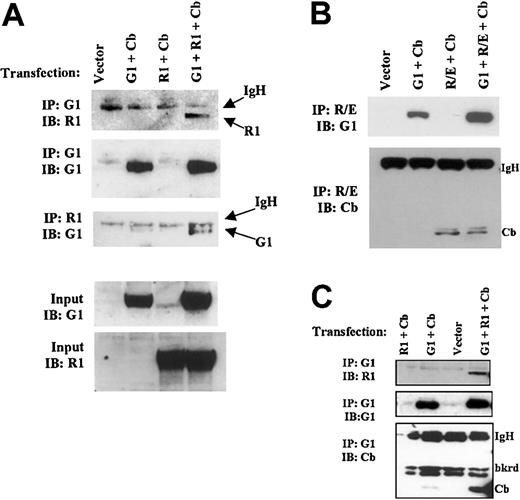
Functional interactions of GATA-1 with RUNX1/CBFβ and RUNX1-ETO raised the possibility of physical interactions among these transcription factors. To test this possibility, we first conducted bidirectional coimmunoprecipitation assays. Because CBFβ induces RUNX1 to adopt an active, stable conformation, experiments were initially performed with the coexpression of CBFβ.18,34 In appropriately transfected cells, immunoprecipitation with anti–GATA-1 or anti-RUNX1 demonstrated specific coprecipitation of RUNX1 and GATA-1 (Figure 6A). Similarly, the immunoprecipitation of RUNX1-ETO demonstrated coprecipitation of GATA-1 (Figure 6B); a significantly weaker signal, seen in the absence of RUNX1-ETO coexpression (Figure 6B, lane 2), was attributed to the previously described background binding of GATA-1 to protein G beads.41 As a positive control, CBFβ manifested coimmunoprecipitation with RUNX1-ETO with or without GATA-1 coexpression (Figure 6B). Further immunoprecipitation experiments were conducted to address whether GATA-1, RUNX1, and CBFβ coexist in a common complex. CBFβ did not significantly coprecipitate with GATA-1 unless RUNX1 was also expressed (Figure 6C), suggesting that RUNX1, CBFβ, and GATA-1 indeed participate in a common complex.
Physical association of GATA-1 with RUNX1 and with RUNX1-ETO. (A) Interaction of GATA-1 and RUNX1 in IP assays. HEK293T cells were cotransfected with pCMV-RUNX1 1-381, pEF-GATA-1, and pCMV-CBFβ, as indicated. Cellular extracts were immunoprecipitated (IP) using either rat anti–GATA-1 (N6) or rabbit anti-RUNX1 (rhd). Immunoprecipitates were immunoblotted (IB) with the indicated antibodies. (B) Interaction of GATA-1 and RUNX1-ETO. Immunoprecipitation–immunoblot assays were performed as in panel A, except that cells were transfected with pCMV-RUNX1-ETO instead of pCMV-RUNX1 1-381. (C) Participation of GATA-1, RUNX1, and CBFβ in a common complex. Immunoprecipitations were performed with rat anti–GATA-1 (N6) as in panel A, followed by immunoblotting with the indicated antibodies.
Physical association of GATA-1 with RUNX1 and with RUNX1-ETO. (A) Interaction of GATA-1 and RUNX1 in IP assays. HEK293T cells were cotransfected with pCMV-RUNX1 1-381, pEF-GATA-1, and pCMV-CBFβ, as indicated. Cellular extracts were immunoprecipitated (IP) using either rat anti–GATA-1 (N6) or rabbit anti-RUNX1 (rhd). Immunoprecipitates were immunoblotted (IB) with the indicated antibodies. (B) Interaction of GATA-1 and RUNX1-ETO. Immunoprecipitation–immunoblot assays were performed as in panel A, except that cells were transfected with pCMV-RUNX1-ETO instead of pCMV-RUNX1 1-381. (C) Participation of GATA-1, RUNX1, and CBFβ in a common complex. Immunoprecipitations were performed with rat anti–GATA-1 (N6) as in panel A, followed by immunoblotting with the indicated antibodies.
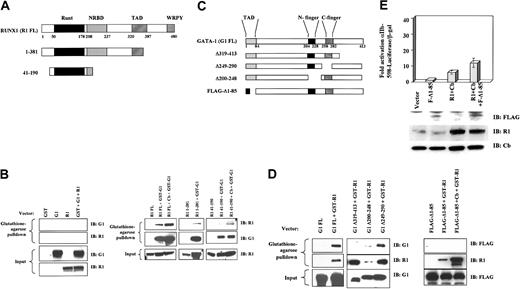
To map domains critical in the physical interaction of GATA-1 and RUNX1, we used a mammalian GST pull-down system. The indicated RUNX1 and GATA-1 mutants (Figure 7A,C) were coexpressed in 293T cells, respectively, with GST–GATA-1 (full-length) or GST-RUNX1 (full-length AML1b). In some experiments, CBFβ was also expressed, as indicated. Proteins pulled-down with glutathione-agarose beads were then subjected to immunoblot detection. As shown in Figure 7B (left panel), neither GATA-1 nor RUNX1 interacted with GST only. GST–GATA-1 interacted with full-length RUNX1 and with the truncation mutants, including the minimal runt domain (R1 41-190) (Figure 7B, left panel). Full-length RUNX1 appeared to bind GST–GATA-1 with the greatest efficiency, particularly in the presence of CBFβ. Notably, the minimal runt domain required the coexpression of CBFβ for appreciable binding to GST–GATA-1.
Mapping of domains important in the interaction of RUNX1 with GATA-1. Protein interactions were analyzed using mammalian GST pull-down. Vectors expressing GST alone, GST-GATA-1 full length, or GST-RUNX1 full length were cotransfected with expression vectors for RUNX1 and GATA-1 mutants into 293T cells. (A) Diagram of RUNX1 proteins analyzed, indicating the following domains: runt, negative regulatory domain for DNA binding (NRBD), transcriptional activation domain (TAD), and WRPY sequence, which binds TLE/groucho corepressors. (B) Absence of RUNX1 and GATA-1 binding to GST (left panel). Interaction of RUNX1 mutants with GST–GATA-1; where indicated, CBFβ (Cb) was coexpressed (right panel). (C) Diagram of GATA-1 proteins analyzed, indicating TAD and zinc fingers (left panel). (D) Interaction of GATA-1 mutants with GST-RUNX1; where indicated, CBFβ (Cb) was coexpressed (right panel). The Δ1-85 GATA-1 mutant was expressed with an amino terminal FLAG tag because of loss of epitopes recognized by the N1 and N6 monoclonal antibodies. (E) The Δ1-85 GATA-1 mutant shows loss of cooperation with RUNX1 plus CBFβ. Transactivation of αIIb-598-Luciferase reporter by FLAG-Δ1-85 GATA-1 (F-Δ1-85), RUNX1 (R1), and CBFβ (Cb). K562 cells were transiently cotransfected with the indicated vectors. Results represent the mean ± SEM of 3 independent experiments. Whole cell lysates from duplicate transfections were immunoblotted with the indicated antibodies.
Mapping of domains important in the interaction of RUNX1 with GATA-1. Protein interactions were analyzed using mammalian GST pull-down. Vectors expressing GST alone, GST-GATA-1 full length, or GST-RUNX1 full length were cotransfected with expression vectors for RUNX1 and GATA-1 mutants into 293T cells. (A) Diagram of RUNX1 proteins analyzed, indicating the following domains: runt, negative regulatory domain for DNA binding (NRBD), transcriptional activation domain (TAD), and WRPY sequence, which binds TLE/groucho corepressors. (B) Absence of RUNX1 and GATA-1 binding to GST (left panel). Interaction of RUNX1 mutants with GST–GATA-1; where indicated, CBFβ (Cb) was coexpressed (right panel). (C) Diagram of GATA-1 proteins analyzed, indicating TAD and zinc fingers (left panel). (D) Interaction of GATA-1 mutants with GST-RUNX1; where indicated, CBFβ (Cb) was coexpressed (right panel). The Δ1-85 GATA-1 mutant was expressed with an amino terminal FLAG tag because of loss of epitopes recognized by the N1 and N6 monoclonal antibodies. (E) The Δ1-85 GATA-1 mutant shows loss of cooperation with RUNX1 plus CBFβ. Transactivation of αIIb-598-Luciferase reporter by FLAG-Δ1-85 GATA-1 (F-Δ1-85), RUNX1 (R1), and CBFβ (Cb). K562 cells were transiently cotransfected with the indicated vectors. Results represent the mean ± SEM of 3 independent experiments. Whole cell lysates from duplicate transfections were immunoblotted with the indicated antibodies.
As indicated in Figure 7D, full-length GATA-1 bound efficiently to GST-RUNX1. Interestingly, the deletion of either N-finger or C-finger did not significantly impair binding of GATA-1 to GST-RUNX1. The diminished signal seen for the N-finger deletion mutant (G1 Δ200-248) was simply a result of the diminished expression of GST-RUNX1 in this particular transfection. Surprisingly, deletion of the carboxy terminal 95 amino acids (G1 Δ319-413) completely abolished the binding of GATA-1 to GST-RUNX1. Furthermore, deletion of the amino terminal transcription activation domain of GATA-1 (FLAG-Δ1-85) also completely abolished binding to GST-RUNX1, despite very high expression levels of both proteins (Figure 7D). FLAG-tagged GATA-1 was used in this assay because the amino terminal deletions eliminate the epitope recognized by the N1 and N6 antibodies. In summary, mapping studies have shown that the runt domain, in the presence of CBFβ coexpression, is sufficient for RUNX1 interaction with GATA-1. In the absence of CBFβ, an additional sequence within RUNX1 is required to bind GATA-1. Within GATA-1, the amino and carboxy termini (amino acids 1-85 and 319-413, respectively) are critical for binding RUNX1, whereas the N- and C-finger domains appear to be dispensable.
To examine the functional importance of the GATA-1 amino terminal domain required for RUNX1 binding, we examined the ability of the GATA-1 mutant FLAG-Δ1-85 to cooperate with RUNX1 in the activation of the αIIb promoter. As shown in Figure 7E, FLAG-Δ1-85 showed minimal functional cooperation with wild-type RUNX1. The combination of FLAG-Δ1-85 plus RUNX1 barely enhanced the activation of the αIIb promoter over that seen with RUNX1 alone and was far less potent than the combination of wild-type GATA-1 plus RUNX1: 12-fold versus 40- to 50-fold (compare Figures 7E and 4A,C).
RUNX1 influences the endogenous megakaryocytic differentiation program
To verify a role for RUNX1 in megakaryopoiesis, we examined the effects of enforced RUNX1 expression on megakaryocytic differentiation. K562 cells were transduced with a RUNX1 retroviral construct (MIG-RX1-FL) or with control vector (MIG). Transduced GFP+ cells were sorted and subjected to megakaryocytic induction with TPA. At 48 hours of induction, cells transduced with RUNX1 manifested 2-fold enhancement of gpIIb up-regulation and more than 10-fold enhancement of α2 integrin up-regulation compared with cells transduced with control vector (Figure 8). Cells transduced with MIG-RX1-FL expressed 2- to 3-fold more RUNX1 than cells transduced with control vector (Figure 8). These results support the participation of RUNX1 in the activation of endogenous megakaryocytic integrin promoters during megakaryocytic differentiation. Notably, though, the overexpression of RUNX1 in K562 cells was not sufficient for gpIIb and α2 integrin up-regulation in the absence of megakaryocytic induction.
Enforced expression of RUNX1 is associated with enhanced up-regulation of the megakaryocytic integrins αIIb and α2. K562 cells were transduced with a RUNX1 retroviral construct (MIG-RX1-FL) or a control vector (MIG). GFP+ cells were sorted and induced to undergo megakaryocytic differentiation as described in “Materials and methods.” Whole cell lysates were immunoblotted with the indicated antibodies.
Enforced expression of RUNX1 is associated with enhanced up-regulation of the megakaryocytic integrins αIIb and α2. K562 cells were transduced with a RUNX1 retroviral construct (MIG-RX1-FL) or a control vector (MIG). GFP+ cells were sorted and induced to undergo megakaryocytic differentiation as described in “Materials and methods.” Whole cell lysates were immunoblotted with the indicated antibodies.
Discussion
The unique expression pattern of RUNX1 and CBFβ in Figures 2 and 3 suggests participation in the divergence of the megakaryocytic lineage from the erythroid lineage. Virtually all transcription factors previously implicated in megakaryocytic development have demonstrated coexpression and function in erythroid development. For example, GATA-1 and GATA-2, along with their cofactor FOG-1, clearly play critical roles in megakaryocytic and erythroid maturation.8 NF-E2, originally identified as an erythroid transcription factor, has been found in knock-out mice to program late phases of megakaryocytic development, including platelet release.5 SCL/tal and its cofactor LMO2 display expression in erythroid and megakaryocytic lineages,39,42 and transduction of human CD34+ progenitors with SCL/tal expression vector jointly enhances erythroid and megakaryocytic development.43 The zinc-finger onco-protein Gfi-1b manifests high-level coexpression in erythroid and megakaryocytic lineages, and Gfi-1b—/— mice have selective defects in erythroid and megakaryocytic maturation, analogous to those in GATA-1—/— mice.44 The ets-domain factor Fli1 bears several features of a megakaryocytic transcription factor—high-level expression in megakaryocytes, binding to megakaryocytic promoters, knock-out associated with early arrest in megakaryocytic development, and hemizygosity in patients with congenital thrombocytopenia.45,46 However, Fli1 also shows expression in erythroid progenitors, and Fli1—/— mice show defective erythroid maturation with marked diminution in fetal liver erythroid colony-forming unit (CFU-E) and BFU-E.47
In addition to its unique expression pattern, other features of RUNX1 make it an attractive candidate for a promegakaryocytic transcription factor. Consensus DNA-binding sites for RUNX1, TGTGGT, occur in many megakaryocytic promoters and enhancers—the αIIb integrin promoter (Figure 4),48 the β3 integrin promoter (—544),49 the α2 integrin megakaryocyte-specific enhancer element (—1899),50 the c-MPL promoter,51 the platelet basic protein (PBP) promoter,52 and the thromboxane synthase enhancer.53 Our data in Figure 8 reinforce the possibility of RUNX1 regulation of megakaryocytic target genes, in particular those encoding the integrins αIIb and α2. In the αIIb promoter, at least 3 RUNX sites appear to contribute to responsiveness to RUNX1/CBFβ plus GATA-1; notably, one of these sites is a variant, TGGGGT (Figure 4C). However, it is also possible that RUNX1 could act on some of its target genes through non-DNA–binding mechanisms, such as by modulating the transcriptional function of GATA-1. Of potential importance in this regard, RUNX1 physically interacts with a known megakaryocytic transcription factor, c-Ets-1,51 leading to cooperative DNA binding by both factors.54,55 Virtually all megakaryocytic promoters have ets binding sites adjacent to GATA binding sites. Thus, one function of RUNX1 could be to assemble a megakaryocyte-specific transcriptional complex juxtaposing GATA-1 and c-Ets-1 factors.
The finding of germline monoallelic RUNX1 mutations in a human hereditary disease, familial platelet disorder with predisposition to acute myeloid leukemia (FPD-AML), provides further evidence for RUNX1 involvement in megakaryopoiesis.21,22 Patients with this disorder manifest moderate thrombocytopenia associated with defective platelet aggregation; their marrow specimens show diminished numbers of CFU-MK, with abnormally small colony sizes.21 In animal models, RUNX1—/— mice display a block in definitive hematopoiesis but notably retain intact primitive erythropoiesis.17 RUNX1+/— mice show altered timing and distribution of hematopoietic stem cells during embryogenesis but do not recapitulate the phenotype of FPD-AML.56,57 Therefore, the role of RUNX1 in megakaryopoiesis may differ between mice and humans, or the specific RUNX1 mutations occurring in FPD-AML may contribute to the phenotype.22 However, inducible biallelic targeting of RUNX1 in adult mice resulted in selective thrombocytopenia with markedly impaired megakaryocyte development,58 a phenotype that recapitulates the megakaryocyte-specific knock-down of GATA-1 expression in mice.13
The functional interplay between RUNX1 and GATA-1 seen in Figure 4 corroborates the high-level coexpression of these factors in megakaryocytes. Interestingly, RUNX1 and GATA-1 have primordial homologs in Drosophila hematopoiesis, lozenge (lz) and serpent (srp), which function in a coordinate fashion to program crystal cell development.59 In humans, hereditary germline mutations in GATA-1 recapitulate some of the phenotypic features of FPD-AML, namely familial thrombocytopenia with defective megakaryocyte maturation.15 Acquired GATA-1 mutations leading to diminished transactivation function specifically associate with megakaryocytic leukemias in Down syndrome (M7-DS), in which RUNX1 overexpression (caused by trisomy 21) may also play a role.23 Compelling genetic evidence in flies and humans points to the cooperation of RUNX1 and GATA-1 in overlapping developmental pathways. Our data in Figures 6 and 7 suggest that physical interaction may provide one basis for this functional cooperation.
Our mapping studies have shown that the highly conserved runt domain within RUNX1 is sufficient for binding to GATA-1. Unexpectedly, the N- and C-fingers of GATA-1 appear not to play a role in binding to RUNX1. Rather, the amino and carboxy terminal portions of GATA-1 appear to be critical for this function. Although less highly conserved within the GATA family, these terminal regions of GATA-1 appear to play key roles in GATA-1 function. The amino terminal region of GATA-1 (amino acids 1-84) has transcription activation functions that may be relevant in normal human megakaryopoiesis; mutations causing deletion of this domain were found in 100% of cases of the megakaryocytic leukemia M7-DS.23 Two possible models exist. First, the truncated forms of GATA-1 associated with M7-DS form aberrant megakaryocytic transcriptional complexes lacking RUNX1. Second, RUNX1, which may be up-regulated as a consequence of trisomy 21, forms aberrant megakaryocytic transcriptional complexes lacking GATA-1. Future experiments will test each of these possibilities. Our data in Figure 7E suggest that the loss of physical interaction between GATA-1 mutant Δ1-85 and RUNX1 results in impaired transcriptional cooperation. GATA-1 binding to RUNX1 may be required for efficient recruitment of specific transcriptional complexes to megakaryocytic promoter elements.
The carboxy terminal region of GATA-1 (amino acids 319-413) is important in regulation of the endogenous GATA-1 gene. Whereas full-length GATA-1 does not activate the expression of endogenous GATA-1 in 416B myeloid cells, carboxy terminal deletions of GATA-1 potently activate endogenous GATA-1 expression.24 Along these lines, we have shown that the up-regulation of GATA-1 expression appears to be a feature of erythroid rather than megakaryocytic differentiation in human CD34+ cells (Figure 3). Thus, an interesting possibility is that GATA-1–RUNX1 complexes might serve to repress endogenous GATA-1 expression during megakaryocytic differentiation.
The ability of RUNX1-ETO to repress GATA function may elucidate several previously unexplained observations. RUNX1-ETO has been postulated to promote leukemia through the blockade of normal RUNX1 function—that is, programming of myeloid maturation. However, leukemias with t(8;21) frequently manifest myeloid maturation, and leukemias with inv(16), which generates a dominant-negative CBFβ fusion protein, also are characterized by extensive myeloid maturation.60 Most notably, these leukemias show significant interference with erythroid and megakaryocytic maturation,60 an effect possibly attributable to the inhibition of GATA-1 function. Similarly, forced expression of RUNX1-ETO or truncated RUNX1 in zebrafish or Xenopus embryos inhibits normal erythroid development.61,62 Likewise, enforced RUNX1-ETO expression in human primary erythroid progenitors interferes with normal maturation.63 Because RUNX1 does not appear to play a direct role in the regulation of erythroid genes,17 it is possible that these RUNX1 mutants could be blocking erythroid development through the repression of GATA-1 function. In summary, the functional and physical interaction of RUNX1 and GATA-1 may underlie normal developmental programs, such as megakaryocytic-erythroid divergence, and pathologic development, as in acute megakaryocytic leukemia (M7-DS) and acute myeloid leukemias with abnormalities of the core-binding factor complex.
Prepublished online as Blood First Edition Paper, February 6, 2003; DOI 10.1182/blood-2002-09-2708.
Supported by National Institutes of Health grants RO1 CA93735 and RO1 CA100057 (National Cancer Institute) (A.N.G.) and K08 HL04017 (National Heart, Lung, and Blood Institute) (F.K.R.).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
We thank Dr Henry Frierson and Sharon Birdsall for expertise in immunohistochemistry; Drs Scott Hiebert, Alan Friedman, Jane Visvader, John Bushweller, and Janet Cross for providing plasmids; Drs Richard Mulligan and Takashi Tsuji for providing cell lines; Dr Dan Rosson for providing antibody; and Dr John Bushweller for helpful comments and advice.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal