Abstract
SMAD proteins are downstream signal transducers of the transforming growth factor β (TGF-β) superfamily, which serve as pleiotropic regulators in embryonic and adult hematopoiesis. SMAD5, initially considered to mediate bone morphogenetic proteins (BMPs) signals, can also transduce the inhibitory signal of TGF-β1 on proliferation of hematopoietic progenitors derived from human bone marrow. To define its specific role in regulation of primitive multipotential progenitors during early embryonic hematopoiesis, we examined Smad5−/− yolk sacs at E9.0 to 9.5 and detected an elevated number of high-proliferative potential colony-forming cells (HPP-CFCs) with enhanced replating potential. To exclude the possible influence of microenvironmental deficit on embryonic hematopoiesis in vivo, we performed in vitro embryonic stem (ES) cell differentiation assay and investigated the HPP-CFCs in particular. Smad5−/− embryoid bodies (EBs) contained an elevated number of blast colony-forming cells (BL-CFCs), the in vitro equivalent of hemangioblast, in contrast to reduced proliferation of primitive erythroid precursors (Ery/Ps) within the mutant EBs. More importantly, profoundly increased frequency of HPP-CFCs, featured with a gene-dosage effect, was detected within day 6 Smad5−/−EBs compared with the wild type. In addition, Smad5−/−HPP-CFCs displayed enhanced self-renewal capacity and decreased sensitivity to TGF-β1 inhibition, suggesting a critical role of Smad5 in TGF-β1 regulation of embryonic HPP-CFCs. Consistently, reverse transcription-polymerase chain reaction analysis detected alterations of the transcription factors including GATA-2 and AML1 as well as cytokine receptors in Smad5−/− HPP-CFC colonies. Together, these data define an important function of SMAD5 in negative regulation of high-proliferative potential precursors during embryonic hematopoiesis.
Introduction
The transforming growth factor β (TGF-β) superfamily, a large group of highly conserved growth factors including TGF-β, activins, and bone morphogenetic proteins (BMPs), regulate a wide variety of cellular functions such as proliferation, differentiation, apoptosis, and migration. Signals from these growth factors are transduced by a group of SMAD proteins.1,2 To date, there are 9 vertebrate SMADs, including the receptor-activated pathway-specific SMADs (R-SMADs), SMAD1-3, 5, and 8, the common mediator SMAD4 and SMAD4β, and the inhibitory SMADs, SMAD6 and 7. Signaling is initiated when the ligand induces assembly of a heteromeric complex of type II TGF-β receptors (TβRII) and type I TGF-β receptors (TβRI). Then R-SMADs are directly phosphorylated by activated TβRI. On phosphorylation, R-SMADs interact with SMAD4 to form heteromeric complexes that translocate to the nucleus, where they act as transcription factors to regulate the transcriptional response of the target genes. Original in vitro studies suggest that SMAD2 and 3 act downstream of the TGF-β and activin receptors, whereas SMAD1 and 5 respond to BMP signals.3-7
Previous studies have strongly shown that TGF-β can either in vivo or in vitro preferentially inhibit proliferation of the most primitive types of hematopoietic cells, long-term culture-initiating cells (LTC-ICs) and their presumed immediate progeny, high-proliferative potential colony-forming cells (HPP-CFCs), but spare more mature low-proliferative potential colony-forming cells (LPP-CFCs).8-11 Although conventionally thought to respond to BMP signals, a recent finding demonstrates that SMAD5 is also capable of transducing the inhibitory signal of TGF-β1 and TGF-β2 on proliferation of hematopoietic progenitor cells from human adult bone marrow.12
Whether SMAD5 can also play a role in negative regulation of primitive multipotential progenitors by TGF-β during embryonic hematopoiesis is particularly investigated in this study. Targeted disruption of theSmad5 gene results in multiple defects and embryonic lethality at E10.5 to E11.5.13,14 Preliminary analysis has shown that Smad5−/− yolk sacs can give rise to an increased number of granulocyte-macrophage colony-forming units (CFU-GMs); therefore, it is highly probable that SMAD5 may also mediate the inhibitory signal of TGF-β on proliferation of hematopoietic progenitor cells during embryonic development. Considering the selective modulation of TGF-β on primitive hematopoietic progenitors from bone marrow and cord blood, the appropriate choice of the precursors to be focused may be critical. Previous in vitro assays have defined the presence of quiescent bone marrow progenitors that represent cells close to, or actually within, the hematopoietic stem cell (HSC) compartment. HPP-CFCs, first described as murine bone marrow cells giving rise to macroscopic colonies and quiescent cells with relative resistance to 5-fluorouracil treatment, are the earliest multipotential hematopoietic precursors that can be cultured in vitro in the absence of stromal cells.15 During embryonic hematopoiesis, it is detected first at early somite stages (E8.25) in mouse yolk sac and can be recapitulated using the differentiation model of embryonic stem (ES) cells.16 Whereas CFU-GMs detected in 7-day CFC assay represent hematopoietic progenitors at a relatively mature stage, accordingly, the specific concern for HPP-CFCs is more relevant in terms of the primitiveness (stemness).
Moreover, because the defective microenvironment, including angiogenesis defect and mesenchyme apoptosis, is detected in early Smad5−/− embryos, direct analysis of the embryonic hematopoiesis in these mutant embryos would not give the most reliable results.13 Alternatively, the in vitro hematopoietic differentiation of ES cells has been shown to recapitulate the early embryonic hematopoiesis while circumventing the drawbacks of defective development and early death of some mutant embryos.17Hence, in this study, we capitalized on this system and analyzed hematopoietic precursors such as blast colony-forming cells (BL-CFCs), primitive erythroid precursors (Ery/Ps), and definitive HPP-CFCs, sequentially initiating within embryoid bodies (EBs), to identify the roles of SMAD5 in regulation of these cells.
Here we demonstrated increased frequency and self-renewal capacity of HPP-CFCs from Smad5−/− E9.0 to 9.5 yolk sac and ES cells with the latter characterized by a gene-dosage effect and more profoundly affected. Moreover, reduced sensitivity of the mutant HPP-CFC to TGF-β1 inhibition further supported the involvement of the Smad5 gene in the TGF-β1/HPP-CFC regulatory pathway. In contrast to enhanced frequency of BL-CFCs within Smad5−/− EBs, reduced proliferation of Ery/P was observed. These data may suggest essential and pleiotropic involvements of SMAD5 and its upstream TGF-β superfamily members during embryonic hematopoiesis.
Materials and methods
ES cell lines and culture conditions
ES cells in vitro hematopoietic differentiation
The methods used for the hematopoietic differentiation of ES cells were essentially as described by Keller et al.17 For the generation of EBs, ES cells (6000 cells/3.5-cm Petri dish) were plated into methylcellulose medium without extra cytokines and after 7 days of differentiation c-kit ligand (KL; 20 ng/mL) and interleukin 11 (IL-11; 10 ng/mL) were added. At day 12 of ES cell differentiation, hematopoietic EBs were scored according to a previously described standard.19
BL-CFC assay
To assess BL-CFCs, blast colonies were generated as described previously.20 Briefly, blast colonies were generated from dispersed day 3.5 EBs cells in the presence of vascular endothelial growth factor (VEGF; 5 ng/mL) and KL (50 ng/mL) and scored after 4 to 6 days of incubation.
HPP-CFC assay
Disaggregated cells from day 6 EBs as well as E9.0 to 9.5 yolk sac were replated into methylcellulose HPP-CFC cultures containing KL (50 ng/mL), IL-3 (20 ng/mL), IL-11 (20 ng/mL), granulocyte-macrophage colony-stimulating factor (GM-CSF; 20 ng/mL), and erythropoietin (Epo; 6 U/mL) in 3.5-cm Petri dishes. Epo and TGF-β2 were purchased from Kirin Brewery (Tokyo, Japan) and R & D Systems (Minneapolis, MN), respectively, and the other cytokines were obtained from Peprotech (Rocky Hill, NJ). For HPP-CFC identification, compact colonies (> 0.5 mm) or more diffuse colonies (> 1.0 mm) were scored as HPP-CFCs by an inverted microscope after incubation at 37°C for 2 weeks. Other small colonies, such as CFU-mix and CFU-GM, were scored after 7 to 10 days of culture. For detection of Ery/Ps, disaggregated EB cells at the indicated time were cultured in medium containing Epo (6 U/mL) and scored after 5 to 7 days of incubation.
Replating experiments
At 14 days of culture, plucked HPP-CFC colonies were either individually or bulk resuspended in 200 μL Iscove modified Dulbecco medium (IMDM) to form single-cell suspensions, then replated into HPP-CFC culture. Secondary HPP-CFCs or other more committed colonies were scored after 14 and 7 days of culture. The tertiary cultures were performed by replating 1 × 105 cells from secondary HPP-CFC culture into HPP-CFC medium after 2 weeks of incubation. For yolk sac, the indicated number of HPP-CFC colonies derived from 3 yolk sacs were individually replated into HPP-CFC culture and secondary colonies were scored after 7 days of incubation.
Flow cytometry
Cells in HPP-CFC culture were resuspended at 1 × 106/100 μL phosphate-buffered saline (PBS) containing 1% fetal bovine serum (FBS), stained for 30 minutes at 4°C with the phycoerythrin-conjugated CD11b mouse antibodies (Pharmingen, San Diego, CA), and analyzed.
Semiquantitative RT-PCR
Total RNA was isolated from EBs or HPP-CFCs using Trizol (Gibco BRL, Gaithersburg, MD) according to the manufacturer's instructions and then treated with DNase (Promega, Madison, WI). Reverse transcription-polymerase chain reaction (RT-PCR) was performed by using the mRNA selective PCR kit (Takara Shuzo, Japan). A 10-fold dilution of each sample was PCR amplified to achieve signals within the linear amplification range. Cycles for each primer pair were empirically determined so as to yield product within the early exponential phase of synthesis to ensure comparative analyses. The gene primers, selected to cross introns where possible, are listed in Table 1. All the genes were analyzed on more than one occasion using cDNA from independently derived RNA samples.
Oligonucleotide primers used for RT-PCR
| Gene . | Primer sequence . | T, °C* . | Size, bp . | Reference no. . |
|---|---|---|---|---|
| Brachyury | 5′-TGCTGCCTGTGAGTCATAAC-3′ | 52 | 947 | Keller et al17 |
| 5′-TCCAGGTGCTATATATTGCC-3′ | ||||
| Flk-1 | 5′-TAGGTGCCTCCCCATACCCTGG-3′ | 60 | 398 | Fennie et al56 |
| 5′-TGGCCGGCTCTTTCGCTTACTG-3′ | ||||
| SCL | 5′-TATGAGATGGAGATTTCTGATG-3′ | 55 | 396 | Begley et al57 |
| 5′-GCTCCTCTGTGTAACTGTCC-3′ | ||||
| GATA-1 | 5′-GGAATTCGGGCCCCTTGTGAGGCCAGAGAG-3′ | 60 | 404 | Tsai et al58 |
| 5′-CGGGGTACCTCACGCTCCAGCCAGATTCGACCC-3′ | ||||
| GATA-2 | 5′-CTCCAGCTTCACCCCTAAGCAG-3′ | 60 | 254 | SD |
| 5′-CATAAGGTGGTGGTTGTCGTCT-3′ | ||||
| EKLF | 5′-TCGCCGGAGACGCAGGCT-3′ | 55 | 359 | Keller et al17 |
| 5′-CCCAGTCCTTGTGCAGGA-3′ | ||||
| AML1 | 5′-GACCGCAGCATGGTGGAGGTACT-3′ | 60 | 269 | SD |
| 5′-ACTCAGTGAGAAGGACCAGAGACTTCTA-3′ | ||||
| NF-E2 | 5′-GAGCCCTGGCCATGAAGATTCC-3′ | 62.5 | 391 | Andrews et al59 |
| 5′-CACCATCAGCAGCCTGTTGCAG-3′ | ||||
| βH1 globin | 5′-CTCAAGGAGACCTTTGCTCA-3′ | 55 | 265 | Keller et al17 |
| 5′-AGTCCCCATGGAGTCAAAGA-3′ | ||||
| ζ-globin | 5′-GCTCAGGCCGAGCCCATTGG-3′ | 55 | 371 | Weiss et al60 |
| 5′-TAGCGGTACTTCTCAGTCAG-3′ | ||||
| Epo R | 5′-TTGACGCTGTCTCTCATTCTGG-3′ | 62 | 356 | Kuramochi et al61 |
| 5′-CAATACCAAGTAGGTGTCCTGG-3′ | ||||
| IL-3 R | 5′-TTCCTTTGGGCTCTTCTATCGC-3′ | 60 | 554 | Schmitt et al62 |
| 5′-CCAGGAGCAAGGTGAAGATGAG-3′ | ||||
| GM-CSF Rα | 5′-GCGGGCGACACGAGGATGAAGCAC-3′ | 60 | 264 | Park et al63 |
| 5′-CTAGGGCTGCAGGAGGTCCTTCCT-3′ | ||||
| c-kit | 5′-TGTCTCTCCAGTTTCCCTGC-3′ | 50 | 765 | Keller et al17 |
| 5′-TTCAGGGACTCATGGGCTCA-3′ | ||||
| Smad5 | 5′-CTTGGATGGACGTCTGCAAG-3′ | 60 | 880 | SD |
| 5′-CATGGTGAAAGTTGCAGTTC-3′ | ||||
| TGF-β1 | 5′-CACCATCCATGACATGAACCG-3′ | 55 | 375 | SD |
| 5′-TTGCGACCCACGTAGTAGACG-3′ | ||||
| TβRI | 5′-CGTTACAGTGTTTCTGCCACCT-3′ | 52 | 314 | Roelen64 |
| 5′-AGACGAAGCACACTGGTCCAGC-3′ | ||||
| TβRII | 5′-GGAAGTCTGCGTGGCCGTGTGG-3′ | 52 | 298 | Roelen64 |
| 5′-CTATGGCAATCCCCAGCGGAGG-3′ | ||||
| HPRT | 5′-GCTGGTGAAAAGGACCTCT-3′ | 55 | 249 | Keller et al17 |
| 5′-CACAGGACTAGAACACCTGC-3′ |
| Gene . | Primer sequence . | T, °C* . | Size, bp . | Reference no. . |
|---|---|---|---|---|
| Brachyury | 5′-TGCTGCCTGTGAGTCATAAC-3′ | 52 | 947 | Keller et al17 |
| 5′-TCCAGGTGCTATATATTGCC-3′ | ||||
| Flk-1 | 5′-TAGGTGCCTCCCCATACCCTGG-3′ | 60 | 398 | Fennie et al56 |
| 5′-TGGCCGGCTCTTTCGCTTACTG-3′ | ||||
| SCL | 5′-TATGAGATGGAGATTTCTGATG-3′ | 55 | 396 | Begley et al57 |
| 5′-GCTCCTCTGTGTAACTGTCC-3′ | ||||
| GATA-1 | 5′-GGAATTCGGGCCCCTTGTGAGGCCAGAGAG-3′ | 60 | 404 | Tsai et al58 |
| 5′-CGGGGTACCTCACGCTCCAGCCAGATTCGACCC-3′ | ||||
| GATA-2 | 5′-CTCCAGCTTCACCCCTAAGCAG-3′ | 60 | 254 | SD |
| 5′-CATAAGGTGGTGGTTGTCGTCT-3′ | ||||
| EKLF | 5′-TCGCCGGAGACGCAGGCT-3′ | 55 | 359 | Keller et al17 |
| 5′-CCCAGTCCTTGTGCAGGA-3′ | ||||
| AML1 | 5′-GACCGCAGCATGGTGGAGGTACT-3′ | 60 | 269 | SD |
| 5′-ACTCAGTGAGAAGGACCAGAGACTTCTA-3′ | ||||
| NF-E2 | 5′-GAGCCCTGGCCATGAAGATTCC-3′ | 62.5 | 391 | Andrews et al59 |
| 5′-CACCATCAGCAGCCTGTTGCAG-3′ | ||||
| βH1 globin | 5′-CTCAAGGAGACCTTTGCTCA-3′ | 55 | 265 | Keller et al17 |
| 5′-AGTCCCCATGGAGTCAAAGA-3′ | ||||
| ζ-globin | 5′-GCTCAGGCCGAGCCCATTGG-3′ | 55 | 371 | Weiss et al60 |
| 5′-TAGCGGTACTTCTCAGTCAG-3′ | ||||
| Epo R | 5′-TTGACGCTGTCTCTCATTCTGG-3′ | 62 | 356 | Kuramochi et al61 |
| 5′-CAATACCAAGTAGGTGTCCTGG-3′ | ||||
| IL-3 R | 5′-TTCCTTTGGGCTCTTCTATCGC-3′ | 60 | 554 | Schmitt et al62 |
| 5′-CCAGGAGCAAGGTGAAGATGAG-3′ | ||||
| GM-CSF Rα | 5′-GCGGGCGACACGAGGATGAAGCAC-3′ | 60 | 264 | Park et al63 |
| 5′-CTAGGGCTGCAGGAGGTCCTTCCT-3′ | ||||
| c-kit | 5′-TGTCTCTCCAGTTTCCCTGC-3′ | 50 | 765 | Keller et al17 |
| 5′-TTCAGGGACTCATGGGCTCA-3′ | ||||
| Smad5 | 5′-CTTGGATGGACGTCTGCAAG-3′ | 60 | 880 | SD |
| 5′-CATGGTGAAAGTTGCAGTTC-3′ | ||||
| TGF-β1 | 5′-CACCATCCATGACATGAACCG-3′ | 55 | 375 | SD |
| 5′-TTGCGACCCACGTAGTAGACG-3′ | ||||
| TβRI | 5′-CGTTACAGTGTTTCTGCCACCT-3′ | 52 | 314 | Roelen64 |
| 5′-AGACGAAGCACACTGGTCCAGC-3′ | ||||
| TβRII | 5′-GGAAGTCTGCGTGGCCGTGTGG-3′ | 52 | 298 | Roelen64 |
| 5′-CTATGGCAATCCCCAGCGGAGG-3′ | ||||
| HPRT | 5′-GCTGGTGAAAAGGACCTCT-3′ | 55 | 249 | Keller et al17 |
| 5′-CACAGGACTAGAACACCTGC-3′ |
SD indicates self-designed primer sequence; Epo R, erythropoietin receptor; and HPRT, hypoxanthine phosphoribosyl transferase.
Annealing temperature.
Statistical analyses
Values shown are the mean ± SD. Significant differences between groups were evaluated using the Student t test andP < .05.
Results
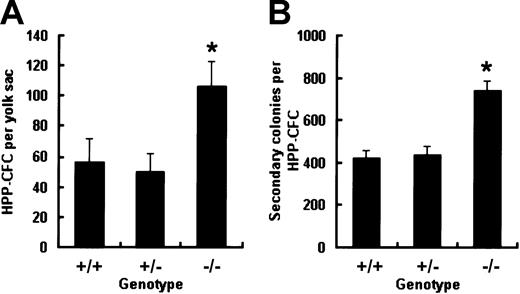
Increased frequency and replating potential of HPP-CFCs within E9.0 to E9.5 Smad5−/− yolk sac
It has been previously reported that when assayed in 7-day CFC cultures containing KL, IL-3, IL-6, and Epo, E9.5 Smad5−/− yolk sac can give rise to an elevated number of CFU-GMs compared with wild-type littermates.13 To further evaluate the influence of Smad5 disruption on more primitive progenitors in vitro, HPP-CFCs within the yolk sac were identified with a distinct cytokine cocktail composed of KL, IL-3, IL-11, GM-CSF, and Epo proven to favor the macroscopic colony growth. Single-cell suspensions made from E9.0 to E9.5 yolk sac were plated into the HPP-CFC culture and macroscopic colonies were scored according to described criteria after 14 days of incubation. As shown in Figure1A, the total number of HPP-CFCs from Smad5−/− yolk sac was twice that from wild-type and heterozygous embryos. Thereafter, single-cell suspensions made from individual HPP-CFC colonies plucked after 14 days of primary culture were replated into secondary HPP-CFC cultures to compare the replating potential among the 3 genotypes. After 7 days of secondary culture, a similarly elevated number of colonies was found in the Smad5−/− group (Figure 1B). Cells from these secondary colonies were examined by May-Giemsa staining and showed no morphologic abnormalities. These results indicate that the Smad5 gene may be involved in negative regulation of frequency and regeneration capacity of HPP-CFCs during early embryonic hematopoiesis.
Increased frequency and replating potential of HPP-CFCs from Smad5−/− yolk sacs at E9.0 to 9.5.
Dispersed yolk sac cells were cultured in HPP-CFC medium and the number of macroscopic colonies was counted after 14 days of culture (A). Then individual plucked macroscopic colonies were resuspended and plated into secondary replating system and the number of secondary colonies was scored after 7 days of culture (B). Results of HPP-CFC assay were from 5 wild-type, 8 Smad5+/−, and 6 Smad5−/−embryos and are expressed as means ± SEMs. Results of replating assay were obtained from 36 wild-type, 36 heterozygous, and 72 Smad5−/− HPP-CFC macroscopic colonies of 3 yolk sac cultures, respectively, and are expressed as means ± SEMs. *represents data found to be significantly different from the corresponding control values (P < .05).
Increased frequency and replating potential of HPP-CFCs from Smad5−/− yolk sacs at E9.0 to 9.5.
Dispersed yolk sac cells were cultured in HPP-CFC medium and the number of macroscopic colonies was counted after 14 days of culture (A). Then individual plucked macroscopic colonies were resuspended and plated into secondary replating system and the number of secondary colonies was scored after 7 days of culture (B). Results of HPP-CFC assay were from 5 wild-type, 8 Smad5+/−, and 6 Smad5−/−embryos and are expressed as means ± SEMs. Results of replating assay were obtained from 36 wild-type, 36 heterozygous, and 72 Smad5−/− HPP-CFC macroscopic colonies of 3 yolk sac cultures, respectively, and are expressed as means ± SEMs. *represents data found to be significantly different from the corresponding control values (P < .05).
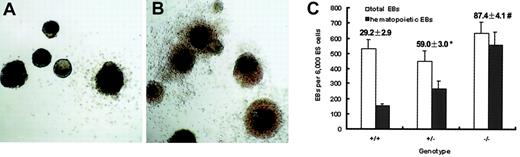
Elevated incidence of hematopoietic EBs from Smad5−/−ES cells
Dissection of the direct effect of Smad5 disruption on hematopoietic development in vivo is complicated by its influence on nonhematopoietic tissues. In E9.5 Smad5−/− yolk sac, mesenchyme apoptosis and defects in angiogenesis have been reported, the latter contributing to the observed perturbations of embryonic circulation and ultimately interfering with the migration of hematopoietic progenitors between extraembryonic yolk sac and embryo proper. To get more reliable information in this study, we next used the in vitro ES cell differentiation system validated as a well-defined model of early embryonic hematopoiesis. Initially, the effect of Smad5 disruption on hematopoietic EB formation was examined. On differentiation in semisolid culture containing KL and IL-11, proven critical for efficient hematopoietic commitment of EBs, ES cells of all the 3 genotypes could form hematopoietic EBs. After 7 days of culture, these EBs contained primitive erythrocytes and by 12 days macrophages were observed at the periphery of the EBs. Strikingly at this time, Smad5−/− hematopoietic EBs showed a densely aggregated hematopoietic halo around the central cell mass, whereas only a few erythrocytes and myeloid cells were scattered at the periphery of the wild-type hematopoietic EBs (Figure2A,B). More intriguingly, the incidences of hematopoietic EBs were 29%, 59%, and 87% within wild-type, heterozygous, and homozygous EB-forming cultures, respectively, therefore, demonstrating a gene-dosage effect (Figure 2C). These data may implicate the Smad5 gene as an inhibitor on hematopoietic EB generation.
Increased incidence of hematopoietic EBs from Smad5−/− ES cells.
ES cells were plated into methylcellulose medium containing KL and IL-11 to form EBs. Note the extending hematopoietic halo surrounding the central cell mass of 85% Smad5−/− EBs (A), whereas approximately 29% wild-type EBs ruptured with a few scattered hematopoietic cells (B) after 12 days of differentiation. Incidence of hematopoietic EBs was scored and values shown above each column represent the means ± SEMs from 3 independent experiments (C). Significance was determined using the Student t test: *represents data found to be significantly different from wild-type control values (P < .05); #, data found to be significantly different from heterozygous values (P < .05). Original magnification for panels A and B, × 40.
Increased incidence of hematopoietic EBs from Smad5−/− ES cells.
ES cells were plated into methylcellulose medium containing KL and IL-11 to form EBs. Note the extending hematopoietic halo surrounding the central cell mass of 85% Smad5−/− EBs (A), whereas approximately 29% wild-type EBs ruptured with a few scattered hematopoietic cells (B) after 12 days of differentiation. Incidence of hematopoietic EBs was scored and values shown above each column represent the means ± SEMs from 3 independent experiments (C). Significance was determined using the Student t test: *represents data found to be significantly different from wild-type control values (P < .05); #, data found to be significantly different from heterozygous values (P < .05). Original magnification for panels A and B, × 40.
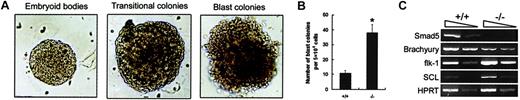
Enhanced BL-CFC number along with SCL and flk-1 transcripts within day 3.5 Smad5−/−EBs
To further determine the effect of Smad5 loss on distinct hematopoietic precursors, EBs differentiated for various periods of time were analyzed. BL-CFCs, the in vitro counterpart of hemangioblast with hematopoietic and endothelial potential, were first investigated. Two previous independent observations of Smad5 null mutant embryos suggest the possible involvement of Smad5 gene in BL-CFC development. One group has detected increased and ectopic expression of flk-1, a VEGF receptor and necessary for expansion and migration of hemangioblast in mutant embryos.14 In the other report, simultaneous abnormalities of hematopoietic progenitors' pool and angiogenesis are documented.13 Hence, Smad5−/− ES cells were assayed for their potential to generate the VEGF-responsive blast colonies. Secondary plating of dissociated day 3.5 EBs cells gave rise to 3 types of colonies: secondary EBs, transitional colonies, and blast colonies as previously reported by other groups.21,22 As shown in Figure3A, these colonies could be readily detected and distinguished according to established morphologic criteria. In comparison to the previous studies we saw a reduced efficiency of blast colony formation because we did not add the medium conditioned by the EB-derived endothelial cell line, D4T, to our assay. This medium has been shown to reproducibly increase the growth potential of BL-CFCs, and this omission was therefore likely to be responsible for the reduction of blast colonies we observed.23 However, as shown in Figure 3B, we clearly found that Smad5−/− EBs generated approximately a 3-fold higher number of blast colonies than wild-type EBs (38 ± 5.7 from Smad5−/− EBs versus 10 ± 1.9 from wild-type EBs).
Increased BL-CFC number and altered expression of hemangioblast-related genes within day 3.5 Smad5−/− EBs.
(A) Day 3.5 EBs were dissociated and then plated in BL-CFC assay culture containing VEGF and KL. After 5 days of growth, 3 types of colonies, secondary EBs, transitional colonies, and blast colonies were observed. These colonies could be readily detected and distinguished according to established morphology criteria. Original magnification for panel A, × 100. (B) The number of blast colonies shown represents the means ± SEMs from 3 independent experiments. *represents data found to be significantly different from the wild-type control values (P < .05). (C) Expression of Smad5 and hemangioblast-related genes were defined by RT-PCR analysis of day 3.5 EBs. The arrows on the top of the photo in panel C represent the 10-fold dilution of samples.
Increased BL-CFC number and altered expression of hemangioblast-related genes within day 3.5 Smad5−/− EBs.
(A) Day 3.5 EBs were dissociated and then plated in BL-CFC assay culture containing VEGF and KL. After 5 days of growth, 3 types of colonies, secondary EBs, transitional colonies, and blast colonies were observed. These colonies could be readily detected and distinguished according to established morphology criteria. Original magnification for panel A, × 100. (B) The number of blast colonies shown represents the means ± SEMs from 3 independent experiments. *represents data found to be significantly different from the wild-type control values (P < .05). (C) Expression of Smad5 and hemangioblast-related genes were defined by RT-PCR analysis of day 3.5 EBs. The arrows on the top of the photo in panel C represent the 10-fold dilution of samples.
To further define the molecular change within Smad5−/−EBs, the gene expression patterns of day 3.5 EBs were investigated (Figure 3C). In agreement with the ubiquitous expression pattern ofSmad5 gene from gastrulation onward in embryos, its transcripts were detected in day 3.5 wild-type EBs. Brachyury, coding for a transcription factor expressed in mesoderm cells along the primitive streak,24 was detected at a lower level in Smad5−/− EBs. In contrast, flk-1 was detected at a higher level in mutant EBs, consistent with that in the Smad5−/−yolk sac. Furthermore, SCL, a helix-loop-helix transcription factor expressed in both endothelial and hematopoietic lineages and essential for hemangioblast and hematopoietic specification,22,25,26 was also up-regulated. It is suggested from previous findings that mesoderm cells fated to hematopoietic and endothelial lineages may down-regulate Brachyury and up-regulate flk-1 and SCL as they migrate to extraembryonic sites.27 The collective alterations of Brachyury, flk-1, and SCL that we have observed here in the Smad5−/− EBs may suggest an accelerated switch from mesoderm toward hematopoietic lineage and possibly contribute to the enhanced frequency of BL-CFCs within Smad5−/− EBs. Thus, these findings suggest that the negative role of Smad5 gene in regulating BL-CFC development may be important at the point of mesodermal cell specification.
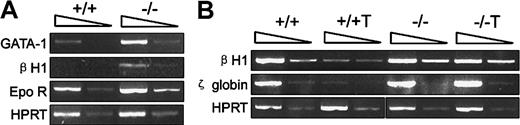
Smad5 gene was required for inhibition of embryonic globin expression by TGF-β1
As shown in Figure 4A, precocious expression of embryonic βH1 globin was detected within Smad5−/− day 3.5 EBs. Furthermore, the mutant EBs also displayed enhanced expression of both GATA-1, indispensably required for yolk sac primitive erythropoiesis, and Epo receptor.28It has been reported that strengthened TGF-β signaling by either enforced expression or additional TGF-β1 can significantly delay and decrease the embryonic globin expression in cystic EBs formed in liquid cultures.29 To determine if the precocious globin expression within mutant EBs was due to the loss of the inhibitory effect by TGF-β1, we added TGF-β1 to the EB-forming cultures and compared such inhibition at day 4 of differentiation, the exact time point for initial appearance of embryonic globin within wild-type EBs. As shown in Figure 4B, TGF-β1 was found to greatly decrease the expression of βH1-globin and ζ-globin within wild-type EBs, whereas Smad5−/− cells were not affected, demonstrating that Smad5 gene was required for the negative regulation by TGF-β1 on embryonic globin expression at the transcription level during primitive erythropoiesis.
Investigation of embryonic globin gene expression by RT-PCR within day 3.5 and 4.0 EBs.
The arrows on the top of each photo represent the 10-fold dilution of samples. Hematopoietic-related genes were defined in collected day 3.5 EBs (A) and day 4 EBs (B). +/+T and −/−T in panel B denote samples in which additional TGF-β1 (2 ng/mL) was supplemented to wild-type and mutant EB-forming cultures.
Investigation of embryonic globin gene expression by RT-PCR within day 3.5 and 4.0 EBs.
The arrows on the top of each photo represent the 10-fold dilution of samples. Hematopoietic-related genes were defined in collected day 3.5 EBs (A) and day 4 EBs (B). +/+T and −/−T in panel B denote samples in which additional TGF-β1 (2 ng/mL) was supplemented to wild-type and mutant EB-forming cultures.
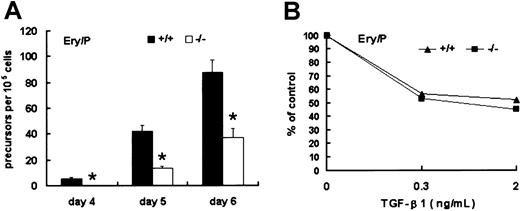
Loss of Smad5 gene resulted in defective proliferation of Ery/Ps
The first detectable unilineage hematopoietic precursors within EBs are Epo-responsive Ery/Ps, a population comparable to the blood cells detected in mouse E7.5 yolk sac. To investigate the effect of Smad5 disruption on this population, day 4, 5, and 6 EBs were dissociated into single cells and cultured in methylcellulose medium containing Epo to grow Ery/P colonies. At day 4, Ery/P colonies were detectable only within wild-type EBs. At day 5 and day 6, such colonies appeared within Smad5−/− EBs, but the number was significantly lower than that of wild type (Figure5A). These results demonstrate thatSmad5 gene may be required for proliferation of Ery/Ps.
Identification of Ery/Ps within wild-type and Smad5−/− EBs.
(A) EBs were dissociated at day 4 to 6 and replated into cultures containing Epo favoring the growth of primitive erythroid colonies. (B) The inhibitory effect of TGF-β1 added to Ery/P colony assays at final concentrations of 0.3 and 2 ng/mL. Numbers of Ery/P colonies were scored after 5 to 7 days of culture. Results shown (means ± SEMs) are from 3 independent experiments. Significance was determined using the Student t test. *represents data found to be significantly different from wild-type control values (P < .05).
Identification of Ery/Ps within wild-type and Smad5−/− EBs.
(A) EBs were dissociated at day 4 to 6 and replated into cultures containing Epo favoring the growth of primitive erythroid colonies. (B) The inhibitory effect of TGF-β1 added to Ery/P colony assays at final concentrations of 0.3 and 2 ng/mL. Numbers of Ery/P colonies were scored after 5 to 7 days of culture. Results shown (means ± SEMs) are from 3 independent experiments. Significance was determined using the Student t test. *represents data found to be significantly different from wild-type control values (P < .05).
To determine the effect of TGF-β1 on the colony formation by Ery/Ps, TGF-β1 was directly added to Ery/P cultures containing Epo. As shown in Figure 5B, wild-type and Smad5−/− Ery/Ps displayed a similar response to the inhibition of TGF-β1. Thus, Smad5gene was probably not involved in mediating the inhibitory signal of TGF-β1 on Ery/P colony generation.
Disruption of Smad5 gene led to increased frequency of HPP-CFC within day 6 EBs
Next, HPP-CFCs within day 6 EBs were examined. As illustrated in Figure 6C for the results averaged from 3 representative experiments, the frequency of HPP-CFCs and CFU-mix was profoundly elevated within day 6 Smad5−/− EBs compared with that of wild-type EBs. A gene-dosage effect was also noted on these 2 progenitor populations, with Smad5+/− EBs (10.0 HPP-CFCs and 17.9 CFU-mix) producing more colonies than wild type (4.2 HPP-CFCs and 8.9 CFU-mix) but fewer than Smad5−/− EBs (20.7 HPP-CFC and 50.0 CFU-mix). However, the frequency of CFU-GMs seemed unaffected. Another interesting observation was the precocious emergence of megacaryocyte CFUs within Smad5−/− day 6 EBs when cultured in medium supplemented with only thrombopoietin (Tpo). However, terminal differentiation of cells within the HPP-CFC cultures appeared unaffected. Specific lineages such as erythroid, granulocyte, and macrophage lineage of Smad5−/− origin were similar in morphology to those of wild-type origin examined by May-Giemsa staining. To confirm the visual impression, the cellular content of HPP-CFC culture was analyzed by flow cytometry. Similar percentage of CD11b+ myeloid cells was detected in wild-type and mutant cultures. Together, these results suggest that disruption of Smad5 gene resulted in greatly enhanced proliferation of definitive hematopoietic progenitors at an early stage but the subsequent differentiation and maturation were not blocked.
Identifications of HPP-CFC and primitive progenitor-related gene expression within day 6 wild-type and Smad5−/− EBs.
HPP-CFCs derived from day 6 EBs of wild-type (A) and Smad5−/− (B) ES cells were able to generate macroscopic colonies in vitro. Macroscopic colonies by HPP-CFCs are shown by white arrows; secondary EBs are shown by white arrowheads. Grid lines on the dishes are spaced 2 mm apart. Note the solid and round morphology of secondary EBs, whereas the HPP-CFCs displayed a looser morphology and often with red hemoglobin. The actual number of HPP-CFC, erythroid mix (CFU-mix), and CFU-GM colonies derived from day 6 EBs are shown in panel C. The results are shown as means ± SEM from 3 representative experiments; the other 2 experiments demonstrated similar results. Significance was determined using the Studentt test. *represents data found to be significantly different from wild-type control values (P < .05); #, data found to be significantly different from Smad5+/− values (P < .05). The patterns of gene expression were defined using day 6 EBs by RT-PCR (D). The arrows on the top of the photo in panel D represent the 10-fold dilution of samples.
Identifications of HPP-CFC and primitive progenitor-related gene expression within day 6 wild-type and Smad5−/− EBs.
HPP-CFCs derived from day 6 EBs of wild-type (A) and Smad5−/− (B) ES cells were able to generate macroscopic colonies in vitro. Macroscopic colonies by HPP-CFCs are shown by white arrows; secondary EBs are shown by white arrowheads. Grid lines on the dishes are spaced 2 mm apart. Note the solid and round morphology of secondary EBs, whereas the HPP-CFCs displayed a looser morphology and often with red hemoglobin. The actual number of HPP-CFC, erythroid mix (CFU-mix), and CFU-GM colonies derived from day 6 EBs are shown in panel C. The results are shown as means ± SEM from 3 representative experiments; the other 2 experiments demonstrated similar results. Significance was determined using the Studentt test. *represents data found to be significantly different from wild-type control values (P < .05); #, data found to be significantly different from Smad5+/− values (P < .05). The patterns of gene expression were defined using day 6 EBs by RT-PCR (D). The arrows on the top of the photo in panel D represent the 10-fold dilution of samples.
Thereafter, the gene expression patterns of day 6 EBs were examined with particular regard to stem cell–related transcription factors. GATA-2 is a zinc-finger transcription factor with probable functions in driving early progenitor expansion,30,31 whereas AML1, encoding the DNA-binding α subunit of core binding factor (CBF, also known as PEBP2), is required for generation of all definitive hematopoietic lineages.32 33 As shown in Figure 6D, day 6 Smad5−/− EBs displayed increased expression of SCL and AML1, which were likely responsible for increased numbers of HPP-CFCs within mutant EBs. Nevertheless, expression of GATA-2 was greatly decreased. The implication of this contradictory alteration of these stem cell–related transcription factors within mutant EBs remains to be determined.
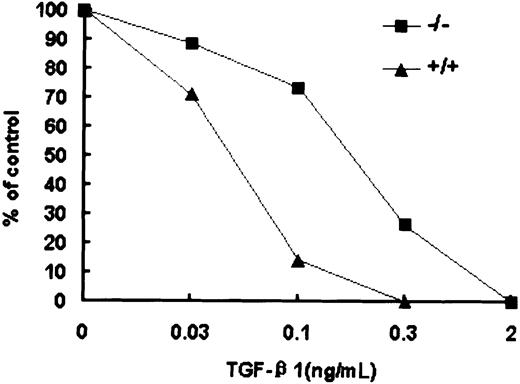
Reduced sensitivity of Smad5−/− HPP-CFCs to inhibition by TGF-β1
The increased number of macroscopic colonies generated by early progenitors within day 6 EBs largely resembled previous findings using TGF-β1 antisense oligonucleotides or anti–TGF-β1 serum in human bone marrow and cord blood.34 35 Therefore, the response of wild-type and Smad5−/− HPP-CFCs to TGF-β1 was tested (Figure 7). At concentrations of between 0.03 and 0.3 ng/mL TGF-β1, macroscopic colony formation was inhibited within wild-type EBs in a concentration-dependent manner and this inhibition was partly but significantly abrogated by the loss of Smad5. However, whereas the Smad5−/− HPP-CFCs were significantly resistant to TGF-β1–induced inhibition, higher doses (up to 2 ng/mL TGF-β1) could completely inhibit the formation of macroscopic colonies in Smad5−/− cultures. These results suggest that in addition to Smad5, there are other signaling cascades by which TGF-β1 may exert the inhibitory effect on HPP-CFCs. Similar but less efficient inhibition was also observed when the same dosage of TGF-β2 was added (data not shown).
Reduced sensitivity of Smad5−/− HPP-CFCs to TGF-β1 inhibition.
The inhibitory effect of TGF-β1, added to HPP-CFC assay at final concentrations of 0.03, 0.1, 0.3, and 2 ng/mL, is shown. These results are from a representative experiment; 4 other independent experiments demonstrated similar results.
Reduced sensitivity of Smad5−/− HPP-CFCs to TGF-β1 inhibition.
The inhibitory effect of TGF-β1, added to HPP-CFC assay at final concentrations of 0.03, 0.1, 0.3, and 2 ng/mL, is shown. These results are from a representative experiment; 4 other independent experiments demonstrated similar results.
Profoundly elevated replating potential of day 6 Smad5−/− EB-derived HPP-CFCs
Qualitatively, HPP-CFCs are capable of forming secondary colonies. To identify such functional features of ES cell–derived HPP-CFCs, we carried out preliminary investigations on replating potential of day 6 wild-type EB-derived HPP-CFCs. All the HPP-CFCs we individually plucked were capable of forming secondary CFU-GM colonies composed mainly of neutrophils, mast cells, and macrophages. About 60% of HPP-CFC colonies could form secondary HPP-CFC colonies, whereas approximately 35% of HPP-CFC colonies could produce secondary definitive erythroid colonies (BFU-Es and CFU-Es). The ratio of secondary CFU-mix colonies was relatively lower, nearly 30%. The cellular content of 24 secondary HPP-CFC colonies was examined by May-Giemsa staining and all the colonies uniformly contained neutrophils, mast cells, and macrophages resembling in constitution those from E9.5 yolk sac–derived HPP-CFCs described previously.16 These results showed that ES cell–derived HPP-CFCs were multipotential.
Next, to investigate the effect of Smad5 disruption on the regeneration capacity of HPP-CFCs, we plated cells from day 6 EB-derived individual or bulk HPP-CFCs into secondary HPP-CFC culture and secondary HPP-CFCs were scored after a further 14 days of incubation. As shown in Table2, each primary wild-type HPP-CFC could give rise to 3.4 secondary HPP-CFCs on average. In contrast, 19.7 secondary HPP-CFCs were generated from each primary Smad5−/− HPP-CFC, approximately 4- fold higher than that of wild type. Thereafter, the secondary HPP-CFCs were individually picked and mixed and 1 × 105 cells were plated for tertiary HPP-CFC assay. Although no tertiary HPP-CFC colonies were detected in wild-type culture, a significant number of tertiary macroscopic colonies were observed within the Smad5−/−culture.
Replating potential of HPP-CFCs derived from day 6 EBs
| . | Wild type . | Smad5−/− . |
|---|---|---|
| Secondary replating | ||
| Individual replating | ||
| No. of primary HPP-CFCs replated | 24 | 48 |
| No. of secondary HPP-CFCs* | 3.4 | 19.7 |
| Bulk replating | ||
| No. of secondary HPP-CFCs† | 8 ± 3 | 37 ± 8 |
| Tertiary replating | ||
| No. of tertiary HPP-CFCs† | 0 | 51 ± 4 |
| . | Wild type . | Smad5−/− . |
|---|---|---|
| Secondary replating | ||
| Individual replating | ||
| No. of primary HPP-CFCs replated | 24 | 48 |
| No. of secondary HPP-CFCs* | 3.4 | 19.7 |
| Bulk replating | ||
| No. of secondary HPP-CFCs† | 8 ± 3 | 37 ± 8 |
| Tertiary replating | ||
| No. of tertiary HPP-CFCs† | 0 | 51 ± 4 |
Results are from a representative experiment and reproduced in 3 independent experiments.
Average number of secondary HPP-CFCs per replated primary HPP-CFCs.
Results (mean ± SD) are from 3 independent cultures in a representative experiment.
Another discrepancy was detected in an aspect of constitution of secondary HPP-CFC colonies. Most secondary Smad5−/−HPP-CFCs were of multilineage potential consisting of erythrocytes, granulocytes, mast cells, and macrophages (Figure8B,F,J). In contrast, erythrocytes were absent from about 80% of secondary wild-type HPP-CFC colonies (Figure8A,E,I). The tertiary mutant HPP-CFC colonies were also composed of multilineage cells (Figure 8C,D,G,H,K,L). Together, these results indicate that Smad5−/− HPP-CFCs from day 6 EBs harbored extensive self-renewal capacity, whereas this capacity of wild-type HPP-CFC was relatively limited.
Secondary and tertiary colonies derived from primary wild-type and Smad5−/− HPP-CFCs in replating assay.
Most of the Smad5−/− HPP-CFC colony was able to form secondary HPP-mix consisting of erythrocytes, granulocytes, mast cells, and macrophages (B,F,J), whereas wild type generated almost all HPP-GMs (A,E,I). In tertiary cultures, no macroscopic HPP-CFC colony was detected in wild-type culture (C,G,K), whereas the tertiary macroscopic HPP-CFC colonies, most of which were of mixed cellular content, were readily observed in mutant culture (D,H,L). Original magnifications: panels A-D, × 40; panels E-H, × 200; and panels I-L, × 1000.
Secondary and tertiary colonies derived from primary wild-type and Smad5−/− HPP-CFCs in replating assay.
Most of the Smad5−/− HPP-CFC colony was able to form secondary HPP-mix consisting of erythrocytes, granulocytes, mast cells, and macrophages (B,F,J), whereas wild type generated almost all HPP-GMs (A,E,I). In tertiary cultures, no macroscopic HPP-CFC colony was detected in wild-type culture (C,G,K), whereas the tertiary macroscopic HPP-CFC colonies, most of which were of mixed cellular content, were readily observed in mutant culture (D,H,L). Original magnifications: panels A-D, × 40; panels E-H, × 200; and panels I-L, × 1000.
To define genetic correlation with the increased regeneration capacity of mutant HPP-CFCs, we performed RT-PCR on primary HPP-CFCs. Compared with EBs, the gene expression profile of HPP-CFCs was far more specific to define the status of primitive multipotential progenitors. As shown in Figure 9, wild-type and mutant HPP-CFCs expressed elements of TGF-β signaling cascade including TGF-β1, TβRI, and TβRII, thereby suggesting a possible regulation loop of hematopoietic progenitors by autocrine TGF-β1. Smad5−/− HPP-CFCs contained profoundly enhanced transcripts of GATA-2, AML1, GATA-1, EKLF, and comparable NF-E2. In addition, IL-3 receptor and GM-CSF receptor were found at significantly higher expression levels in Smad5−/− HPP-CFCs, although c-kit and Epo receptor were unaffected. Together, the implications of these molecular findings were generally in agreement with the increased regeneration capacity of Smad5−/− HPP-CFCs.
Altered gene expression within Smad5−/−HPP-CFCs.
Macroscopic HPP-CFC colonies derived from day 6 wild-type and Smad5−/− EBs were individually plucked and mixed for RT-PCR examination. The arrows on the top of each photo represent the 10-fold dilution of samples.
Altered gene expression within Smad5−/−HPP-CFCs.
Macroscopic HPP-CFC colonies derived from day 6 wild-type and Smad5−/− EBs were individually plucked and mixed for RT-PCR examination. The arrows on the top of each photo represent the 10-fold dilution of samples.
Discussion
In this study, we showed that disruption of Smad5 gene led to increased frequency and regeneration capacity of HPP-CFCs within yolk sac and EBs, demonstrating that SMAD5 may negatively regulate the proliferation and self-renewal of these early progenitors during embryonic hematopoiesis. The involvement of Smad5 gene in TGF-β signaling pathway has been raised by previous genetic and biochemical experiments. First, the resemblance of the Smad5 mutant yolk sac phenotype to the TGF-β1, TβRI, and TβRII knockouts is fairly striking.36-38 Second, Smad5 has a unique DNA-binding property that is similar to Smad3, which specifically transduces signals for TGF-β receptors.39 The most direct investigation using antisense oligonucleotides has shown that Smad5 is required for proliferative inhibition by TGF-β on hematopoietic progenitors from human bone marrow. Several lines of evidence demonstrated here suggest that the escaping from TGF-β1 modulation may contribute, at least in part, to the aberrant behaviors of Smad5−/− HPP-CFCs during embryonic hematopoiesis. It has been reported that differentiating EBs express a set of components involved in TGF-β1 signaling pathway including TGF-β1, TβRI, and TβRII, suggesting that an autocrine regulation exerted by TGF-β1 may exist in embryonic development.29,40,41 Here, we showed that these genes as well as Smad5 gene were expressed in EB-derived HPP-CFC colonies, thus, specifying such regulation loop to embryonic hematopoiesis. As reported, TGF-β1 is preferentially either in vivo or in vitro active on LTC-ICs and HPP-CFCs. Such a specific effect was also observed on EB-derived HPP-CFC. However, sensitivity of Smad5−/− HPP-CFCs to inhibition by TGF-β1 was significantly reduced, partly in line with the finding using antisense oligonucleotide to Smad5 gene in human bone marrow. Nevertheless, it seems that Smad5 is not the sole signal transducer of TGF-β1 during embryonic hematopoiesis because a high concentration of TGF-β1 still can abrogate the proliferation of Smad5−/−HPP-CFCs. In addition, an elevated number of HPP-CFC within Smad5−/− yolk sacs and EBs was similar to an alternative approach using blocking antibodies or antisense oligonucleotides to neutralize TGF-β1 secreted by hematopoietic cells, and consequently leading to increased numbers of macroscopic colonies in human bone marrow and cord blood.34,35 Finally, the up-regulated expression of IL-3 and GM-CSF receptors in Smad5−/−HPP-CFCs fit well with the ability of TGF-β1 to modulate the expression of a variety of cytokine receptors.42
Commonly, the inactivation of the TGF-β signaling cascade in early hematopoietic cells, which are normally quiescent, has been proposed to result in their escaping from cell cycling inhibition and finally malignant transformation. For instance, abnormalities in the expression of TGF-β receptors have been described in proliferative syndromes including both early myeloid43,44 and lymphocytic leukemia.45,46 Likewise, either Smad mutations or Smad functional inactivation have also been associated with malignant transformation. Chimeric transcription factors, such as AML1/Evi-1 and AML1/ETO resulting from t(3;21) and t(8;21) chromosomal translocations in chronic and acute myelogenous leukemia, have been documented to block TGF-β signaling by repression of Smad3 activity.47,48Smad5 gene has been located at human chromosome 5q31, which is commonly deleted in myelodysplastic syndromes and acute myeloid leukemias (MDS/AML), thus is suspected as a leukemia suppressor gene.49 50 Nevertheless, the aberrant behavior of Smad5−/− primitive progenitors was found mainly in significantly increased self-renewal capacity with the terminal differentiation unaffected, and, therefore differed to some extent from that of AML1/Evi-1 and AML1/ETO, which ultimately cause abnormal and blocked differentiation. More intriguingly, a gene-dosage effect was observed in hematopoietic EB incidence and HPP-CFC assay, which in some sense strengthened the specificity of Smad5gene. This haploinsufficiency of Smad5 gene may be an important clue for the speculation of Smad5 as a potent leukemia repressor. Previous observation that no intragenic mutations are detected in the remaining allele of leukemic patients and several human leukemia cell lines prescreened for hemizygously loss ofSmad5 gene may suggest the possible correlation between such haploinsufficiency and leukemogenesis. Hence, the hematopoietic characteristics of heterozygous Smad5+/− ES cells and mouse deserve more comprehensive analysis.
Another interesting finding demonstrated that loss of Smad5gene resulted in increased BL-CFC frequency concomitant with elevated transcriptional level of SCL and flk-1 gene, which are considered to be specific markers of the hemangioblast.21,22 Which signal mediated by SMAD5 is responsible for the negative regulation on BL-CFC development? The importance of BMP-4, the initially proposed upstream ligand of SMAD5, for hematopoietic development in the mouse has been established from effects of its genetic deletion, which disrupts mesoderm and blood cell formation in the yolk sac.51 It is further verified in an ES cell differentiation as well as Xenopus model, demonstrating the function of BMP-4 in inducing or up-regulating the expression of specific transcription factors including SCL, GATA-1, and GATA-2 together with embryonic globins.52-54 Generally, BMP-4 may continuously play pivotal and positive roles throughout the course of hematopoietic commitment. Particularly, some previous data implicate its positive involvement during BL-CFC commitment. In rhesus monkey ES cell differentiation assay, BMP-4 is able to induce the expression of SCL, indispensably required for hemangioblast commitment from mesoderm cells, and to elevate the expression of flk-1 that is necessary for expansion and migration of hemangioblast.53 Furthermore, flk-1 is not detectable in EBs stably transfected with dominant-negative BMP receptors.54 Unlike BMP-4, TGF-β appears to be a successive inhibitor on hematopoietic differentiation revealed by its negative roles in primitive/definitive hematopoiesis together with vasculogenesis. However, its direct regulation on BL-CFC development has not been identified. Thus, the result may simply imply the negative signal that SMAD5 mediated cannot be from the BMP-4 molecule and the corresponding upstream ligand remains to be determined.
A striking contradiction existed between the precocious expression of globin and delayed appearance and decreased frequency of the Ery/P colony within Smad5−/− EBs. How can these disparate observations be reconciled? SMAD5 may relay signals from distinct ligands in the proliferation of Ery/Ps and the embryonic hemoglobin synthesis. It has been observed that the induction of erythroid cells by GATA-1 requires the normal BMP signaling cascade to function.55 As a result, Smad5−/−Ery/Ps, although harboring increased transcripts of GATA-1, failed to form typical colonies efficiently as wild-type ones because the intact BMP pathway might be partially blocked due to loss ofSmad5 gene. On the other hand, it was clearly demonstrated that Smad5 gene was required for inhibition of TGF-β1 on embryonic globin production at least on the transcriptional level.
It is to be noted that the conclusion concerning the regulatory roles of TGF-β during embryonic hematopoiesis should be drawn by an appropriate approach and overall analysis. To date, disruptions of any element involved in TGF-β1 signaling pathway universally result in a defect of vasculogenesis or angiogenesis, which may interfere with embryonic hematopoiesis. For instance, the role of TGF-β1 as a positive regulator on primitive erythropoiesis has been raised for detection of very few red blood cells in the yolk sacs of TGF-β1 and TβRII null mouse embryos.34,35 However, the similar anemic appearance of TβRI knockout embryos contrasts strikingly with large numbers of erythroid colonies and the normal number of CFU-GM and CFU-mix colonies detected by in vitro colony assay of mutant yolk sacs.36 So it may indicate there is no intrinsic defect in hematopoiesis but rather the environment in the yolk sac, with malformed vascular structure, does not support proper hematopoiesis. Another approach, in vitro differentiation model of ES cells, was first used to study the role of TβRII in embryonic hematopoiesis.29 Results indicate that increased or precocious TGF-β signaling by ectopic wild-type TβRII expression inhibits hematopoiesis, which virtually resembled the fewer contained benzidine-positive cells within EBs in the presence of exogenous TGF-β1. Therefore, the ES cell differentiation model used here allowed precise dissections of distinct hematopoietic precursors and could be considered as a complementary approach to gene-targeting embryos. As such, a variety of Smad-specific signaling pathways in diverse stages of hematopoietic commitment can be more clearly defined by comprehensive applications of knockout and ES cell differentiation in vitro.
We thank Dr Gengsheng Feng, Dr Joanne Mountford, and Dr Sheng Zhou for critical review of the manuscript. We thank Dr Shengkun Sun, Dr Ye Yuan, and Chunmei Hou for expert technical assistance; Xuan Cheng and Yaxin Lu for embryo preparation; and Dr Delin Du and Dr Zikuan Guo for helpful discussions.
Prepublished online as Blood First Edition Paper, July 12, 2002; DOI 10.1182/blood-2002-02-0398.
Supported by the National Sciences Fund for Distinguished Young Scholars (30025028) and the Natural Science Foundation of Beijing (5002012).
B.L. and Y.S. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ning Mao, Department of Cell Biology, Institute of Basic Medical Sciences, Tai Ping Rd 27, Beijing 100850, Peoples' Republic of China; e-mail:maoning@nic.bmi.ac.cn.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal