Abstract
Factor V (FV) deficiency is a rare bleeding disorder whose genetic basis has been described in a relatively small number of cases. Among a total of 12 genetic defects reported in severely or moderately severe deficient patients, 3 were missense mutations and in no case was the mechanism underlying the deficiency explored at the molecular level. In this study, a homozygous missense mutation at cDNA position 6394 in exon 23 of the FV gene was identified in a 22-year-old Italian patient. This mutation causes the replacement of arginine 2074 with a cysteine residue (Arg2074Cys) in the C2 domain of the protein. The effect of the Arg2074Cys mutation on FV secretion, stability, and activity was investigated. Site-directed mutagenesis of FV cDNA was used to introduce the identified mutation, and wild-type as well as mutant FV proteins were expressed by transient transfection in COS-1 cells. An enzyme immunoassay detected low FV antigen levels both in the conditioned media of cells expressing the mutant protein and in cell lysates. Metabolic labeling and pulse-chase experiments confirmed that the mutation caused an impaired secretion of FV associated with rapid intracellular degradation. In addition, evaluation of wild-type and mutant coagulant activity demonstrated that the FV molecules carrying the Arg2074Cys mutation have reduced activity. These findings, beside confirming the structural and functional importance of the arginine 2074 residue, demonstrate that its substitution with a cysteine impairs both FV secretion and activity.
Introduction
Factor V (FV) is a high-molecular-weight glycoprotein (330 kd), synthesized mainly by hepatocytes and megakaryocytes as a single polypeptide chain, circulating in plasma as an inactive nonenzymatic procofactor (15%-20% of total FV is in the alpha granules of platelets). FV shares with the highly homologous factor VIII (FVIII) the domain structure, characterized by an overall organization of A1-A2-B-A3-C1-C2 protein domains.1-3
Proteolytic removal of the large B domain by thrombin or factor Xa converts FV into its active form (FVa), consisting of a heavy chain (A1-A2 domains) and a light chain (A3-C1-C2 domains) in noncovalent association, stabilized by a single Ca++ ion.4FVa is an essential cofactor in the conversion of prothrombin to thrombin by factor Xa in the presence of Ca++ and phospholipids. The A and C domains of FV and FVIII exhibit a 35% to 40% identity; conversely, extensive divergency exists between the B domains of the 2 proteins.5 6
FV deficiency causes a hemorrhagic phenotype, first described by Owren in 1947.7 It is inherited as an autosomal recessive trait (Mendelian Inheritance in Man *227400), with an incidence of about 1 in 1 million. Congenital FV deficiency can be classified as either CRM− (cross-reacting material negative) (type I deficiency), with low or unmeasurable antigen levels, or CRM+ (type II deficiency), showing normal or mildly reduced antigen levels associated with reduced coagulant activity. The FV gene (F5) was mapped to chromosome 1q23, spans more than 80 kb and contains 25 exons, the B domain being encoded by the large exon 13.5 The genetic basis of severe or moderately severe type I FV deficiency is still poorly explored (up to now, a total of 12 mutations scattered through F5 have been identified).8-13 Most mutations are nonsense, frameshift, or splicing mutations, giving rise to null alleles, whereas only 3 missense mutations have been identified. An additional 8 mutations causing FV deficiency have been reported in the heterozygous state, 5 of which were found in patients who carried the FV Leiden mutation on the second allele, leading to the “pseudohomozygous APC-resistance” condition.14-18 Only one genetic defect associated with type II deficiency has been so far reported.19
The molecular characterization by transient expression in COS-1 cells of the Arg2074Cys missense mutation in F5 causing moderately severe FV deficiency in an Italian patient is reported here for the first time.
Materials and methods
Materials
pMT2/FV expression plasmid, containing the full-length FV complementary DNA (cDNA), was kindly provided by Dr R. J. Kaufman (Howard Hughes Medical Institute, University of Michigan Medical School). Oligonucleotides were purchased from MWG-biotech (Ebersberg, Germany). REDTaq DNA polymerase was from Sigma (St Louis, MO). Sheep anti–human FV polyclonal antibody was from The Binding Site (Birmingham, United Kingdom).
Coagulation tests
FV was measured in plasma using a functional assay based on the prothrombin time with human recombinant relipidated tissue factor (Recombiplastin; Ortho Diagnostic System, Milan, Italy) and FV-deficient plasma from a congenitally deficient patient with unmeasurable FV plasma levels. FV antigen levels were determined using a sandwich enzyme immunoassay (EIA) based on a sheep anti–human polyclonal antibody.20 FV levels were expressed in both tests as the percentage of normal plasma pooled from 40 healthy individuals. Normal ranges for FV activity (FV:C) and antigen (FV:Ag) levels were 58% to 140% and 64% to 139%, respectively.
DNA extraction
The institutional review board of the University of Milan approved this study and appropriate informed consent was obtained from all analyzed individuals. Genomic DNA was extracted from whole blood by using the Nucleon BACC1 kit (Amersham Pharmacia Biotech, Uppsala, Sweden).
DNA sequence analysis
Polymerase chain reaction (PCR) amplifications of exons 1 through 25 of F5 were performed under standard conditions by using genomic DNA as template and REDTaq DNA polymerase. Primers were designed on the basis of the known genomic sequence ofF5 (GenBank accession number Z99572) and their sequences, as well as the specific PCR conditions for each primer couple, are available on request. Sequencing reactions were performed directly on PCR products purified by ammonium acetate precipitation. Sequencing primers were the same as in amplification reactions, except those used for the long exon 13, which was sequenced using additional internal primers. Sequence analysis was carried out on both strands by means of the BigDye Terminator Cycle Sequencing Kit and of an automated ABI-3100 DNA sequencer (Applied Biosystems, Foster City, CA). Factura and Sequence Navigator software (Applied Biosystems) were used for mutation detection.
Site-directed mutagenesis
Site-directed mutagenesis of pMT2/FV plasmid was performed using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The mutagenic primers 5′-CCCTTCCGTGCCTGTCTGAATGCCC-3′ and 5′-GGGCATTCAGACAGGCACGGAAGGG-3′ (underlined letters indicate the mismatch), corresponding to nucleotide positions 6382-6406 (according to Jenny et al1; GenBank accession numberM16967), were used to replace the arginine 2074 codon with a cysteine codon. The resulting mutant plasmid (pMT2/FV-Arg2074Cys) was checked by sequencing. Both the wild-type and the mutant plasmids, used in transfection experiments, were prepared by the EndoFree Plasmid Maxi Kit (Qiagen, Hilden, Germany).
Cell cultures and transfections
COS-1 cells (SV40 transformed green monkey kidney cells) were maintained in Dulbecco modified Eagle medium (DMEM), supplemented with 10% fetal calf serum, 2 mM l-glutamine, and antibiotics (100 μg/mL streptomycin and 100 IU/mL penicillin) in a humidified atmosphere of 5% CO2 and 95% air at 37°C.
Cells were plated at a density of 3 × 105 cells/well into 6-well plates. On the following day, for each well, 15 μL Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA) was diluted into 25 μL DMEM and used within 5 minutes. This mixture was added to 4 μg of wild-type (pMT2/FV) or mutant (pMT2/FV-Arg2074Cys) plasmid per well or to a mixture of equimolar amounts of pMT2/FV and pMT2/FV-Arg2074Cys, and incubated at room temperature for 20 minutes. The mixture was added to the cells in 2 mL DMEM (supplemented withl-glutamine and antibiotics, without serum) and the plates were incubated at 37°C in a 5% CO2 humidified incubator. After 5 hours, the culture medium was replaced with fresh medium, supplemented with fetal calf serum, l-glutamine, and antibiotics. After 24 hours, the medium was removed, cells were washed twice with phosphate-buffered saline (PBS) and cultured for additional 24 or 48 hours in serum-free medium supplemented withl-glutamine, antibiotics, 2.5 mM CaCl2, and 5 mg/mL bovine serum albumin. For each transfection experiment a mock, with the unrelated pUC18 plasmid as a negative control, was set up. In parallel, all transfections were performed in 10-cm diameter dishes (all volumes were scaled up by a factor of 5.8), to obtain sufficient FV protein for functional assays on conditioned media.
FV antigen and activity measurements in conditioned media and cell lysates
FV antigen was evaluated by EIA, as described above, both on conditioned media and on cell lysates. Conditioned media were collected in prechilled tubes containing a protease inhibitor mixture (Complete; Roche, Basel, Switzerland), centrifuged to remove cell debris, and stored at −80°C. Cell lysates were prepared as previously described.21 EIA standard curves were constructed with reference plasma diluted 1:100 to 1:6400 in Tris-buffered saline (50 mM Tris, 150 mM NaCl), pH 7.5; 1% of normal pooled plasma antigen level was set equal to 80 ng/mL. The lower limit of the assay was 80 ng/dL.
FV activity was measured either in the medium as such or in about 50 × concentrated medium as described (see “Coagulation tests”). One unit is defined as the amount of FV activity present in 1 mL of pooled normal plasma and represents the lower limit of the assay. Media were concentrated by Centricon Plus-20 centrifugal filters, containing a cellulose membrane with a molecular weight cut-off of 10 kd (Millipore, Bedford, MA).
The FV-specific activity (expressed as U/μg) was calculated as the ratio between FV:C (expressed as U/dL) and FV:Ag (expressed as μg/dL) levels, both measured in concentrated media.
Metabolic labeling and immunoprecipitation analysis
For metabolic labeling and immunoprecipitation analysis, COS-1 cells were transfected in 6-well plates, as described above. The only difference was that 24 hours after transfection the replacing medium was supplemented with 10% fetal calf serum. At 72 hours after transfection, cells were washed twice with DMEM lacking methionine and cysteine (ICN Biomedicals, High Wycombe, Berks, United Kingdom) and incubated in 1.5 mL/well of methionine- and cysteine-free DMEM supplemented with 200 μCi (7.4 MBq) [35S]-labeled methionine and cysteine (Translable; ICN Biomedicals), 10% dialyzed fetal calf serum, 2 mMl-glutamine, 2.5 mM CaCl2, and 5 mg/mL bovine serum albumin. After 2 hours of labeling, cells were washed with PBS and chase was performed by adding 1 mL/well of DMEM supplemented with a 10-fold excess methionine and a 5-fold excess cysteine, 10% fetal calf serum, 2 mM L-glutamine, 2.5 mM CaCl2, and 5 mg/mL bovine serum albumin. Labeled proteins were analyzed at 0, 30, 60, 120, and 180 minutes after removal of the labeled medium. At each time point, media and cell lysates were processed as described above.
Sheep anti–human FV antibodies, previously adsorbed for 4 hours with protein-A (Sigma; product number P 7155) at room temperature, were added to cell lysates and media, and incubated for 16 hours on ice. Pellets were collected by centrifugation for 5 minutes and washed 3 times with lysis buffer (1 × PBS, 1% Triton X-100, 1 × Complete). The immunoprecipitated proteins were released from protein-A by boiling for 5 minutes in sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Samples were analyzed by 4% SDS-PAGE, according to the method of Laemmli,22 with the modification that no reducing agent was added to the loading buffer. Gels were dried under vacuum at 80°C for 1 hour. Labeled proteins were visualized by exposing the gels to a storage phosphor screen overnight (Amersham Pharmacia Biotech) and analyzed using a Typhoon 9200 phosphor imager and the ImageQuant software (Amersham Pharmacia Biotech).
Results
Case report
The proband is a 22-year-old Italian woman; her plasma FV:Ag and FV:C levels were 2.3% and 4%, respectively (Figure1A). FV deficiency was first diagnosed at the age of 10 years, after abnormal coagulation screening tests were found preceding an operation for strabismus. She received prophylactic treatment with fresh-frozen plasma before the operation. She did not bleed after a dental extraction not preceded by plasma infusion. Her menstruation is normal and the only reported bleeding symptom is easy bruising after minor trauma. Her parents, apparently not consanguineous (Figure 1A), are asymptomatic and have FV functional and antigen levels typical of heterozygotes for type I mutations, except for the FV antigen level of the proband's father, which is within the normal range.
Pedigree of the FV-deficient proband and electropherograms showing the identified missense mutation.
(A) Pedigree of the Italian family. Plasma FV coagulant activity (FV:C; % [reference interval 58%-140%]) and antigen level (FV:Ag; %; [reference interval 64%-139%]) are indicated in this order below each symbol. The arrow indicates the proband. (B) DNA sequencing electropherograms showing the mutation identified in the FV-deficient proband. The C-to-T transition is indicated by an arrow (numbering according to Jenny et al1). DNA sequences of a healthy control individual and of the heterozygous father are also reported. The predicted amino acid (aa) sequences are designated by their one-letter codes below the corresponding nucleotide (nt) sequences; in the nucleotide sequence, Y denotes C or T nucleotide.
Pedigree of the FV-deficient proband and electropherograms showing the identified missense mutation.
(A) Pedigree of the Italian family. Plasma FV coagulant activity (FV:C; % [reference interval 58%-140%]) and antigen level (FV:Ag; %; [reference interval 64%-139%]) are indicated in this order below each symbol. The arrow indicates the proband. (B) DNA sequencing electropherograms showing the mutation identified in the FV-deficient proband. The C-to-T transition is indicated by an arrow (numbering according to Jenny et al1). DNA sequences of a healthy control individual and of the heterozygous father are also reported. The predicted amino acid (aa) sequences are designated by their one-letter codes below the corresponding nucleotide (nt) sequences; in the nucleotide sequence, Y denotes C or T nucleotide.
Identification of Arg2074Cys mutation
Sequencing of all 25 F5 exons, including exon-intron boundaries and approximately 300 base pair (bp) of the promoter region, revealed that the proband was homozygous for a 6394C>T transition in exon 23. This resulted in a missense mutation leading to a nonconservative Arg-Cys amino acid substitution at position 2074 (Arg2074Cys) in the C2 domain of FV. Both parents were heterozygous for the same missense mutation (Figure 1B), which was never found in 200 screened haploid genomes from control individuals.
Transient expression of wild-type and mutant recombinant FV in COS-1 cells
To study the molecular basis of FV activity and antigen deficiency observed in the FV-deficient patient, a recombinant mutated FV (Arg2074Cys-FV) was transiently expressed in COS-1 cells, which do not express endogenous FV. The 6394C>T nucleotide substitution was introduced in pMT2/FV expression plasmid by site-directed mutagenesis to obtain the mutated construct pMT2/FV-Arg2074Cys. Cells were transfected with either the wild-type or the mutant construct; an equimolar mixture of both plasmids was also transfected to mimic the heterozygous condition. Serum-free media as well as cell extracts were analyzed for the presence of FV antigen by EIA.
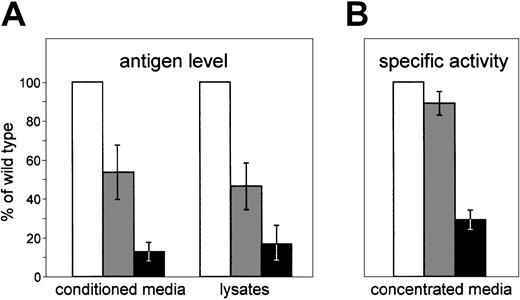
FV antigen in media conditioned for 48 hours by cells expressing the wild-type construct ranged from 3.3 μg/dL to 5.9 μg/dL. In conditioned media of cells transfected with pMT2/FV-Arg2074Cys plasmid, an about 8-fold reduction in FV antigen was observed. Cotransfection of wild-type and mutant FV cDNAs increased extracellular FV antigen levels to about 50% of the wild type (Figure2A). Similar results were obtained on media conditioned for 24 hours, even though in this case, FV antigen levels were one-half those measured after 48 hours of expression (data not shown). In cell lysates (Figure 2A), FV antigen was reduced to 46.5% and 16.9% of the wild type in cells transfected with equimolar amounts of pMT2/FV and pMT2/FV-Arg2074Cys or with the sole mutant plasmid, respectively. Altogether these data suggest that the Arg2074Cys mutation prevents FV secretion and causes an intracellular degradation of the mutant protein.
Transient expression of wild-type and mutant FV protein in COS-1 cells.
pMT2/FV (wild type; ■), pMT2/FV-Arg2074Cys (mutant; ▪), or equimolar amounts of both plasmids (heterozygous condition; ░) were transiently transfected in COS-1 cells. Equal numbers of cells and equal amounts of plasmids were used in transfection experiments, as described in “Materials and methods.” (A) Antigen levels of recombinant FV were measured both in conditioned media and in the corresponding cell lysates by an EIA assay. (B) The specific activities of recombinant FV were determined by calculating the ratio between FV activity (measured by a functional assay based on the prothrombin time) and FV antigen levels, both assayed in about 50 × concentrated media. Bars represent means ± standard deviations of 3 independent experiments, each performed in duplicate. The mean value of wild-type FV was set as 100%.
Transient expression of wild-type and mutant FV protein in COS-1 cells.
pMT2/FV (wild type; ■), pMT2/FV-Arg2074Cys (mutant; ▪), or equimolar amounts of both plasmids (heterozygous condition; ░) were transiently transfected in COS-1 cells. Equal numbers of cells and equal amounts of plasmids were used in transfection experiments, as described in “Materials and methods.” (A) Antigen levels of recombinant FV were measured both in conditioned media and in the corresponding cell lysates by an EIA assay. (B) The specific activities of recombinant FV were determined by calculating the ratio between FV activity (measured by a functional assay based on the prothrombin time) and FV antigen levels, both assayed in about 50 × concentrated media. Bars represent means ± standard deviations of 3 independent experiments, each performed in duplicate. The mean value of wild-type FV was set as 100%.
FV coagulant–specific activity was measured on concentrated conditioned media by an assay based on prothrombin time, and the results are reported in Figure 2B. The average specific activity estimated for the wild-type recombinant protein was 0.48 U/μg, which is comparable to levels of activity previously reported.23 24 The activity of the Arg2074Cys-FV (0.14 U/μg) was 29% of wild type, suggesting that the low quantities of FV antigen measured in conditioned media are constituted by dysfunctional molecules. FV molecules secreted by cells coexpressing wild-type and mutant cDNAs had a specific activity similar to the wild-type recombinant FV (0.43 U/μg). To exclude possible artifacts due to the concentration step, the coagulant activity of the wild-type recombinant FV was also measured in conditioned medium as such. The calculated specific activity resulted to be in good agreement with data obtained from the concentrated medium (data not shown).
Pulse-chase analysis of wild-type and mutant FV molecules
To evaluate whether the observed reduction of mutant FV in cultured media and cell lysates resulted from an impairment of Arg2074Cys-FV secretion and/or from increased intracellular or extracellular degradation, pulse-chase experiments of the Arg2074Cys and the wild-type FV were performed. Transfected COS-1 cells were labeled for 2 hours with [35S]-methionine and [35S]-cysteine, and subsequently chased with an excess of the corresponding cold amino acids for 30 to 180 minutes. Immunoprecipitated proteins from cell lysates and culture media were separated on SDS-PAGE under nonreducing conditions. To normalize the results, equal numbers of cells were present at the time of transfection, and equal proportions of the labeled cell lysates and conditioned media were immunoprecipitated and loaded for SDS-PAGE analysis.
Both recombinant proteins were detected as a single band of about 330 kd in the cell extracts (Figure 3A-B). Wild-type FV was exported from the cells to the medium immediately, and nearly all labeled FV was found in the medium 3 hours after labeling (Figure 3C). Conversely, only trace amounts of Arg2074Cys-FV were detectable in conditioned media (Figure 3D). The mutant FV molecules, however, did not accumulate inside the cells as demonstrated by the marked reduction of the FV specific band after 120 to 180 minutes of chase (Figure 3B). The results of mock-transfected COS-1 cells (pUC18) demonstrated that the higher-molecular-weight protein, coimmunoprecipitating with FV from culture media, is likely to represent an aspecific band (Figure 3C-D).
Pulse-chase experiments of wild-type and mutant FV protein in COS-1 cells.
COS-1 cells, transiently transfected with pMT2/FV (wild type) or pMT2/FV-Arg2074Cys (mutant), were pulse-labeled with [35S]-methionine and [35S]-cysteine for 2 hours, and then chased by cold methionine and cysteine for various periods of time up to 180 minutes. At the specified chase period (0, 30, 60, 120, and 180 minutes) radiolabeled FV was immunoprecipitated from cell lysates (A,B) and from the corresponding conditioned media (C,D), electrophoresed on 4% SDS-PAGE gels under nonreducing conditions, and then detected by a phosphor imager (Typhoon 9200). The arrowheads indicate the 330-kDa FV molecule. In all panels, the pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid. Below each lane, a densitometric analysis of the band corresponding to immunoprecipitated FV is shown. Bar graphs are expressed as arbitrary densitometry units (y-axis), as calculated by the ImageQuant software by integrating intensities of all the pixels in the band excluding the background.
Pulse-chase experiments of wild-type and mutant FV protein in COS-1 cells.
COS-1 cells, transiently transfected with pMT2/FV (wild type) or pMT2/FV-Arg2074Cys (mutant), were pulse-labeled with [35S]-methionine and [35S]-cysteine for 2 hours, and then chased by cold methionine and cysteine for various periods of time up to 180 minutes. At the specified chase period (0, 30, 60, 120, and 180 minutes) radiolabeled FV was immunoprecipitated from cell lysates (A,B) and from the corresponding conditioned media (C,D), electrophoresed on 4% SDS-PAGE gels under nonreducing conditions, and then detected by a phosphor imager (Typhoon 9200). The arrowheads indicate the 330-kDa FV molecule. In all panels, the pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid. Below each lane, a densitometric analysis of the band corresponding to immunoprecipitated FV is shown. Bar graphs are expressed as arbitrary densitometry units (y-axis), as calculated by the ImageQuant software by integrating intensities of all the pixels in the band excluding the background.
Discussion
None of the missense mutations underlying severe FV deficiency identified so far has been analyzed by in vitro expression.11,12,25 This study reports the identification of a homozygous missense mutation (Arg2074Cys) in an Italian family with FV deficiency and the molecular characterization of the consequences on FV secretion and activity by expression of the recombinant mutant protein in COS-1 cells. During the preparation of this manuscript, the same mutation was reported but not further characterized by Bossone et al12 in an unrelated Italian family.
The proband had low functional and antigen levels of FV in plasma. The proband's parents were genotypically heterozygous, and had slightly reduced FV plasma levels. For both of the proband's parents, functional FV levels were lower than antigen levels, suggesting the presence of small circulating quantities of dysfunctional Arg2074Cys-FV. For the proband, the markedly reduced level of FV coagulant activity (4%) was slightly higher than the antigen level (2.3%). No clear explanation can be put forward for this discrepancy, except methodologic variability, taking into consideration that the FV activity levels measured in the proband's plasma are close to the sensitivity limit of the assay.
The identified Arg2074Cys mutation involves a strictly conserved residue located in the C2 domain of FV. Alignment of mammalian FV and FVIII sequences in the region surrounding the mutation showed an almost complete amino acid conservation of the whole region (data not shown). The FV C2 domain is composed of 8 antiparallel strands arranged in a jelly-roll β-barrel structure, whose lower surface exhibits 3 adjacent loops (spikes 1, 2, and 3). The 3 spikes have been proposed to mediate binding of activated FV to phospholipid membranes.26 The arginine 2074 residue, located between spike 1 and spike 2, is considered critical for proper folding of the C2 domain.26 Moreover, investigation of the functional role of charged amino acids within the C2 domain of FV by in vitro alanine-scanning mutagenesis27 demonstrated that a different amino acid substitution involving the same codon (Arg2074Ala) causes the synthesis and secretion of an FV molecule with a coagulant activity less than 15% of wild type and with reduced phospholipid binding affinity. Recently, a spontaneous mutation at the same residue (Arg2074His) was identified in 2 unrelated FV-deficient Tunisian patients.11 The role of this substitution in causing type I FV deficiency was postulated on the basis of computer-assisted structural predictions and of the existence of hemophilia A–causing mutations in the corresponding residue of FVIII (Arg2209Gln/Leu/Gly).28 However, depending on the amino acid change, mutation of arginine 2209 in FVIII has been reported to cause either the secretion of a stable dysfunctional FVIII or the production of a destabilized protein.28 It is therefore difficult to predict the pathogenetic effect of the reported Arg2074Cys mutation merely on the basis of molecular modeling.
The occurrence in unrelated FV-deficient patients of missense mutations affecting the same residue (2074) contrasts with the observed high level of allelic heterogeneity underlying the disease.8-13A possible explanation for the recurrence of the Arg2074Cys mutation is the involvement of a CpG mutation hot spot. On the other hand, the presence of a different mutation not involving a CpG dinucleotide at the same residue in FV (Arg2074His), as well as of several hemophilia A–causing mutations in the corresponding residue of FVIII, points to the important role of this amino acid for FV synthesis and secretion. Therefore, to investigate the functional role of the Arg2074Cys mutation, the mutant protein was expressed in COS-1 cells. Expression studies showed that, in cells transfected with the pMT2/FV-Arg2074Cys plasmid expressing the mutant protein, FV was secreted at very low levels into culture media (12.7% of wild type; Figure 2A). Furthermore, Arg2074Cys-FV protein accumulated in lower amounts inside the cell compared with wild type (16.9% of wild type; Figure 2A). These data demonstrated that the Arg2074Cys mutation is sufficient to impair FV secretion and that the mutant protein undergoes a rapid intracellular degradation. Pulse-chase experiments confirmed that only very low amounts of Arg2074Cys-FV are secreted from transfected cells and that the mutant FV molecules do not accumulate intracellularly.
Evaluation of the coagulant activity of secreted wild-type and mutant FV was possible after concentration of conditioned media of transfected cells. These experiments showed that the Arg2074Cys mutation reduced by two-thirds the coagulant activity of the cofactor. Interestingly, coexpression of wild-type and mutant FV caused only a slight reduction of the coagulant activity of secreted molecules compared with the wild type (89% vs 100%), confirming that the wild-type protein is secreted much more efficiently than the mutant one. The replacement of Arg2074 with a cysteine residue in FV C2 domain seems therefore to affect both secretion and activity of FV. These conclusions fit well with plasma FV levels (FV:Ag, 14%; FV:C, 5%) reported for the other Italian FV-deficient patient carrying the Arg2074Cys mutation in the homozygous state.12 It is worth noticing that a thiol group at position 2074 may form a new disulfide bridge with other cysteines, for example with the predicted free cysteine residue located at position 2113 of FV,29 within the spike 3 of C2 domain. This would probably cause a complete refolding of this region of the C2 domain, which mediates the interaction with phospholipid membranes. In conclusion, these data represent the first molecular characterization of the pathogenetic mechanism responsible for moderately severe FV deficiency caused by a missense mutation, that, as demonstrated by transfection and subsequent pulse-chase experiments, affects both the secretion and coagulant activity of FV.
We thank all family members for their participation in this study. We thank Dr R. J. Kaufman for kindly providing the pMT2/FV expression plasmid. We wish to acknowledge Dr A. Rocino (Centro Emofilia e Trombosi Divisione Ematologia, Ospedale Nuovo Pellegrini, Napoli, Italy) for the clinical identification of the proband, family history, and blood collection, and Dr Rossella Bader and Maria Teresa Bajetta (Angelo Bianchi Bonomi Hemophilia and Thrombosis Center, Maggiore Hospital, Milan, Italy) for FV antigen quantitation by EIA.
Prepublished online as Blood First Edition Paper, August 15, 2002; DOI 10.1182/blood-2002-06-1928.
Supported by grant no. 2001057917 from MURST (Ministero dell'Università e della Ricerca Scientifica e Tecnologica) and by IRCCS Maggiore Hospital, Milan, Italy. Funded also in part by a grant of Fondazione Italo Monzino to F.P.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Maria Luisa Tenchini, Department of Biology and Genetics for Medical Sciences, via Viotti, 3/5-20133 Milano, Italy; e-mail: marialuisa.tenchini@unimi.it.

![Fig. 1. Pedigree of the FV-deficient proband and electropherograms showing the identified missense mutation. / (A) Pedigree of the Italian family. Plasma FV coagulant activity (FV:C; % [reference interval 58%-140%]) and antigen level (FV:Ag; %; [reference interval 64%-139%]) are indicated in this order below each symbol. The arrow indicates the proband. (B) DNA sequencing electropherograms showing the mutation identified in the FV-deficient proband. The C-to-T transition is indicated by an arrow (numbering according to Jenny et al1). DNA sequences of a healthy control individual and of the heterozygous father are also reported. The predicted amino acid (aa) sequences are designated by their one-letter codes below the corresponding nucleotide (nt) sequences; in the nucleotide sequence, Y denotes C or T nucleotide.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/1/10.1182_blood-2002-06-1928/6/m_h80133603001.jpeg?Expires=1767739390&Signature=Eot9tvL6Ww7o1iKUwIG5mT1fjxZC9j-ipd8n8ZSOOriMgSplXp~9gp7360eP9yY~99Px5bosJkODx8ltcGidKfWDoSzaTTa5rJnRgIRvWdQHLzXUxWicqKELd0SxMyE1GgQ0nF9V9SXWPPTNzb6VRHLQEf1zuT3ZlANpG4IoUGywnnKxz5mDV4BtG3w7zIYsAKiAJPQ40xp1kraqoKWqIR6kvbYa1WvGFXpiSYg7v~n5XfF9X5esPXHrL6I~mpRqd2v4h3AqKAENqXX7ErWoYHFDHPvn71oEk-RE8XS6fr48kG6VFGTjwgnupCa9q0TbWOc5V7Y1K8QO5kaIprwpVA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Fig. 3. Pulse-chase experiments of wild-type and mutant FV protein in COS-1 cells. / COS-1 cells, transiently transfected with pMT2/FV (wild type) or pMT2/FV-Arg2074Cys (mutant), were pulse-labeled with [35S]-methionine and [35S]-cysteine for 2 hours, and then chased by cold methionine and cysteine for various periods of time up to 180 minutes. At the specified chase period (0, 30, 60, 120, and 180 minutes) radiolabeled FV was immunoprecipitated from cell lysates (A,B) and from the corresponding conditioned media (C,D), electrophoresed on 4% SDS-PAGE gels under nonreducing conditions, and then detected by a phosphor imager (Typhoon 9200). The arrowheads indicate the 330-kDa FV molecule. In all panels, the pUC18 lane contains immunoprecipitable proteins at the end of the pulse period from COS-1 cells transfected with the unrelated pUC18 plasmid. Below each lane, a densitometric analysis of the band corresponding to immunoprecipitated FV is shown. Bar graphs are expressed as arbitrary densitometry units (y-axis), as calculated by the ImageQuant software by integrating intensities of all the pixels in the band excluding the background.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/101/1/10.1182_blood-2002-06-1928/6/m_h80133603003.jpeg?Expires=1767739390&Signature=gOKrrlhr5mIjBNtPApXoxkDqy5iZ8bFNb8f0hcQd1Te8UXkSM4wFtlpUoCMplN4odRTvg4STFcs7r82kwjgMzg-0R3Ldd75qxheEnOhXNmBZHLvM~-DiK34RZUyMXwRkdc3-EqIQBYYWwqR5RMLgbSbyrKYq0xzc9B0KZN8-uiIFrhtMr5acC0RiklJRBanSqsgcU4bUCwICDWWP~TEs9KYaPfl5TZQIuutlqVN7~wLkdj9ETCdmAv2faIfWSRy1qnI-43v0cUpSOTUqSy27FdcYNC0g7SwQjqLPvYJwQbYYlUo453viWzNtYfun3hBGBE0wdb6qnjITY8Z54Uyt~g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal