Recently, it was reported that the TF-1 cell line is derived from K562 cells,1 and in this context, we cytogenetically characterized several erythroid leukemia cell lines (the SAM-1 and K562 cell lines) using spectral karyotyping (SKY) and fluorescence in situ hybridization (FISH) analysis. The SAM-1 cell line was established from a patient with chronic myelogenous leukemia (CML) in blast crisis. It was described as having the Philadelphia chromosome and a BCR-ABL rearrangement pattern that was identical to the patient's original cells, although the full karyotype was not reported.2 K-562 is a CML cell line established in 1970 from a female patient with CML in blast crisis,3 and several groups have completely characterized this cell line using FISH (with BCR/ABL probes), multicolor FISH, and comparative genomic hybridization.4 5
We received the SAM-1 cell line from the original investigators (University Medical Center, Georgetown, Washington, DC); K-562 cells were in house (Sloan Kettering Institute, New York, NY). Both cell lines were cultured in RPMI-1640 medium (Biological Industries, Beit Haemek, Israel) with 10% fetal bovine serum. Cells were harvested using standard methods. SKY was performed according to the manufacturer's protocol (Applied Spectral Imaging, Migdal Haemek, Israel). FISH was done using standard techniques with the locus-specific identifier BCR/ABL Dual Color, Dual Fusion Translocation Probe (Vysis, Downers Grove, IL). The karyotype description of SAM-1 based on SKY and FISH is as follows: 68-69 < 3n >, XX,-X, 2ish.add(2)(q37)(BCR/ABL+),der(3)del(3)(p?)t(3;10)(q1?;q24), +der(3)del(3)(p12)t(3;5)(q1?;q?), der(5)t(5;6)(q12-13;?), +del(7)(q21), +der(7)t(7;7)(p?;q?),-9, del(9)(p13), 9ish.der(9)t(9;9)(p1?3;q22)(ABL+;ABL+),der(10)t(3;10)(p21;q24), der(12)t(12;21)(p11.2;q11.2),-13, der(13)t(9;13) (?::13p11—> 13qter)(BCR/ABL+),-14, der(17)t(9;17)(?;p11.2)x2, der(17)t(5;17)(?;q12-21), der(18)t(1;18)(?;q21), dic(6;20)(?;p?), del(20)(q11), der(21)t(1;21)(?;p?), der(22)t(9;22)(q34;q11.2)(BCR/ABL+),+ der(22)t(22pter-> 22q11.2::hsr9q34;22q11.2))(BCR/ABL+)(Figure 1A).
Cytogenetic and molecular characterization of the SAM-1 and K-562 cell lines.
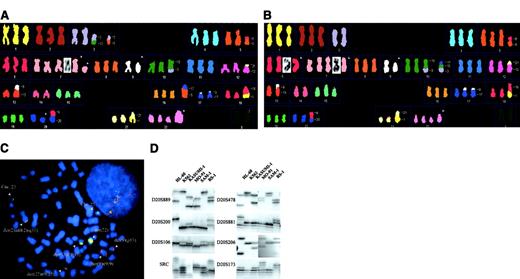
SKY analysis of SAM-1 (A) and K-562 (B) cells. Translocations are identified with numbers, and structural abnormalities (observed by G-banding) are indicated with arrows. See text for complete description. (C) Using SAM-1 cells, dual-color FISH probe forBCR, (green signal), was seen on 22q11 on 2 normal chromosomes 22, ABL, (red signal), was seen on the 9q34 band on the del(9)(p13) and on both ends of der(9)t(9;9)(p1?3;q22). Simple fusion signals were observed in Ph chromosome and in add(2). Multiple fusion signals were seen in the der(13) and in the der(22). (D) PCR-based analysis of 8 dinucleotide repeat loci in 6 myeloid leukemia cell lines. The D20S889 marker is on p-arm, whereas all others are on the q-arm of chromosome 20. SAM-1 and K562 cells exhibit an identical pattern of PCR amplification.
Cytogenetic and molecular characterization of the SAM-1 and K-562 cell lines.
SKY analysis of SAM-1 (A) and K-562 (B) cells. Translocations are identified with numbers, and structural abnormalities (observed by G-banding) are indicated with arrows. See text for complete description. (C) Using SAM-1 cells, dual-color FISH probe forBCR, (green signal), was seen on 22q11 on 2 normal chromosomes 22, ABL, (red signal), was seen on the 9q34 band on the del(9)(p13) and on both ends of der(9)t(9;9)(p1?3;q22). Simple fusion signals were observed in Ph chromosome and in add(2). Multiple fusion signals were seen in the der(13) and in the der(22). (D) PCR-based analysis of 8 dinucleotide repeat loci in 6 myeloid leukemia cell lines. The D20S889 marker is on p-arm, whereas all others are on the q-arm of chromosome 20. SAM-1 and K562 cells exhibit an identical pattern of PCR amplification.
The karyotype of K562 cells, based on SKY and FISH, shows the same modal number and 14 identical marker chromosomes to SAM-1. It differs from SAM-1 by the presence of –3, dup(6)(pter-> p12::p22qter), and der(10)t(3;10;17)(?;p11.2;q22), and by the absence of der(3)del(3)(p?)t(3;10)(q1?;q24), der(3)del(3)(p12)t(3;5)(q1?;q?)x2, del(7)(q21), der(17)t(5;17)(?;q12-21), and del(20)(q11) (Figure 1B). However, several chromosomal abnormalities found in SAM-1 cells and absent from our K-562 cell line [del(7)(q21), and the del(20)(q11)] have been reported previously in K562 cells.4 The only unique changes observed in SAM-1 (and never described in K-562 cells) are the chromosome 3 abnormalities and the der(17)(5;17).
An additional common feature between these 2 cell lines is the presence of extensive amplification of BCR/ABL fusion genes, clustered on marker chromosomes (Figure 1C). This has been previously described for K-562 cells.5 6
In the light of these cytogenetic data, we reviewed our prior polymerase chain reaction–microsatellite analysis of 43 microsatellite loci on chromosome 20 in the K562 and SAM-1 cell lines.7 An identical pattern was observed in these 2 cell lines at 39 loci (Figure 1D).
The presence of many identical and very complex structural aberrations, the presence of a unique cell clone, and the microsatellite pattern on chromosome 20 allowed us to conclude that SAM-1 is a derivative of K-562. Drexler and colleagues8have estimated that 18% of human tumor cell lines have intraspecies cross contamination, and several cases of cross contamination with the K562 cell line have been reported.1,8 9 With this report we wish to further emphasize the need for tight control of the identities of the cell lines used in research settings.
Supported by grant SAF2001-0056 from the Spanish Ministry of Science and Technology.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal