Abstract
Autoimmune phenomena may precede or accompany lymphoid malignancies, especially B-chronic lymphocytic leukemia (B-CLL). We report a patient with a 7-year history of primary (idiopathic) cold agglutinin (CA) disease in whom B-CLL subsequently developed. Immunophenotyping and single-cell reverse transcription–polymerase chain reaction (RT-PCR) were applied to investigate the origin and diversification of leukemic B cells. The obtained data indicate a memory cell-type origin of the B-CLL cells. Remarkably, theIgVκ genes of the B-CLL cells showed intraclonal diversity, whereas the mutational pattern of their paired IgVH genes were invariant. Thus, the light-chain–restricted intraclonal diversity in individual leukemic B cells in this patient strongly indicates a differential regulation or selection of the ongoing mutational process. Of note, our findings suggest that this B-CLL had developed from the patient's CA-producing B-cell population.
Introduction
Several humoral autoimmune phenomena have been reported in B-chronic lymphocytic leukemia (B-CLL), including autoimmune hemolytic anemia.1 However, it remains uncertain whether these phenomena arise from the malignant cells or from residual B cells.2,3 In both possibilities, autoimmunity should develop in association with or following malignancy. On the other hand, autoimmune dysregulation is a well-known risk factor of the development of lymphoid malignancies.4B-CLL developed in a patient with existing chronic B-cell dyscrasia that had become clinically apparent as primary cold agglutinin disease (CAD),5 which is generally regarded as premalignant or low-grade malignant lymphoproliferation.6Thus, a B-CLL origin from the cold agglutinin (CA)–expressing population was likely. Because the respective autoantigens in CAD, mostly belonging to the I/i system, are widely expressed on healthy red blood cells, high antigenic pressure on the proliferating B cells may be hypothesized. Using single-cell reverse transcription–polymerase chain reaction (RT-PCR) technology, the current study examined whether B-CLL cells express immunoglobulin (Ig) receptors that share features of CAs and whether signs of intraclonal diversification and ongoing antigenic selection can be found. Notably, a light-chain–restricted, antigen-independent intraclonal diversification was found in IgM+/CD5+ B-CLL cells expressing mutatedVH4-34/VκA27 gene rearrangements.
Study design
Patient
A 56-year-old man sought treatment for a 1-year history of cold-dependent acrocyanosis and acral numbness. Primary CAD5 was diagnosed by the detection of high-titer (1:4000 at 4°C) monoclonal IgM/κ CA and the exclusion of CA syndrome secondary to another disease, especially lymphoma. CA specificity was determined as anti-I by reactivity against panels of group 0+ erythrocytes. Results of direct antiglobulin testing using anticomplement sera were positive, and mild hemolytic anemia was observed. The patient responded to the prevention of cold exposition, repeated plasma exchange, and administration of low-dose prednisolone.
During a 7-year follow-up, the paraproteinemia level increased slowly, whereas IgG serum levels (10 to 5.5 g/L) decreased. Increasing CA titers (up to 1:512 000) but moderate autoimmune hemolytic anemia (hemoglobin count, 10.4 g/dL) accompanied worsening of the acral symptoms. Platelet count was normal. Increased and sustained absolute levels of small, mature-appearing lymphocytes (6.5 × 109/L-7.2 × 109/L) with coarsely clumped chromatin were observed in the peripheral blood. In the bone marrow, erythroid hyperplasia and nodular infiltrates of CD20+/CD23+/CD5+/IgMκ+lymphoid cells (accounting for approximately 40% of all nuclear cells) were detected. The diagnosis of B-CLL7 was confirmed by flow cytometry and by analysis of IgV gene rearrangements.8 9 Approval was obtained from the institutional review board of the Charité University Hospital for these studies. Informed consent was provided according to the Declaration of Helsinki.
Cytometric analysis
Peripheral blood mononuclear cells (PBMCs) were prepared as described10 from heparinized venous blood samples kept at 37°C. CD19+ B cells were enriched by negative immunomagnetic separation (Miltenyi Biotec, Bergish-Gladbach, Germany). Incubation with antibodies, propidium iodide staining, and flow cytometric analysis were performed as reported previously.10
Single-cell cDNA synthesis and IgV gene analysis
For analysis of IgV gene mRNA transcripts, individual CD19+ B cells from the expanded CD5+/sIgMκ+ population were sorted (FACSVantage, Becton Dickinson, CA) into single wells containing modified 1× RT-PCR buffer (5 mM dithiothreitol, 400 ng oligo-dT18, 0.2 mM dNTP, 1% Triton, 10 U RNAsin, 40 U avian myeloblastosis virus reverse transcriptase). First-strand cDNA was generated at 42°C for 60 minutes. VHand Vκ mRNA transcripts were amplified by family-specific nested PCR protocols4,11,12 using 5 μL cDNA in the first round.VκJκgene rearrangements were amplified from wells with clonally related heavy-chain transcripts. Moreover, the protooncogene bcl-2 mRNA was specifically amplified13 from these wells to further characterize the clonally related cells. To delineate disease stage, before and after diagnosis of B-CLL, cDNAs from metachronously collected enriched CD19+ B cells and single sorted B cells were subjected to specific nested PCR protocols to identify clonalIgVH rearrangements4,11 and bcl-2 and bcl-2/IgH fusion transcripts.13,14 Following column purification, all PCR products were directly sequenced using the BigDye Termination Sequencing Kit (Perkin-Elmer, Foster City, CA) and were analyzed with an automated sequencer (ABI 377; Perkin-Elmer). Sequence alignments were performed by DNAPLOT version 2.0.1 (University of Cologne, Germany) using the VBASE Sequence Directory15 or GenBank. Analysis of mutations was performed as described.16
Results and discussion
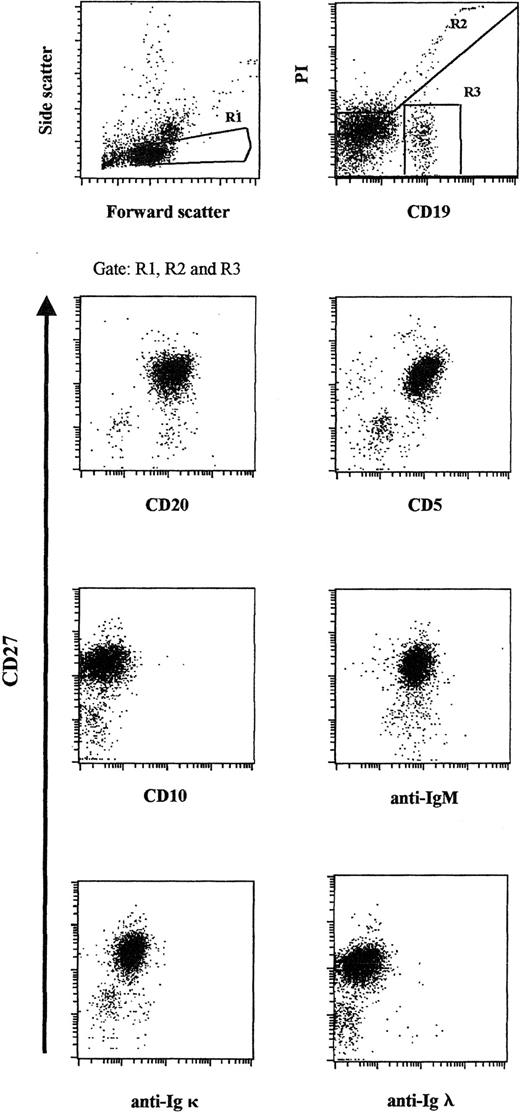
The demonstration of an expanded monotypic B-cell population is central to the diagnosis of B-CLL. In this study, the diagnosis of B-CLL was confirmed by the detection of a markedly expanded CD19+/CD20+/CD5+/CD27+/sIgM+/CD10−peripheral blood population (comprising approximately 98% of all CD19+ cells), including a striking κ light-chain restriction (Figure 1).7-9No sIgG, sIgD, or sIgA was detected.
Flow cytometric analysis of the expanded peripheral blood B-cell population.
Patient PBMCs were stained for CD19 and CD27 and gated according to light-scatter properties. Propidium iodide and CD19 expression were used to identify viable CD19+ cells. Cells were counterstained for CD20, CD5, CD10, sIgM, sIgκ light chain, and sIgλ light chain, respectively, and were plotted against CD27.
Flow cytometric analysis of the expanded peripheral blood B-cell population.
Patient PBMCs were stained for CD19 and CD27 and gated according to light-scatter properties. Propidium iodide and CD19 expression were used to identify viable CD19+ cells. Cells were counterstained for CD20, CD5, CD10, sIgM, sIgκ light chain, and sIgλ light chain, respectively, and were plotted against CD27.
By single-cell RT-PCR, these leukemic B cells were found to express mutatedIgVH/Vκgene pairings, indicating their memory cell-type origin.17,18 Twenty-eight clonally related Cμ-transcripts were analyzed, each encompassing a mutatedVH4-34 gene segment (mutational frequency, 7.7%) rearranged to D3-22 andJH3 segments, with a unique 42–base pair (bp) complementarity determining region (CDR)3. Invariantly, all of theseVH rearrangements shared the same mutations. The ratios of replacement (R) and silent (S) mutations determined for the FRs and CDRs were 4:7 (0.6) and 4:1 (4.0), respectively. A significant scarcity of FR R mutations, as observed in the patient's leukemic heavy-chain transcripts, is regarded to reflect negative selection of R mutations in these regions to maintain the functional stability of the expressed immunoglobulin molecule.16
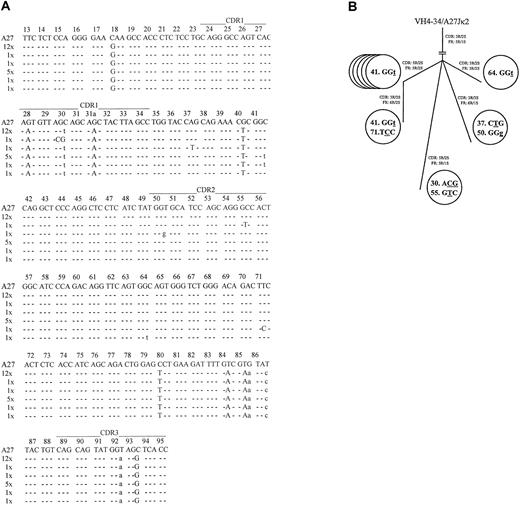
Coexpressed light-chain transcripts were identified in 21 of the 28 analyzed leukemic B cells. All expressed mutatedVκA27-Jκ2 rearrangements, with the same 27-bp CDR3 lacking any indication of TdT activity. Twelve light-chain rearrangements displayed an identical mutational pattern (mutational frequency, 4.8%). The R/S ratios determined for FRs and CDRs were 5:1 (5.0) and 3:2 (1.5), respectively. Most notably, 9 rearrangements displayed additional bp exchanges in their FRs or CDRs (Figure 2a) that were confirmed by several PCR reamplifications from the original wells. These nucleotide exchanges appeared to reflect ongoing mutations because their frequency (13/5271 bp; 2.5 × 10−3/bp) was significantly higher than the estimated PCR error rate12(1 × 10−4/bp; P < .001; χ2analysis), and no variations were found in 7420 bp on the heavy-chain level. The intraclonal relationship of cells expressing diverseVκA27 transcripts is given in a genealogical scheme19 (Figure 2B). To further delineate the origin of these clonally related cells, the presence of the protooncogene bcl-2 mRNA20 was analyzed by specific nested PCR and DNA sequencing. Notably, bcl-2 transcripts were detected in invariant and variant clonally related single cells in the stage of B-CLL, further indicating their relationship to the same population. By contrast, clonally related single-sorted B cells (4 of 96; 4.2% of sorted cells) obtained before the diagnosis of B-CLL did not express bcl-2 transcripts. Moreover, enriched CD19+ B cells obtained at the time of B-CLL were positive for bcl-2 and bcl-2/IgH fusion transcripts, whereas those collected before the time of B-CLL were negative for both products, suggesting that this transformation step occurred between these time points.20
Sequence analysis of theVκ transcripts from individual VH4-34expressing/IgM+/CD5+ B-CLL cells.
(A) Intraclonal light-chain diversity in 21 clonally relatedVκA27-Jκ2transcripts. VκA27 was identified as the closest germline sequence.15 Dashes indicate sequence identity. Somatic mutations are shown either as small letters for silent mutations or as capital letters for replacement mutations. Positions of CDRs and FRs are marked according to Kabat et al.28 The numbers of V rearrangements carrying the same sequences are indicated. AllVκ and correspondingVH sequences have been reported to GenBank (accession numbers AY057999–AY058047). (B) Genealogical tree of cells expressing additional mutations in clonally relatedVκA27-Jκ2transcripts, as demonstrated in panel A. Numbers within each circle represent the codon position of the respectiveVκ rearrangements. Mutated nucleotides are underlined, and silent or replacement substitutions are shown as small or capital letter, respectively. Resultant R/S ratios of CDRs and FRs, respectively, for each of the clonally relatedVκA27-Jκ2transcripts are also given.
Sequence analysis of theVκ transcripts from individual VH4-34expressing/IgM+/CD5+ B-CLL cells.
(A) Intraclonal light-chain diversity in 21 clonally relatedVκA27-Jκ2transcripts. VκA27 was identified as the closest germline sequence.15 Dashes indicate sequence identity. Somatic mutations are shown either as small letters for silent mutations or as capital letters for replacement mutations. Positions of CDRs and FRs are marked according to Kabat et al.28 The numbers of V rearrangements carrying the same sequences are indicated. AllVκ and correspondingVH sequences have been reported to GenBank (accession numbers AY057999–AY058047). (B) Genealogical tree of cells expressing additional mutations in clonally relatedVκA27-Jκ2transcripts, as demonstrated in panel A. Numbers within each circle represent the codon position of the respectiveVκ rearrangements. Mutated nucleotides are underlined, and silent or replacement substitutions are shown as small or capital letter, respectively. Resultant R/S ratios of CDRs and FRs, respectively, for each of the clonally relatedVκA27-Jκ2transcripts are also given.
To our knowledge, this is the first report of intraclonal variable light-chain diversification in IgM+/CD5+B-CLL.21,22 However, given that the common and diverse mutations were randomly scattered throughout theVκ segments lacking a preferential accumulation of R mutations in the CDRs, the ongoing mutational process seemed to be antigen independent. In this regard, our findings are in line with those of a recent study suggesting that antigen may no longer be able to play a significant role in the clonal expansion of CD5+/IgM+ B-CLL cells.22 One explanation for the variable light-chain–restricted intraclonal heterogeneity in our study might be an ongoing antigen-independent but differential mutational process affecting only the light-chain locus—for example, because of differences in accessibility or genetic abnormalities.23In this context, preliminary data show that approximately 10% of patients with B-CLL have mutations in their rearranged variable light-chain genes only.18 Whether these findings belong to a distinct subset of patients, however, remains uncertain.
Remarkably, clinical findings, shared immunoglobulin isotypes with the paraprotein, andVH4-34/VκIIIexpression24-26 suggest that the patient's leukemic cells had developed from his CA-expressing population. This assumption is supported by the detection of the same monoclonal and uniqueVH4-34/CDR3/Cμ sequence in the patient's B cells obtained before diagnosis of B-CLL lacking bcl-2/IgH fusion transcripts and in B cells obtained at the stage of B-CLL with bcl-2/IgH translocation. Notably, VH4-34expression is crucial in the specific recognition of the anti-I/i system,24-26 whereas the HCDR3 and light chain are thought to modify the fine specificity and affinity of binding.26 Moreover, expansions of CD5+/CD20+ B cells in CAD6,27 and, conversely, B-CLL cells producing CA2 have been reported. Although antibodies using VH4-34 do not necessarily have anti-I/i specificity25 and althoughVH4-34 usage in IgM+ B-CLL cells is frequent,17 18 selection by the I-antigen might have contributed to the distinct mutational pattern of the patient's expanded CD5+/sIgMκ+ B-cell population. In conclusion, single-cell RT-PCR provides a powerful tool for analyzing IgV gene expression in cellular (sub)populations of heterogenous lymphoproliferative diseases, such as B-CLL, to enable further insight into the association with known autoimmune phenomena.
S.R. was a fellow of the Competence Network Rheumatology at the Department of Medicine/Rheumatology and Clinical Immunology, Berlin, Germany.
T.D. and A.H. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Arne Hansen, Outpatients Department of Medicine, Charité University Hospital, Schumanstrasse 20/21, 10117 Berlin, Germany; e-mail: arne.hansen@charite.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal