Abstract
Monocytic differentiation of 32DPKCδ cells in response to activation of protein kinase C δ (PKCδ) by phorbol 12-myristate 13-acetate (PMA) was inhibited by exogenous CCAAT/enhancer binding protein α–estradiol receptor (C/EBPα-ER), which impeded morphologic maturation and induction of macrosialin mRNA. Inhibition of monopoiesis was also evident in 32DPKCδ subclones expressing C/EBPαLeu12Val-ER, which cannot dimerize or bind DNA because of mutation of the leucine zipper, C/EBPαGZ-ER, in which the leucine zipper has been replaced by the GCN4 zipper, or C/EBPαΔ3-8-ER, lacking the C/EBPα transactivation domains. In contrast, C/EBPαBR3-ER, containing a mutant basic region, did not inhibit monocytic differentiation. C/EBPα-ER strongly inhibited endogenous AP-1 DNA-binding. Supershift analysis revealed that the major AP-1 complex contains JunB. Activation of C/EBPα-ER specifically reduced endogenous JunB RNA and protein and exogenous JunB levels without affecting endogenous or exogenous c-Jun. The stability of PMA-induced JunB was not affected. Thus, C/EBPα-ER suppresses both JunB transcription and posttranscriptional protein generation or induction. PU.1 levels and activity were increased. The Leu12Val, GZ, and Δ3-8 mutants also inhibited JunB expression, whereas the BR3 mutant was ineffective, indicating that inhibition of JunB expression and monocytic differentiation by C/EBPα-ER depends upon an interaction mediated by its basic region. Exogenous JunB restored AP-1 DNA-binding but did not prevent inhibition of macrosialin expression by C/EBPα-ER, indicating that JunB is not the only target relevant to inhibition of monopoiesis by C/EBPα.
Introduction
Pluripotent hematopoietic stem cells give rise to granulocyte/monocyte progenitors (GMPs) expressing CD34 and Fcγ receptor (II/III).1,2 These cells in turn produce more committed progenitors that develop into granulocytes or monocytes (reviewed in Friedman3). Within hematopoiesis, CCAAT/enhancer binding protein (C/EBP) family members are expressed predominantly in granulocytic and monocytic cells.4C/EBPα is a key regulator of granulopoiesis, as neonatal mice lacking C/EBPα do not develop granulocytic progenitors but develop the other blood lineages, including monocytes.5 In addition, overexpression of C/EBPα in the U937 or 32D cl3 myeloid cell lines or in avian multipotent progenitors enables their neutrophilic maturation.6-8
Several transcription factors have been implicated in monocyte commitment and maturation. PU.1 is required for the formation of B-lymphoid cells, monocytes, and neutrophils.9,10 AP-1 and PU.1 cooperate to bind and activate the promoters of the genes encoding macrosialin (MS) and the scavenger receptor in monocytes,11,12 and c-Jun may activate additional myeloid genes via its ability to interact with PU.1.13,14 In addition, c-Jun or JunB induce monocyte maturation when expressed in myeloid cell lines.15-17 MafB and c-Maf heterodimerize with Jun or Fos family members and also induce monocytic maturation.18,19 Egr-1 and interferon consensus sequence binding protein (ICSBP) have been implicated as regulators of monopoiesis as well.20 21
When expressed in human CD34+ cells, C/EBPα or C/EBPβ favor the development of granulocytes at the expense of monocytes.22 To investigate the mechanism underlying this inhibitory effect of C/EBP proteins on monocyte commitment or maturation we used 32DPKCδ cells, in which phorbol ester–responsive protein kinase C δ (PKCδ) has been introduced into interleukin-3 (IL-3)–dependent 32D cells.23 In contrast to 32D cl3 cells, 32DPKCδ cells do not respond to granulocyte colony-stimulating factor (G-CSF) but differentiate within 24 hours to mature monocyte/macrophages when exposed to phorbol 12-myristate 13-acetate (PMA). Activation of PKC isoforms by phorbol esters induces monocytic differentiation in other hematopoietic cell lines, and signals from the macrophage colony-stimulating factor receptor act similarly.24 In response to estradiol, C/EBPα–estradiol receptor (ER) inhibited morphologic maturation and macrosialin mRNA expression in 32DPKCδ cells. Endogenous JunB protein and RNA expression were reduced, dependent upon the C/EBPα basic region (BR), but not its leucine zipper (LZ), its transactivation domains (TADs), its ability to bind DNA, or its ability to slow G1 to S cell-cycle progression. C/EBPα-ER specifically reduced exogenous JunB protein expression, without affecting endogenous or exogenous c-Jun. In addition, activation of C/EBPα-ER induced the myeloperoxidase (MPO) and C/EBPε genes, which are predominantly expressed in the granulocytic lineage. Our findings indicate that C/EBPα simultaneously induces granulopoiesis and inhibits monopoiesis in a committed myeloid progenitor.
Materials and methods
Cell culture, morphology, and proliferation assays
32DPKCδ and 32D cl3 cells were cultured in phenol red–free Iscoves modified Dulbecco medium with 10% heat-inactivated fetal calf serum (HI-FCS), 1 ng/mL IL-3 (PeproTech, Rocky Hill, NJ), 100 U/mL penicillin, and 100 μg/mL streptomycin. ϕCRE cells25 were cultured in Dulbecco modified Eagle medium (DMEM) with 10% heat-inactivated calf serum. 293T cells were cultured in DMEM with 10% HI-FCS. We used 1 μM estradiol (E2) or 200 nM 4-hydroxytamoxifen (4HT) in ethanol, 65 nM PMA in dimethylsulfoxide (DMSO), 50 μg/mL cycloheximide (CHX), and 0.1% to 0.2% ethanol or 0.1% DMSO as vehicle controls. Zinc chloride was used at 100 μM. Morphology was assessed by Wright-Giemsa staining of cytospins or of cells cultured on glass coverslips that had been acid-etched by exposure to boiling 1 M HCl for 15 minutes. Viable cell counts were obtained by enumerating, by means of a hemocytometer, cells that exclude trypan blue dye. The proportion of cells in each cell-cycle phase was determined by fluorescent-activated cell sorter (FACS) analysis of cells stained with propidium iodide as described.26
Plasmids and transduction
pBabePuro-C/EBPα-ER, and pBabePuro-ER have been described.8,27 pBabePuro-C/EBPαLeu12Val-ER, pBabePuro-C/EBPαBR3-ER, and pBabePuro-C/EBPαΔ3-8-ER were constructed by replacing a 1.1-kilobase (kb) NcoI fragment containing the majority of the C/EBPα open reading frame with the analogous NcoI fragment from p murine sarcoma virus (MSV)–C/EBPαBR3, pMSV-C/EBPαLeu12Val, or pMSV-C/EBPαΔ3-8.28,29 pBabePuro-C/EBPαGZ-ER was constructed by transfering the C/EBPαGZ cDNA from pMSV-C/EBPαGZ29 to pBabePuro and then linking the human estradiol receptor (ER) ligand-binding domain in frame with the C-terminus of the GCN4 leucine zipper (GZ) after inserting a restriction site just upstream of the GCN4 stop codon by polymerase chain reaction (PCR) mutagenesis. As a result, the final GCN4 residue, Arg, was lost, and AKPNL was inserted between the GCN4 and ER segments, as confirmed by DNA sequencing. These retroviral vectors were introduced into ϕCRE cells using Lipofectamine 2000 (Gibco-BRL, Gaithersburg, MD), and pooled transfectants were selected using 2 μg/mL puromycin. The 32DPKCδ, and 32D cl3 cells were transduced by coculture with subconfluent ϕCRE packaging cells irradiated to 3000 cGy and 4 μg/mL polybrene. Subclones were isolated by limiting dilution in 2 μg/mL puromycin. The cDNAs encoding murine JunB, c-Jun, and c-Fos (kindly provided by D. Liebermann, Temple University, Philadelphia, PA) were positioned downstream of the cytomegalovirus (CMV) promoter in pGEM/CMVa and transiently transfected into 293 cells using Lipofectamine 2000. The JunB and c-Jun cDNAs were also inserted into pMTCB6, which was linearized using ScaI and electroporated into 32DPKCδ-αER cells as described.26 Subclones were selected using puromycin and 1.2 mg/mL G418 (total) and isolated by limiting dilution. (PU.1)4TKLUC, (mPU.1)4TKLUC (kindly provided by G. Behre, Ludwig Maximilians University, Munich, Germany), or p(C/EBP)2TKLUC were transiently transfected into 32DPKCδ-αER cells with CMV-βGal by incubation at room temperature in the presence of 250 μg/mL diethylamino ethyl (DEAE)–dextran in 137 mM NaCl, 5 mM KCl, 0.37 mM Na2HPO4, 25 mM Tris (tris(hydroxymethyl)aminomethane, pH 7.5), 0.68 mM CaCl2, and 1 mM MgCl2.
Western, Northern, and gel-shift assays
Preparation of total cellular protein and RNA and Western and Northern blotting were carried out as described.4,26 We used ERα (HC-20), c-Jun (N), JunB (N17), PU.1 (T21), C/EBPβ (C19) antisera (all from Santa Cruz Biotechnology, Santa Cruz, CA), and actin monoclonal antibody AC-15 (Sigma, St Louis, MO). The murine myeloperoxidase (MPO), lactoferrin (LF), macrosialin (MS), C/EBPε, PU.1, β-actin, and JunB cDNAs used as probes have been described.12,15,27 The sense strands of the oligonucleotides used for electrophoretic mobility shift assay (EMSA) were as follows12 30: AP-1(MS), 5′-GATCCAGGTGTCTGAGTCAGGTTTGG-3′; mAP-1(MS), GATCCAGGTGTCCTCGAGAGGTTTGG-3′; PU.1(MS), 5′-GATCCGTTAAGGGAAGTGA-3′; and mPU.1(MS), 5′-GATCCGTTAAGCCTACTGA-3′.
The antisense oligonucleotides had corresponding GATC or TCGA 5′ overhangs. The wild-type and mutant core binding sites are underlined. Preparation of nuclear extracts, gel shift, and supershift assays were carried out as described.30 Antisera listed above and GATA-1 (N6X) were used for supershift assay (Santa Cruz Biotechnology).
Results
Reciprocal effect of C/EBPα and PKCδ on monocytic and granulocytic differentiation
32DPKCδ cells were transduced with pBabePuro retroviral vectors expressing C/EBPα-ER or the ER segment alone (diagrammed in Figure1A), and subclones were isolated by limiting dilution. Expression of C/EBPα-ER in 2 subclones (αER-1 and αER-2) and of the ER segment in 1 subclone was confirmed by Western blotting (Figure 1B). Despite screening multiple lines, we could not express ER at levels similar to C/EBPα-ER.
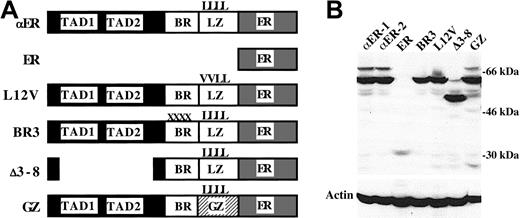
Expression of C/EBPα-ER and mutant variants.
(A) Diagram of C/EBPα-ER (αER), the ER segment alone, and the Leu12Val (L12V), BR3, Δ3-8, and GZ C/EBPα-ER mutant variants. In Leu12Val, the first 2 leucines of the leucine zipper (LZ) are changed to valines (V). In BR3, 4 residues on the basic region (BR) are altered. Δ3-8 deletes C/EBPα's transactivation domains (TAD1, TAD2). In GZ, the GCN4 LZ replaces the C/EBPα LZ. (B) Western blot of PKCδ cell lines stably expressing the indicated proteins. The blot was probed sequentially using an ER antiserum and β-actin antibody.
Expression of C/EBPα-ER and mutant variants.
(A) Diagram of C/EBPα-ER (αER), the ER segment alone, and the Leu12Val (L12V), BR3, Δ3-8, and GZ C/EBPα-ER mutant variants. In Leu12Val, the first 2 leucines of the leucine zipper (LZ) are changed to valines (V). In BR3, 4 residues on the basic region (BR) are altered. Δ3-8 deletes C/EBPα's transactivation domains (TAD1, TAD2). In GZ, the GCN4 LZ replaces the C/EBPα LZ. (B) Western blot of PKCδ cell lines stably expressing the indicated proteins. The blot was probed sequentially using an ER antiserum and β-actin antibody.
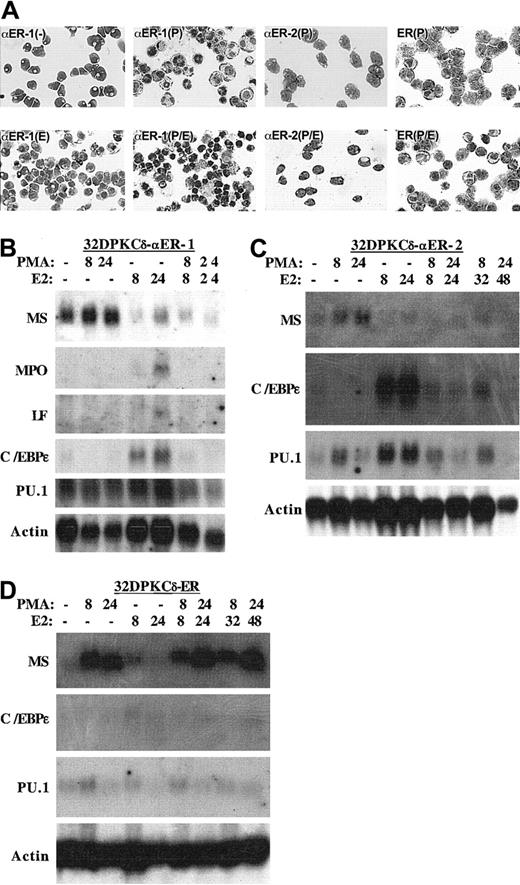
32DPKCδ-αER-1, -αER-2, and -ER cells were cultured in the presence of PMA, estradiol, both, or neither. After 8 hours in PMA, each cell line became adherent and developed a macrophage morphology (Figure 2A). Simultaneous activation of C/EBPα-ER interfered with the morphologic differentiation of αER-1 and αER-2 cells but did not affect the maturation of the ER line. Exposure of αER-1 cells to estradiol alone did not induce a neutrophilic morphology at 24 hours (Figure 2) or at later times (not shown).
C/EBPα inhibits monocytic differentiation.
(A) The morphology of 32DPKCδ-αER-1 cells was assessed with no additions (−), after 24 hours in estradiol (E), 8 hours in PMA (P), or 8 hours in PMA and estradiol (P/E). The morphologies of the αER-2 and ER lines in PMA or PMA + estradiol for 8 hours are also shown. (B-D) Total cellular RNAs were prepared from the indicated cell lines cultured in PMA, estradiol (E2), both, or neither for the indicated number of hours. These RNAs, 10 μg per lane, were then subjected to Northern blotting for macrosialin (MS), myeloperoxidase (MPO), lactoferrin (LF), C/EBPε, PU.1, and β-actin. If no transcripts were detected, the results are not shown.
C/EBPα inhibits monocytic differentiation.
(A) The morphology of 32DPKCδ-αER-1 cells was assessed with no additions (−), after 24 hours in estradiol (E), 8 hours in PMA (P), or 8 hours in PMA and estradiol (P/E). The morphologies of the αER-2 and ER lines in PMA or PMA + estradiol for 8 hours are also shown. (B-D) Total cellular RNAs were prepared from the indicated cell lines cultured in PMA, estradiol (E2), both, or neither for the indicated number of hours. These RNAs, 10 μg per lane, were then subjected to Northern blotting for macrosialin (MS), myeloperoxidase (MPO), lactoferrin (LF), C/EBPε, PU.1, and β-actin. If no transcripts were detected, the results are not shown.
To further assess the effect of C/EBPα-ER on monocytic maturation, total cellular RNAs were prepared from these 3 cell lines 8 or 24 hours after addition of PMA, estradiol, or both. These RNAs were subjected to Northern blotting for macrosialin mRNA (MS), a marker of monocyte differentiation (Figure 2B-D). PMA induced MS in each cell line, estradiol reduced basal MS expression relative to actin in the αER lines but not in the ER cells, and simultaneous exposure to PMA and estradiol prevented induction of MS in the αER lines but not in the control cells. PMA also induced MS in control, ER, cells pretreated with estradiol for 24 hours (Figure 2D, last 2 lanes).
The effect of PKCδ activation on C/EBPα-ER–mediated induction of mRNAs encoding several granulocytic markers, MPO, lactoferrin (LF), and C/EBPε was also assessed (Figure 2B-D). C/EBPα-ER weakly induced the MPO and LF mRNAs in the αER-1 line but not in the αER-2 line (not shown). Induction of MPO and LF was prevented by PMA. C/EBPα-ER, but not ER, induced C/EBPε, and this induction was again prevented by exposure to PMA. Activation of PKCδ by addition of PMA 24 hours after addition of estradiol even reversed the marked induction of C/EBPε mRNA (Figure 2C, last 2 lanes).
Inhibition of monocytic differentiation depends upon the C/EBPα basic region
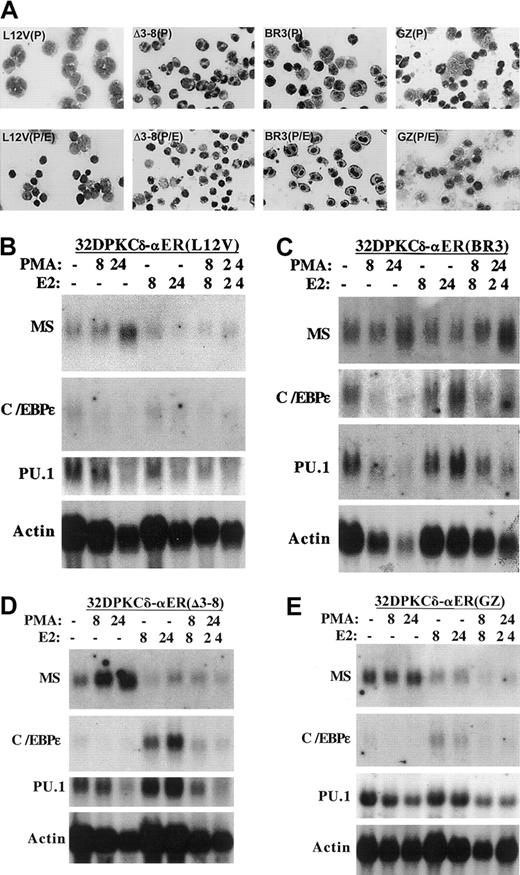
To identify the C/EBPα domain required for inhibition of monocytic maturation, we developed cell lines expressing several C/EBPα-ER mutants (Figure 1A): BR3 harbors mutations in the basic region that prevent DNA-binding; Leu12Val cannot dimerize or bind DNA because of mutation of 2 leucines to valine within the leucine zipper; Δ3-8 lacks the C/EBPα transactivation domains (TAD1 and TAD2); and GZ is a variant in which the C/EBPα leucine zipper is replaced with the leucine zipper from a yeast protein, GCN4. GZ retains the ability to homodimerize, bind DNA, and activate transcription, but is not expected to interact with endogenous basic region leucine zipper (bZIP) proteins via the leucine zipper. Expression of these proteins at levels similar to C/EBPα-ER was documented by Western blotting (Figure 1B). The effect of each C/EBPα-ER variant on PMA-mediated induction of macrophage morphology and MS mRNA expression was assessed (Figure 3A-E). The Leu12Val and Δ3-8 variants inhibited morphologic maturation; the GZ variant had an intermediate effect; and the BR3 mutant did not impede the generation of macrophages. The Leu12Val, Δ3-8, and GZ mutants each inhibited MS induction, especially at 24 hours, whereas C/EBPαBR3-ER was again ineffective. Similar findings were obtained using a second Leu12Val and BR3 subclone (not shown). We developed reverse transcriptase (RT)–PCR assays that specifically detect the exogenous Leu12Val or BR3 RNAs and confirmed that the samples used for the Northern blots in Figure 3B-C contained the expected variant C/EBPα-ER RNAs (not shown).
Consistent with their inability to bind DNA, neither Leu12Val nor BR3 induced MPO (not shown). Leu12Val also did not induce a second granulocytic marker, C/EBPε, while BR3 did so minimally compared with C/EBPα-ER (Figure 3B-C). The GZ variant retained the ability to induce C/EBPε, and PMA inhibited this effect (Figure 3E), as observed for wild-type C/EBPα-ER. Unexpectedly, C/EBPαΔ3-8-ER also induced C/EBPε mRNA expression, suggesting that this variant retains transactivation activity in this cellular context. Both C/EBPε and PU.1 mRNA levels are rapidly induced in 32D cl3 cells by C/EBPα-ER even in cycloheximide, suggesting that they are direct C/EBPα genetic targets.8 27 Consistent with their effects on C/EBPε, C/EBPα-ER and its Δ3-8 variant strongly induced PU.1 mRNA expression in response to estradiol, whereas the BR3 or Leu12Val mutants, or the ER segment alone, were again ineffective (Figures 2-3). C/EBPαGZ-ER did not induce PU.1 RNA, perhaps because of high basal expression of this factor in this subclone.
Inhibition of monocytic differentiation by C/EBPα depends upon its basic region.
(A) The morphology of 32DPKCδ-Leu12Val, -BR3, -Δ3-8, and -GZ cells exposed to PMA (P) or PMA + estradiol (P/E) for 8 hours is shown. (B-E) RNAs prepared from these cell lines cultured in PMA, estradiol (E2), both, or neither for the indicated times were subjected to Northern blotting sequentially for macrosialin (MS), myeloperoxidase (MPO), C/EBPε, PU.1, and β-actin. MPO RNA was not detected (not shown).
Inhibition of monocytic differentiation by C/EBPα depends upon its basic region.
(A) The morphology of 32DPKCδ-Leu12Val, -BR3, -Δ3-8, and -GZ cells exposed to PMA (P) or PMA + estradiol (P/E) for 8 hours is shown. (B-E) RNAs prepared from these cell lines cultured in PMA, estradiol (E2), both, or neither for the indicated times were subjected to Northern blotting sequentially for macrosialin (MS), myeloperoxidase (MPO), C/EBPε, PU.1, and β-actin. MPO RNA was not detected (not shown).
C/EBPα inhibits JunB expression
As the MS promoter is regulated by PU.1 and by AP-1 family members,12 we carried out gel-shift assays for these factors using wild-type and mutant oligonucleotides derived from the MS promoter and extracts from 32DPKCδ-αER-1 cells exposed to PMA, estradiol, both, or neither. PU.l DNA-binding was not affected by PMA, but the most abundant, faster-migrating PU.1 species was increased by estradiol, consisent with increased PU.1 RNA expression evident on Northern blotting (Figure4A). Consistent with these findings, the activity of (PU.1)4TKLUC was increased 2.8-fold by activation of C/EBPα-ER for 24 hours in 32DPKCδ-αER cells, in the absence or presence of PMA, whereas the activity of the analogous plasmid carrying mutant PU.1 sites (mPU.1)4TKLUC was increased approximately only 1.3-fold on average (Figure 4B). By comparison, (C/EBP)2TKLUC was induced 7-fold. In the absence of 4HT or PMA, the (PU.1)4TKLUC was 10-fold more active than (mPU.1)4TKLUC (not shown). The estradiol analog 4HT was used for this assessment as estradiol but not 4HT-induced background reporter activity in parental cells. Supershift assay confirmed that the 3 gel-shift species in Figure 4A contain PU.1. Thus, the faster migrating species (*) are proteolytic fragments of PU.1 that retain the DNA-binding domain. None of these bands supershifted with c-Jun or GATA-1 antisera (not shown). In contrast to these mild increases in PU.1 RNA, DNA-binding, and transactivation activity, activation of C/EBPα-ER markedly reduced both basal and PMA-induced AP-1 DNA-binding (Figure 4C).
PU.1 and AP-1 DNA-binding and PU.1 activity in 32DPKCδ-C/EBPα-ER cells.
(A) Nuclear extracts prepared from 32DPKCδ-αER-1 cells cultured in PMA, estradiol (E2), both, or neither for 8 hours were subjected to gel-shift assay (lanes 1-3, 10) using as probe 1 ng of a radio-labeled PU.l-binding site derived from the macrosialin promoter. Also added to the gel-shift reactions as indicated were 50, 100, or 200 ng wild-type (W) or mutant (M) probe, 2 μL PU.1 antiserum (Ab), or 2 μL normal rabbit serum (RS). Asterisks (*) denote faster migrating species that likely represent proteolytic fragments of PU.1. (B) To assess PU.1 or C/EBP activities, 20 μg (PU.1)4TKLUC, (mPU.1)4TKLUC, or (C/EBP)2TKLUC and 2 μg pCMV-βGal were transfected into 2.0 × 107 32DPKCδ-αER cells, which were then split and cultured ± 4HT or in PMA ± 4HT, as indicated. Luciferase and β-galactosidase activities were determined 24 hours later and used to calculate a normalized luciferase activity. Ratios of these activities in the presence versus the absence of 4HT are shown ([activity in 4HT or 4HT + PMA]/[activity in no inducer or PMA]) for each DNA construct (mean and SE of 2 determinations). (C) The same extracts described in panel A were subjected to gel-shift assay using as probe an AP-1 binding site from the macrosialin promoter (lanes 1, 4, 7, and 10). Included as an unlabeled competitor in the gel-shift reactions when indicated were 50-fold excess of wild-type or mutant probe.
PU.1 and AP-1 DNA-binding and PU.1 activity in 32DPKCδ-C/EBPα-ER cells.
(A) Nuclear extracts prepared from 32DPKCδ-αER-1 cells cultured in PMA, estradiol (E2), both, or neither for 8 hours were subjected to gel-shift assay (lanes 1-3, 10) using as probe 1 ng of a radio-labeled PU.l-binding site derived from the macrosialin promoter. Also added to the gel-shift reactions as indicated were 50, 100, or 200 ng wild-type (W) or mutant (M) probe, 2 μL PU.1 antiserum (Ab), or 2 μL normal rabbit serum (RS). Asterisks (*) denote faster migrating species that likely represent proteolytic fragments of PU.1. (B) To assess PU.1 or C/EBP activities, 20 μg (PU.1)4TKLUC, (mPU.1)4TKLUC, or (C/EBP)2TKLUC and 2 μg pCMV-βGal were transfected into 2.0 × 107 32DPKCδ-αER cells, which were then split and cultured ± 4HT or in PMA ± 4HT, as indicated. Luciferase and β-galactosidase activities were determined 24 hours later and used to calculate a normalized luciferase activity. Ratios of these activities in the presence versus the absence of 4HT are shown ([activity in 4HT or 4HT + PMA]/[activity in no inducer or PMA]) for each DNA construct (mean and SE of 2 determinations). (C) The same extracts described in panel A were subjected to gel-shift assay using as probe an AP-1 binding site from the macrosialin promoter (lanes 1, 4, 7, and 10). Included as an unlabeled competitor in the gel-shift reactions when indicated were 50-fold excess of wild-type or mutant probe.
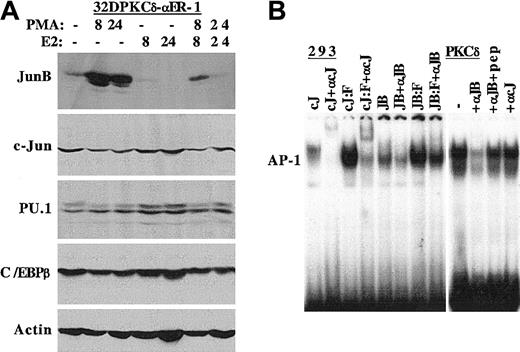
In view of the strong inhibition of AP-1 DNA-binding by C/EBPα-ER, we subjected protein extracts from 32DPKCδ-αER-1 cells to Western blotting for JunB and c-Jun, 2 AP-1 components, as well as PU.1 and C/EBPβ, transcription factors also implicated in monocyte maturation (Figure 5A). PMA potently induced JunB expression but did not strongly affect c-Jun levels. Activation of C/EBPα-ER reduced basal JunB expression and potently inhibited its induction by PMA. In contrast c-Jun levels were only minimally affected. These findings were reproducible upon repetition (not shown). Estradiol induced PU.1 expression, and PMA had little effect on PU.1 levels in the presence or absence of estradiol. Similarly, C/EBPβ levels were nearly identical under the various culture conditions. To confirm that the reduction in AP-1 DNA-binding induced by C/EBPα-ER reflects reduction in JunB levels, we carried out a supershift assay, using extracts from transfected 293 cells as positive controls (Figure5B). The c-Jun (cJ) and c-Jun/Fos (cJ/F) complexes were supershifted by the c-Jun antiserum, and the JunB (JB) and JunB/Fos (JB/F) complexes were supershifted by the JunB antiserum (left panel). A large majority of the AP-1 gel-shift complex detected in parental PKCδ cells was supershifted by the JunB antiserum and was only minimally affected by the c-Jun antiserum. Preincubation of the JunB antiserum with its specific peptide largely prevented the JunB supershift.
C/EBPα inhibits JunB expression.
(A) Total cellular protein extracts prepared from 1 × 106 32DPKCδ-αER-1 cells cultured in PMA, estradiol (E2), or both for 0, 8 or 24 hours were subjected to Western blotting for JunB, c-Jun, PU.1, C/EBPβ, and β-actin. (B) The 293 cells were transfected with 5 μg per 100-mm dish of pCMV-c-Jun (cJ) or pCMV-JunB (JB) alone, or 2.5 μg of these DNAs were transfected with 2.5 μg pCMV-c-Fos (cJ:F, JB:F). Then, 2 days later nuclear extracts were prepared and subjected to gel-shift assay with an AP-1 binding site from the macrosialin promoter. Anti–c-Jun antiserum (αcJ) or anti-JunB antiserum (αJB) was included in several samples (left panel). A nuclear extract from 32DPKCδ cells was subjected to gel-shift assay with no antiserum (−), with 1 μL αJB or αcJ antiserum, or with αJB antiserum that had been preincubated with 0.4 μg JunB peptide (pep).
C/EBPα inhibits JunB expression.
(A) Total cellular protein extracts prepared from 1 × 106 32DPKCδ-αER-1 cells cultured in PMA, estradiol (E2), or both for 0, 8 or 24 hours were subjected to Western blotting for JunB, c-Jun, PU.1, C/EBPβ, and β-actin. (B) The 293 cells were transfected with 5 μg per 100-mm dish of pCMV-c-Jun (cJ) or pCMV-JunB (JB) alone, or 2.5 μg of these DNAs were transfected with 2.5 μg pCMV-c-Fos (cJ:F, JB:F). Then, 2 days later nuclear extracts were prepared and subjected to gel-shift assay with an AP-1 binding site from the macrosialin promoter. Anti–c-Jun antiserum (αcJ) or anti-JunB antiserum (αJB) was included in several samples (left panel). A nuclear extract from 32DPKCδ cells was subjected to gel-shift assay with no antiserum (−), with 1 μL αJB or αcJ antiserum, or with αJB antiserum that had been preincubated with 0.4 μg JunB peptide (pep).
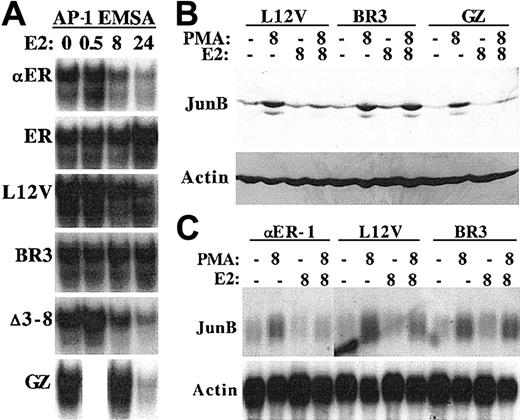
Inhibition of JunB protein and RNA expression depends upon the C/EBPα basic region
C/EBPαBR3-ER was unable to prevent PKCδ-mediated monocytic differentiation induced by PMA. To determine whether this reflects inability to reduce AP-1 DNA-binding and JunB expression, extracts from PKCδ-αER-1 cells and from lines expressing several C/EBPα-ER variants were subjected to gel-shift assay and to Western blotting (Figure 6A-B). C/EBPα-ER, Leu12Val, Δ3-8, and GZ reduced AP-1 DNA-binding, whereas ER and BR3 did not. With the exception of GZ, reduction was evident at 8 hours (and was also seen to a lesser degree at 4 hours; not shown) but was not evident at 30 minutes. Consistent with these findings, the Leu12Val and GZ variants inhibited PMA-induced JunB expression, whereas C/EBPαBR3-ER had no effect. To determine whether inhibition of JunB protein expression might be due, at least in part, to inhibition of JunB transcription, RNA samples from αER-1, Leu12Val, and BR3 cells exposed to PMA, estradiol, or both for 8 hours were subjected to Northern blotting (Figure 6C). PMA induced JunB RNA; C/EBPα-ER strongly inhibited JunB RNA expression; C/EBPαLeu12Val-ER had an intermediate effect; and C/EBPαBR3-ER was ineffective. PMA also induced JunB RNA at 4 hours, and C/EBPα-ER potently inhibited JunB RNA expression in αER-1 cells exposed to PMA and estradiol for 4 hours (not shown).
The C/EBPα basic region is required to inhibit JunB protein and RNA expression.
(A) Nuclear extracts prepared from the indicated 32DPKCδ cell lines 0, 0.5, 8, or 24 hours after addition of estradiol (E2) were subjected to gel-shift assay using the AP-1 oligonucleotide. A 0.5-hour GZ sample was not prepared. (B) Total cellular protein extracts from 1 × 106 Leu12Val, BR3, or GZ cells cultured for 8 hours in PMA, estradiol, both, or neither were subjected to Western blotting for JunB and β-actin. (C) Total cellular RNAs (10 μg) from αER-1, Leu12Val, or BR3 cells were subjected to Northern blotting for JunB and β-actin.
The C/EBPα basic region is required to inhibit JunB protein and RNA expression.
(A) Nuclear extracts prepared from the indicated 32DPKCδ cell lines 0, 0.5, 8, or 24 hours after addition of estradiol (E2) were subjected to gel-shift assay using the AP-1 oligonucleotide. A 0.5-hour GZ sample was not prepared. (B) Total cellular protein extracts from 1 × 106 Leu12Val, BR3, or GZ cells cultured for 8 hours in PMA, estradiol, both, or neither were subjected to Western blotting for JunB and β-actin. (C) Total cellular RNAs (10 μg) from αER-1, Leu12Val, or BR3 cells were subjected to Northern blotting for JunB and β-actin.
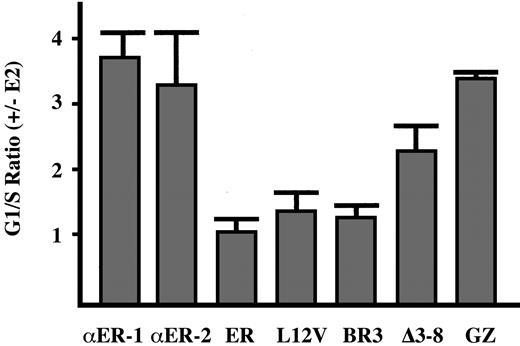
C/EBPα-ER inhibits the G1 to S cell-cycle transition in 32D cl3 cells.8 To determine whether inhibition of monocytic differentiation and JunB expression by C/EBPα-ER and its variants correlates with their ability to slow G1 progression, the PKCδ cell lines were subjected to cell-cycle analysis ± estradiol (Figure7). C/EBPα-ER and C/EBPαGZ-ER potently inhibited G1 to S progression, and the Δ3-8 variant had an intermediate effect. The Leu12Val and BR3 variants both slowed G1 progression mildly, even though Leu12Val had a far greater effect on monocytic differentiation and on JunB expression. In addition, activation of PKCδ by PMA led to a rapid G1 cell-cycle arrest that was unaffected by C/EBPα-ER or its variants (not shown), consistent with the conclusion that inhibition of monocytic differentiation by C/EBPα-ER does not depend upon its ability to inhibit proliferation.
G1 cell-cycle arrest induced by C/EBPα and its variants.
The indicated 32DPKCδ cell lines were cultured in the absence or presence of estradiol (E2) for 48 hours, and then the proportion of cells in the G1, S, and G2/M cell-cycle phases was determined by staining with propidium iodide followed by FACS analysis. The G1/S ratio observed in the presence of estradiol divided by the G1/S ratio observed in its absence is shown (mean and SE from 2 determinations). An increased G1/S ratio reflects slowed G1 to S progression.
G1 cell-cycle arrest induced by C/EBPα and its variants.
The indicated 32DPKCδ cell lines were cultured in the absence or presence of estradiol (E2) for 48 hours, and then the proportion of cells in the G1, S, and G2/M cell-cycle phases was determined by staining with propidium iodide followed by FACS analysis. The G1/S ratio observed in the presence of estradiol divided by the G1/S ratio observed in its absence is shown (mean and SE from 2 determinations). An increased G1/S ratio reflects slowed G1 to S progression.
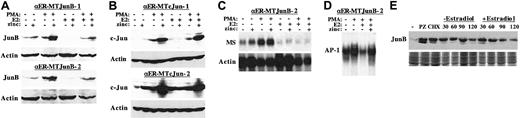
Exogenous JunB is reduced by C/EBPα and does not overcome inhibition of monopoiesis
pMT-JunB and pMT-c-Jun were introduced into 32DPKCδ-αER-2 cells by electroporation, and stable cell lines expressing JunB or c-Jun in response to activation of the metallothionein (MT) promoter by zinc were isolated. Selected for further analysis were 2 lines expressing similar levels of JunB or c-Jun. As shown in Figure8A, zinc induced JunB to levels similar to those obtained with PMA in clone 1, and zinc potentiated a marked induction of exogenous JunB by PMA in both clones. Zinc did not induce JunB in αER-2 cells lacking pMT-JunB (not shown). Notably, activation of C/EBPα-ER reduced expression of JunB in the absence of zinc and markedly attenuated induction of exogenous JunB by zinc, in the absence or presence of PMA. Nevertheless, JunB levels in the presence of PMA, estradiol, and zinc were still similar to those obtained when the cells were exposed to PMA alone. Apparently the high level of exogenous JunB obtained with the combination of PMA and zinc overwhelms the ability of C/EBPα-ER to reduce its expression below that obtained with PMA alone. In sharp contrast, activation of C/EBPα-ER with estradiol did not reduce endogenous, zinc-, PMA-, or zinc plus PMA–induced c-Jun levels in the 2 32DPKCδ-αER-MTcJun subclones (Figure 8B). Thus, C/EBPα-ER markedly reduces JunB expression without affecting the related protein c-Jun, and C/EBPα-ER does not suppress induction of the MT promoter by zinc.
Exogenous JunB does not overcome inhibition of monopoiesis by C/EBPα.
(A) Total cellular proteins prepared from 1 × 10632DPKCδ-αER-MTJunB-1 or -2 cells cultured under the indicated conditions for 16 hours were subjected to Western blotting for JunB and β-actin. (B) Total cellular proteins from 32DPKCδ-αER-MTcJun-1 and -2 cells were analyzed similarly for c-Jun and β-actin. (C) Total cellular RNAs, 10 μg per sample, prepared from αER-MTJunB-2 cells cultured under the indicated conditions for 16 hours were subjected to Northern blotting for macrosialin (MS) and β-actin. (D) Nuclear extracts prepared from αER-MTJunB-2 cells cultured with no inducer, PMA, PMA + estradiol, or PMA + estradiol + zinc for 8 hours were subjected to gel-shift analysis using a radiolabeled AP-1 binding site derived from the macrosialin promoter. (E) Total cellular protein derived from 1 × 106 αER-MTJunB-2 cells exposed to no inducer (−), PMA + zinc for 4 hours (PZ), PMA + zinc followed by cycloheximide for 15 minutes (CHX), or the latter cells exposed to estradiol or the vehicle control for 30, 60, 90, or 120 minutes were subjected to Western blot analysis for JunB (top panel). As a control for protein content, each extract was electrophoresed on a second gel and visualized with Coomassie blue dye (bottom panel).
Exogenous JunB does not overcome inhibition of monopoiesis by C/EBPα.
(A) Total cellular proteins prepared from 1 × 10632DPKCδ-αER-MTJunB-1 or -2 cells cultured under the indicated conditions for 16 hours were subjected to Western blotting for JunB and β-actin. (B) Total cellular proteins from 32DPKCδ-αER-MTcJun-1 and -2 cells were analyzed similarly for c-Jun and β-actin. (C) Total cellular RNAs, 10 μg per sample, prepared from αER-MTJunB-2 cells cultured under the indicated conditions for 16 hours were subjected to Northern blotting for macrosialin (MS) and β-actin. (D) Nuclear extracts prepared from αER-MTJunB-2 cells cultured with no inducer, PMA, PMA + estradiol, or PMA + estradiol + zinc for 8 hours were subjected to gel-shift analysis using a radiolabeled AP-1 binding site derived from the macrosialin promoter. (E) Total cellular protein derived from 1 × 106 αER-MTJunB-2 cells exposed to no inducer (−), PMA + zinc for 4 hours (PZ), PMA + zinc followed by cycloheximide for 15 minutes (CHX), or the latter cells exposed to estradiol or the vehicle control for 30, 60, 90, or 120 minutes were subjected to Western blot analysis for JunB (top panel). As a control for protein content, each extract was electrophoresed on a second gel and visualized with Coomassie blue dye (bottom panel).
We then sought to determine whether exogenous JunB would prevent inhibition of monopoiesis. 32DPKCδ-αER-MTJunB cells were subjected to Northern blotting for MS and β-actin after exposure to E2, PMA, both, or neither in the absence or presence of zinc for 16 hours (Figure 8C and data not shown). In both subclones, induction of JunB alone increased MS RNA expression in the absence of PMA but not in its presence, but it did not prevent C/EBPα-ER from inhibiting MS expression. One concern is that while zinc induction restored JunB protein levels in PMA + estradiol compared with those obtained with PMA alone, AP-1 DNA-binding may not have been restored. Nuclear extracts were therefore prepared from αER-MTJunB cells cultured in the absence of inducer, in PMA, in PMA + estradiol, or in PMA + estradiol + zinc (Figure 8D and data not shown). In both subclones, AP-1 DNA-binding activity obtained from cells cultured with all 3 agents was similar to that present in cells exposed to PMA alone.
C/EBPα-ER potentially destabilizes the JunB protein. To assess this possibility, αER-MTJunB cells were exposed to zinc and PMA for 4 hours, to maximally induce JunB. Cycloheximide was then added to arrest new protein translation, followed 15 minutes later by addition of estradiol or the vehicle control. The rate of JunB degradation was then assessed by Western blotting (Figure 8E). Activation of C/EBPα-ER with estradiol did not significantly affect the half-life of JunB, taking into account the loading of each lane as indicated by the Coomassie blue stain.
Discussion
C/EBPα is a key mediator of granulocytic commitment and maturation, though several transcription factors have emerged as candidates for specifying the monocytic lineage, including components of the AP-1 complex, c-Maf, MafB, Egr-1, and ICSBP.3Bipotential GMPs are the major source of committed granulocytic and monocytic precursors in adult marrow, and it is of interest to determine how these immature cells commit to a single lineage. While the generation of GMPs requires C/EBPα31 retention of some monocytes in C/EBPα (−/−) mice5 and the finding that exogenous C/EBPα or C/EBPβ favors the generation of granulocytes over monocytes from human CD34+cells22 suggest that C/EBPα plays a role in this decision. Consistent with this idea, we now find that activation of exogenous C/EBPα-ER inhibits the monocytic differentiation of a growth factor–dependent cell myeloid line in response to simultaneous activation of PKCδ. Exogenous C/EBPα had previously been shown to render U937 human leukemia cells insensitive to PMA-mediated monocyte differentiation but only after C/EBPα had been expressed for 7 days, at which point the cells were already committed to granulocytic maturation.6
Inhibition of monocytic differentiation by C/EBPα depended upon integrity of its basic region, as the BR3 variant was inactive. C/EBPαBR3-ER carries mutations in 4 amino acids, residues Arg297, Lys298, Arg300, and Lys302. Of these residues, only Arg300 is expected to contact DNA.32 Neither the BR3 nor the Leu12Val variants bind DNA,33 suggesting that interactions with heterologous proteins via Arg297, Lys298, and/or Lys302 are likely responsible for inhibition of monopoiesis in response to activation of PKCδ. Arg297 was recently shown to participate in interaction between C/EBPα and E2F34; however, the C/EBPαBRM2 mutant carrying mutations in Arg297 and Ile294 retained the ability to reduce JunB levels and inhibit macrosialin induction in 32DPKCδ cells (H.L. and A.D.F., unpublished data, September 2002).
C/EBPβ is capable of interacting with both C/ATF, a cAMP-response element binding protein (CREB) family bZIP protein, and with c-Jun or c-Fos, AP-1 family bZIP proteins, via their respective leucine zipper domains.35,36 C/EBPα also interacts with c-Jun, dependent on the presence of both the C/EBPα and c-Jun leucine zipper domains.37 In addition, members of the C/EBP bZIP family heterodimerize with each other. C/EBPαGZ-ER interfered with monocytic maturation and JunB induction when 32DPKCδ cells were exposed to PMA, indicating that heterodimerization of C/EBPα-ER with endogenous bZIP proteins via their respective leucine zipper domains is not required for these effects. C/EBPαΔ3-8-ER lacks 2 C/EBPα TADs but still interfered with monocytic maturation, presumably because it, like the GZ variant, has an intact basic region.
We examined the effect of C/EBPα-ER on several transcription factors implicated in the regulation of monopoiesis. C/EBPβ was unaffected; PU.1 and basal or PMA-induced c-Jun levels were minimally affected; but basal and PMA-induced expression of JunB were markedly diminished. C/EBPα did reduce basal c-Jun expression in U937 human leukemia cells, perhaps reflecting the different cellular environment.37 Moreover, total AP-1 DNA-binding was strongly reduced by activation of C/EBPα-ER. This effect on AP-1 is explained by our finding that JunB is the major component of the AP-1 DNA-binding activity in 32DPKCδ nuclear extracts. C/EBPαBR3-ER did not affect JunB levels or AP-1 DNA-binding, in keeping with its inability to inhibit monocytic differentiation. These findings suggest that C/EBPα-ER interferes with monopoiesis in part by inhibiting JunB expression. Our finding that C/EBPα-ER stimulates PU.1 activity in 32DPKCδ cells is consisent with the ability of C/EBPα and PU.1 to cooperatively activate the neutrophil elastase promoter in 32D cl3 cells30 and with the proposal that C/EBPα inhibits PU.1 activity specifically in dendritic cells during hematopoiesis.38
Reduced JunB expression could reflect altered gene transcription, protein translation, cytokine-mediated protein stabilization, or the stability of induced JunB. The finding that C/EBPα-ER and C/EBPαLeu12Val-ER, but not the BR3 variant, reduced JunB RNA levels favors direct interaction of the C/EBPα-ER basic region with a protein that regulates the JunB gene. The murineJunB gene is activated by a serum response element, a cyclic AMP response element, AP-1, STAT3, Smad proteins, and NF-κB via several cytokine signaling pathways.39-46 The MT-JunB expression cassette lacks JunB promoter elements and 5′ or 3′ untranslated RNA segments likely to regulate RNA translation or stability. Reduced expression of exogenous JunB upon activation of C/EBPα-ER with estradiol therefore favors a model in which C/EBPα-ER also interferes with induction or stability of JunB protein. Although zinc induction of JunB was reduced in the absence of PMA, we cannot eliminate interference with basal cytokine signaling and PKC activity in this setting. Consistent with this idea, C/EBPα-ER also reduced endogenous JunB protein levels in 32D cl3 cells (not shown).
Our finding that C/EBPα-ER did not alter the stability of JunB in the presence of PMA, in cells exposed to CHX, argues against direct destabilization of JunB, but this may still occur under more physiologic conditions. Whether C/EBP and AP-1 family members can directly “zipper” remains an open question, as the observations that C/EBPα/c-Jun or C/EBPβ/c-Jun interactions depend upon the C/EBPα or C/EBPβ leucine zippers were made using variants in which the leucine zipper was either deleted or substituted with the analogous region from GCN4.36 37 Perhaps these alterations modified the structure of the basic region as well, accounting for our mapping the potential interaction domain in C/EBPα to the basic region instead. In future experiments, we will further address whether direct interaction between JunB and C/EBPα accounts in part for interference with JunB expression and monocytic commitment or maturation.
JunB generally inhibits proliferation while c-Jun stimulates proliferation, and JunB inhibits transactivation mediated by c-Jun.47,48 Thus, we do not expect that c-Jun and JunB are interchangeable during monocytic development. Adding further complexity is the finding that exogenous c-Maf or MafB bind AP-1 sites49 and, like JunB or c-Jun, stimulate monocytic differentiation in myeloid cell lines.18,19 Perhaps more than one AP-1 factor mediates monopoiesis, as hematopoietic cells lacking c-Jun or JunB develop all the hematopoietic lineages.50 51
In addition to inhibiting 32DPKCδ monocytic differentiation, C/EBPα-ER also induced several granulocytic genes when activated in the absence of PMA. On the other hand, induction of MPO was not detected in several subclones and was at best weak in others, and neutrophilic morphology did not emerge. We introduced the G-CSF receptor into a 32DPKCδ-αER subclone but did not observe greater induction of granulocytic genes upon exposure of these cells to G-CSF in the absence or presence of estradiol (not shown). Thus, 32DPKCδ cells lack a factor, other than C/EBPα, essential for granulopoiesis.
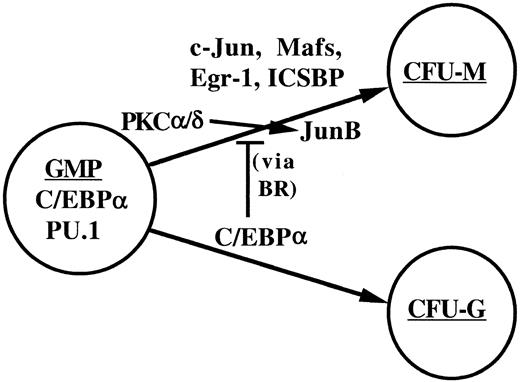
Exogenous JunB did not prevent inhibition of monopoiesis by C/EBPα-ER, and exogenous c-Jun was also ineffective (data not shown), suggesting that the C/EBPα basic region interacts with an additional protein required for monocytic maturation. Together, our findings suggest a model of myelopoiesis (Figure9) in which C/EBPα and PU.1 are required for the formation of GMPs and in which C/EBPα both commits GMPs to granulopoiesis and inhibits monopoiesis via interference of JunB induction and via affects on additional factors, other than PU.1, mediated by its basic region.
A model for commitment to the granulocyte and monocyte lineages.
Bipotent GMPs are specified by PU.1 and C/EBPα. Increased levels of C/EBPα expression specify the granulocyte colony-forming unit (CFU-G), a committed granulocytic progenitor. C/EBPα also inhibits monopoiesis via protein-protein interactions mediated by its basic reigon (BR). These interactions occur with proteins that enable JunB transcription or PKC-mediated JunB induction and with additional mediators of monopoiesis, potentially c-Jun, c-Maf, MafB, Egr-1, or ICSBP.
A model for commitment to the granulocyte and monocyte lineages.
Bipotent GMPs are specified by PU.1 and C/EBPα. Increased levels of C/EBPα expression specify the granulocyte colony-forming unit (CFU-G), a committed granulocytic progenitor. C/EBPα also inhibits monopoiesis via protein-protein interactions mediated by its basic reigon (BR). These interactions occur with proteins that enable JunB transcription or PKC-mediated JunB induction and with additional mediators of monopoiesis, potentially c-Jun, c-Maf, MafB, Egr-1, or ICSBP.
We thank H. Mushinski for 32DPKCδ cells; D. Liebermann for the c-Jun, JunB, and c-Fos cDNAs; O. Quehenberger and C. K. Glass for the macrosialin cDNA; and R. Cleaves for technical assistance.
Prepublished online as Blood First Edition Paper, January 9, 2003; DOI 10.1182/blood-2002-07-2212.
Supported by National Institutes of Health grant R01 HL62274 to A.D.F. A.D.F. is a Leukemia and Lymphoma Society Scholar and is also supported by the Children's Cancer Foundation.
H.L. and J.R.K. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
A. D. Friedman, Johns Hopkins University, Cancer Research Building, Room 253, 1650 Orleans St, Baltimore, MD 21231; e-mail: afriedm2@jhem.jhmi.edu.

![Fig. 4. PU.1 and AP-1 DNA-binding and PU.1 activity in 32DPKCδ-C/EBPα-ER cells. / (A) Nuclear extracts prepared from 32DPKCδ-αER-1 cells cultured in PMA, estradiol (E2), both, or neither for 8 hours were subjected to gel-shift assay (lanes 1-3, 10) using as probe 1 ng of a radio-labeled PU.l-binding site derived from the macrosialin promoter. Also added to the gel-shift reactions as indicated were 50, 100, or 200 ng wild-type (W) or mutant (M) probe, 2 μL PU.1 antiserum (Ab), or 2 μL normal rabbit serum (RS). Asterisks (*) denote faster migrating species that likely represent proteolytic fragments of PU.1. (B) To assess PU.1 or C/EBP activities, 20 μg (PU.1)4TKLUC, (mPU.1)4TKLUC, or (C/EBP)2TKLUC and 2 μg pCMV-βGal were transfected into 2.0 × 107 32DPKCδ-αER cells, which were then split and cultured ± 4HT or in PMA ± 4HT, as indicated. Luciferase and β-galactosidase activities were determined 24 hours later and used to calculate a normalized luciferase activity. Ratios of these activities in the presence versus the absence of 4HT are shown ([activity in 4HT or 4HT + PMA]/[activity in no inducer or PMA]) for each DNA construct (mean and SE of 2 determinations). (C) The same extracts described in panel A were subjected to gel-shift assay using as probe an AP-1 binding site from the macrosialin promoter (lanes 1, 4, 7, and 10). Included as an unlabeled competitor in the gel-shift reactions when indicated were 50-fold excess of wild-type or mutant probe.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/10/10.1182_blood-2002-07-2212/4/m_h81034296004.jpeg?Expires=1763526005&Signature=aegn5nDAvwVj0Z96kTv5YQXWF6MsEML77DGWvcY3Y1onequ6MFEeziPd6UkmMp5pElaDCtFa2V9zmRzBKtVo1Kp1WqXtaHX8mKVF7DGRzyPb76yqAuPv1R3ggLPB4zvcxp8GGQbdquz-2otVfpS3Qm2N9Y6vpad5Ks0JHwBVjdpx2tUtsnufCZPmBAETlKhtKWNwMC8vLOQLlsdg-Elt6CV~e0yreboLAuwzdWb~pB4q2kCnDaEVFpQ-25pjkjD~tfvg1km1ZDIcVhhKhLsrAapIKV3oLYArMkbxbI--ktbW8XGoCeafKE6DG62nl3Tg3J5UIzmyPdLddMnod-oXHg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal