Abstract
Activated transcription of the urokinase-type plasminogen activator (uPA) gene depends on the enhancer, located approximately 2 kb from the start of transcription. The proximal promoter, driving basal transcription, contains a GC-/GA-rich sequence immediately upstream of the TATA box. We have investigated the role played by this element in the transcription of the uPA gene in HeLa and PC3 cells, which do not express or constitutively express the gene, respectively. This region binds either Sp1 or Sp3, as monomers or multimers, but not a combination of the 2 proteins. The more efficient binding of Sp1 to the proximal promoter in PC3 cells is correlated to its phosphorylation state. Polymerase chain reaction (PCR)–coupled, chromatin immunoprecipitation experiments with anti-Sp1 antibodies indeed show an enrichment of proximal promoter sequences in PC3 cells and support the observed difference in transcription levels from proximal promoter constructs in HeLa versus PC3 cells. Furthermore, overexpression of Sp1 increases transcription from the reporter construct in HeLa cells, whereas in PC3 cells, overexpression of Sp3 does not reduce transcription from the same construct, indicating that the Sp1/Sp3 balance cannot be shifted. We conclude that the GC-/GA-rich element of the uPA regulatory region is an independent functional element, regulated by Sp family proteins. Phosphorylation of Sp1 determines the presence in vivo and the functionality of this element in PC3 cells. Thus, the cellular context determines the relevance of the GC-/GA-rich region in uPA gene transcription, which contributes to constitutive gene expression, related, in turn, to the invasive phenotype.
Introduction
Urokinase-type plasminogen activator (uPA) is a serine protease involved in normal tissue remodeling and also in pathological events such as tumor invasion and metastasis.1 Several human tumors have been found to overexpress uPA, and inhibition of the expression or of the enzymatic activity of this protease reduces the invasive and metastatic phenotype.1 A number of physiological, pathological, and chemical stimuli activate transcription of the uPA gene.2Their effect is mediated by the uPA enhancer, located approximately 2000 bp upstream of the transcriptional start site.3 This element contains an Ets-2 site juxtaposed to an octameric AP-1A site at the 5′ end and an eptameric AP-1Bsite at the 3′ end.4 The region between the AP-1 sites is defined as cooperativity mediator (COM) and is necessary for the combined action of the AP-1–binding transcription factors.5 The COM region is subdivided, in turn, in an upstream COM (uCOM) and a downstream COM (dCOM). By electrophoretic mobility shift assays, it was shown that uCOM can form 4 different complexes6-8 with individual proteins, such as Oct-1,9 with heterodimers of the Prep-1 and Pbx proteins10,11 and with an as-yet-unidentified UEF-1 protein. The dCOM region appears to bind specifically only UEF-1 (M.P.C. et al; unpublished data, May 2000). Interestingly, although some of the factors binding to the uCOM region are known transcriptional activators (ie, Oct-112 and Prep-1/Pbx with HoxB111,13), they do not display any transactivating activity in the uPA enhancer context.6However, both uCOM and dCOM are required for full enhancer activity.6
It is thus the proteins binding to the AP-1 sites that behave as transactivators in the uPA enhancer. AP-1 family transcription factors activate many genes in response to a large number of stimuli.14 Since members of the c-Jun family can form homodimers or heterodimers with members of the c-Fos family and with ATF-2,15 it is evident that the number of transcription factors potentially involved in transcriptional activation is rather large. In the case of the uPA enhancer, Cirillo et al16have shown that, besides the transcription factor ATF-2, at least 2 members of the c-Jun family and 2 of the c-Fos family are involved in the response of HepG2 cells to induction by interleukin 1 (IL-1) and tetradecanoyl phorbol acetate (TPA). Furthermore, it was shown that the phosphorylation state of these proteins also is relevant to transcriptional activation.16
Traditionally, the proximal promoter of the human uPA gene has been considered as spanning approximately 86 bp upstream of the transcription start site.3 This region contains 5 high- (3 × GGGCGG) and low- (2 × GGGAGG) affinity binding sites for the Sp1 family transcription factors, immediately upstream of the TATA box. This family is composed of 4 members, with a high degree of structural but not functional similarity.17 Sp1 and Sp4 are transcriptional activators, whereas Sp3 represses Sp1-mediated transcription.18-22 Sp3 contains a portable repression domain, conferring to a fusion protein the ability to repress transcription from reporter promoters containing multiple binding sites.23,24 Both repression and activation functions have been suggested to occur via protein-protein interactions with components of the basal transcriptional machinery.23 24
Sp1 and Sp3 have the same affinity for GC-/GA-rich binding sites19,25 and therefore transcriptional repression by Sp3 involves the competition with Sp1 for the common binding sites.19
Unlike Sp1, of which a single polypeptide of approximately 100 kDa is known, Sp3 is found in 3 different isoforms, the largest of approximately 115 kDa, while the smaller 2 are about 70 kDa. The large form is initiated at a non-AUG codon,25 whereas the smaller forms originate from different internal translation start sites.26 All 3 forms carry the repression domain, while the largest form carries 2 glutamine-rich activation domains and the smaller forms carry only one; however, at least one of them displays the same activity as the 115-kDa polypeptide.26
Here we have investigated the role played by the GC-/GA-rich region located immediately upstream of the TATA box in the human uPA gene transcription in a noninvasive cell line (HeLa)27 and in an invasive cell line (PC3).28 Northern blot analyses reveal that in HeLa cells no steady-state mRNA is detectable, whereas PC3 cells display the conspicuous presence of uPA mRNA. EMSAs show that the transcription factors Sp1 and Sp3 can bind the GC-/GA-rich region of the uPA gene in vitro, albeit at different levels in the 2 cell lines. In particular, the more efficient binding of Sp1 to this region can be ascribed to its phosphorylated state in PC3 versus HeLa cells, although the levels of this protein, and of Sp3, are comparable in the 2 cell lines. Moreover, immunoprecipitation experiments on in vivo cross-linked chromatin show that Sp1 is present on the uPA proximal promoter in PC3 but not HeLa cells.
Transient transfections with reporter constructs, spanning the GC-/GA-rich region of the uPA gene driving a luciferase reporter gene, indicate that this portion of the proximal promoter is essential for expression in PC3 cells, with the enhancer being a dispensable element for basal expression of uPA in this cell line. Finally, in PC3 cells the proximal promoter reporter construct is insensitive to overexpression of Sp3, whereas transcription from the same construct can be increased by overexpression of Sp1 in HeLa cells.
We conclude that occupancy of the GC-/GA-rich region of the uPA promoter by Sp1, possibly in its phosphorylated form, is at least one of the elements that determines the constitutive expression of the uPA gene in PC3 cells, and we suggest a correlation with the invasive phenotype of this cell line.
Materials and methods
Cell culture, mRNA preparation, and transient transfections
HeLa and PC3 cells were grown in Dulbecco modified Eagle medium (DMEM) with the addition of 10% (vol/vol) fetal calf serum (FCS).
Transient transfections were performed using lipofectin (Invitrogen, Milano, Italy) according to the manufacturer's instructions, at a total 1 μg of DNA per 80 000 cells in 24-well plates. Luciferase reporter vectors contained either the region of the uPA gene between −30 and +32 (henceforth called −30) or the region between −86 and +32 (henceforth called −86) in a pGL2Basic (Promega, Madison, WI) backbone. In the same constructs, the uPA enhancer (−1977/−1880) was cloned as a cassette upstream of the −30 or −86 inserts. The eukaryotic and Drosophila Sp1 and Sp3 expression vectors were kindly provided by Profs L. Lania (Naples) and G. Suske (Huddinge), respectively. In all cases, cells were cotransfected with a cytomegalovirus (CMV)–driven β-galactosidase reporter vector (Stratagene, La Jolla, CA) for normalization. The results shown are the average of at least 2 experiments in triplicate.
Detection of DNase I hypersensitive sites
DNase I hypersensitivity assays and probing of the Southern blots was performed as previously described.29
Nuclear extracts, EMSAs, and Western blots
Nuclear extracts were obtained by the method of Dignam et al.30 Aliquots were frozen and kept at −80°C.
EMSAs were performed as described31 using approximately 10 μg nuclear extract proteins/lane. The oligonucleotides used were (only one strand shown):
SpPP: 5′-AAGACAGGGGAGGGAGCCGGGCGGGAGAGGGAGGGGCGGCGCCGGGGCGGGCCCT-3′.
Sp1 consensus: 5′-GATCGATCGGGGCGGGGCGATC-3′.
Unrelated oligo: 5′-CTAGTGATGAGTCAGCCGGATC-3′.
Interactions of the EMSA mix with anti-Sp1 (Santa Cruz, sc 59X), Sp3 (Santa Cruz, sc 644X), or unrelated polyclonal antibodies was for 3 hours prior to the addition of the probe on ice. Dried EMSA gels were exposed for the appropriate time for autoradiography at −80°C, using a β-Max film (Amersham, Milano, Italy).
For Western blots, 8% (Figure 5) or 10% (Figure 4) (wt/vol) polyacrylamide-sodium dodecyl sulfate gels were loaded with 50 μg of nuclear extracts from the different cell lines and the proteins fractionated as detailed in the legends to Figures 4 and 5. Transfer to polyvinylidenefluoride (PVDF) membrane (Millipore, Bedford, MA) was performed in a semidry transfer apparatus (Sigma, Milano, Italy) for 2 hours at 0.8 mA/cm2. Overlay of the blot with the anti-Sp1 and anti-Sp3 antibodies (Santa Cruz, see above) and with the anti–HMGB-1 (formerly named HMG-132) polyclonal antibody (kindly provided by Dr M. Bustin, Bethesda, MD) used to normalize the results was overnight at 4°C in phosphate-buffered saline (PBS) containing 3% (wt/vol) dried milk as quencher. Secondary antibody reaction (anti-rabbit IgG, Amersham) was for 1 hour at 4°C in the same buffer. Bands were revealed by chemoluminescence using the SuperSignal kit (Pierce, Rockford, IL). Quantitation was performed with a SigmaGel program (Sigma). The values for Sp1, the long and the combined short isoforms in HeLa and PC3 cells, were divided by the values obtained for HMGB1 in the 2 cell lines. The numbers in Table1 represent the relative ratio obtained in HeLa cells divided by that obtained in PC3 cells for the same protein.
Chromatin cross-linking, immunoprecipitation assay, and PCR
In vivo cross-linking of cells with formaldehyde for 1 hour was performed as described.33 After 5 sonication cycles (35 seconds sonication in an Ultrasonic Processor XL Sonicator, Miosonix, (Farmingdale, NY) at 60-70 W, followed by a 1 minute rest on ice), the material was fractionated on a CsCl gradient, as described.33 Cross-linked chromatin-containing fractions were pooled and stored at −80°C. Cross-linked chromatin (200 μg) was incubated overnight with 1 μg anti-Sp1 or anti–urokinase-type plasminogen activator receptor (uPAR, used as the unrelated antibody in Figure 6) antibodies in a total volume of 1 mL radioimmunoprecipitation assay (RIPA) buffer.33 Prior to the immunoprecipitation step of the antibody-adsorbed, cross-linked chromatin with 25 μL protein A-sepharose beads (Pharmacia, Uppsala, Sweden), the beads were coated with poly-(dI-dC), poly-(dG-dC), and poly-(dA-dT) at 10 μg/mL and with 100 μg/mL of bovine serum albumin (BSA) in RIPA buffer to reduce nonspecific interactions. The immunoprecipitated material was treated with RNase A (50 μg/mL) for 30 minutes at 37°C followed by proteinase K (500 μg/mL) in 0.5% (wt/vol) sodium dodecyl sulfate (SDS) at the same temperature. Formaldehyde cross-links were reverted by heating the samples at 65°C for 5 hours and the DNA purified. This material represented the “stock” solution, from which serial dilutions (1:2, 1:5, and 1:10) were made.
The polymerase chain reaction mix contained 4 μL DNA (from either the stock or the serial dilutions) and the following oligonucleotides for the amplification of the GC-/GA-rich-containing region:
FpMP2: 5′-AATCTTTGTGAGCGTTGCGG-3′ starting at position −132.
RpMP2: 5′-CTCTGCAAAGGAAGGAGAAGTCAG-3′ ending at position +397.
PCR was performed as follows: 95°C, 3 minutes; 95°C, 1 minute; 57°C, 1 minute; 72°C, 1 minute (repeated for 33 cycles); and 72°C, 3 minutes (final extension).
For the amplification of the negative control region (exon XI of the uPA gene), the following primers were used:
FpEXO11: 5′-TTGTATCTTTGGCGTCACAGG-3′ starting at position 5262.
RpEXO11: 5′-CATTCTCTTCCTTGGTGTGAC-3′ ending at position 5444.
For the negative control region, PCR was performed as follows: 95°C, 3 minutes; 95°C, 1 minute; 58°C, 1 minute; 72°C, 1 minute (repeated for 33 cycles); and 72°C, 3 minutes (final extension).
PCR products were analyzed on a 2% (wt/vol) agarose gel in 0.5 × TBE buffer.31
Results
Different uPA mRNA levels in HeLa and PC3 correlate with DNase I hypersensitivity of the uPA regulatory region
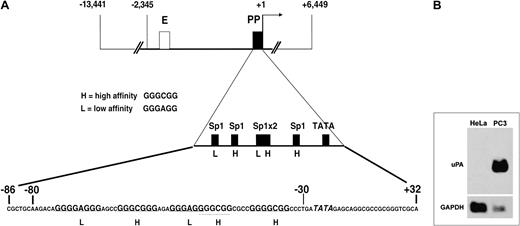
The region analyzed in this report is schematically shown in Figure 1A. High- (GGGCGG) and low- (GGGAGG) affinity Sp family member binding sites are depicted in the first enlargement, whereas the actual sequence is shown in the second enlargement.
Human uPA gene regulatory region and Northern blot analysis of the steady-state uPA mRNA level in HeLa and PC3 cells.
(A) Scheme of the uPA regulatory region, depicting enhancer (E, ■) and proximal promoter (PP, ▪). The first enlargement shows the schematic position of the low- and high-affinity Sp1 sites and of the TATA box in the PP region. The second enlargement shows the actual sequence of the −86/+32 region, where the Sp1 sites are indicated in bold and the TATA box in italics. The sequence comprised between −80 and −30 represents the oligonucleotide used for mobility shift assays (see below). (B) Total mRNA of 30 μg was fractionated on a 1% (wt/vol) agarose/formaldehyde gel. Following transfer to a positively charged nylon membrane, the Northern blot analysis was sequentially probed with uPA cDNA and GAPDH probes. uPA mRNA is detectable only in PC3 cells, which constitutively express the uPA gene.
Human uPA gene regulatory region and Northern blot analysis of the steady-state uPA mRNA level in HeLa and PC3 cells.
(A) Scheme of the uPA regulatory region, depicting enhancer (E, ■) and proximal promoter (PP, ▪). The first enlargement shows the schematic position of the low- and high-affinity Sp1 sites and of the TATA box in the PP region. The second enlargement shows the actual sequence of the −86/+32 region, where the Sp1 sites are indicated in bold and the TATA box in italics. The sequence comprised between −80 and −30 represents the oligonucleotide used for mobility shift assays (see below). (B) Total mRNA of 30 μg was fractionated on a 1% (wt/vol) agarose/formaldehyde gel. Following transfer to a positively charged nylon membrane, the Northern blot analysis was sequentially probed with uPA cDNA and GAPDH probes. uPA mRNA is detectable only in PC3 cells, which constitutively express the uPA gene.
The invasive potential of tumor cells often correlates with overexpression of extracellular matrix proteases, such as uPA. Noninvasive HeLa cells, grown in monolayer, do not produce uPA, whereas invasive PC3 cells produce the enzyme in monolayer cultures. This difference stems from the fact that the uPA gene is not expressed in HeLa cells, whereas it is constitutively expressed in PC3 cells, as revealed by a Northern blot analysis with total RNA from the 2 cell lines, in which the presence of steady-state uPA mRNA is detected in the latter cell line only (Figure 1B).
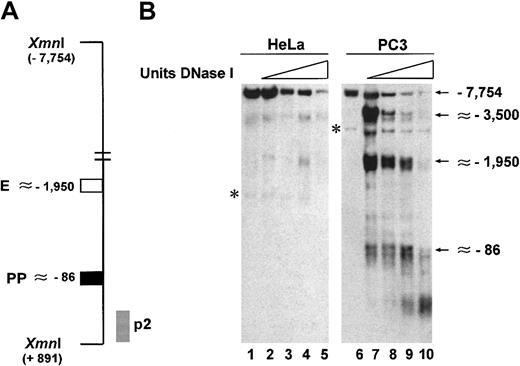
We have also analyzed the chromatin structure of the uPA regulatory region by DNase I hypersensitivity analysis in the 2 cell lines, schematically shown in Figure 2A. Figure2B shows that in HeLa cells, a faint DNase I hypersensitive site is present in correspondence of the enhancer and no site is visible on the proximal promoter (Figure 2B). In contrast, in PC3 cells, 2 clear DNase I hypersensitive sites are detected in correspondence of these 2 regulatory elements (Figure 2B). Interestingly, another hypersensitive site is present approximately 3.5 kb from the start of transcription in PC3 cells (Figure 2B). In HeLa cells, this site is also discernible at high concentrations of DNase I (Figure 2B). Although this finding has not yet been investigated, our observations are in agreement with similar results in murine and porcine cell lines, where DNase I hypersensitive sites upstream of the uPA enhancer have been reported.34-36
Detection of DNase I hypersensitive sites in the regulatory region of the uPA gene.
(A) Schematic representation of the region analyzed by DNase I hypersensitivity. The location of the enhancer (■), proximal promoter (▪), and probe (p2, ░) used are indicated; the location of the restriction sites (Xmn I) is also indicated. Purified, DNase I–digested and restricted DNA was fractionated on a 1% agarose gel in 0.5 × TBE, transferred to positively charged nylon membrane, and probed as described.29 (B) In PC3 cells the regulatory region of the uPA gene displays strong hypersensitivity to DNase I in proximity of the enhancer (∼−1950) and the proximal promoter (∼−86). The latter is completely absent in HeLa cells. The asterisks indicate nonspecific bands also present in lanes 1 and 6, where no DNase I was used.
Detection of DNase I hypersensitive sites in the regulatory region of the uPA gene.
(A) Schematic representation of the region analyzed by DNase I hypersensitivity. The location of the enhancer (■), proximal promoter (▪), and probe (p2, ░) used are indicated; the location of the restriction sites (Xmn I) is also indicated. Purified, DNase I–digested and restricted DNA was fractionated on a 1% agarose gel in 0.5 × TBE, transferred to positively charged nylon membrane, and probed as described.29 (B) In PC3 cells the regulatory region of the uPA gene displays strong hypersensitivity to DNase I in proximity of the enhancer (∼−1950) and the proximal promoter (∼−86). The latter is completely absent in HeLa cells. The asterisks indicate nonspecific bands also present in lanes 1 and 6, where no DNase I was used.
The DNase I hypersensitivity data correlate with the expression of uPA mRNA in the 2 cell lines and indicate that chromatin structure alterations may be involved in the onset of transcription of the uPA gene.
The observed differences in DNase I hypersensitivity of the regulatory region and uPA mRNA levels in PC3 versus HeLa cells may stem from a differential involvement of transcriptionally relevant sequences in the 2 cell lines. Little is known about the role played by the GC-/GA-rich region located upstream of the TATA box (Figure 1A), thus we investigated the role played by these sites in uPA transcription in the 2 cell lines.
In vitro binding of Sp1 and Sp3 to the GC-/GA-rich region of the human uPA gene
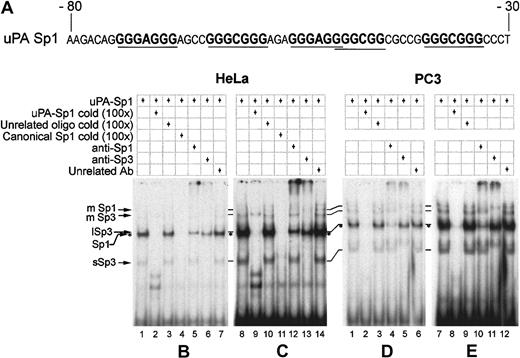
We tested if the region immediately upstream of the TATA box in the uPA promoter (Figure 3) was bound by specific proteins by performing EMSAs with an oligonucleotide spanning all the putative Sp1 binding sites (from −80 to −30 from the start of transcription, Figure 3A) and nuclear extracts from HeLa and PC3.
EMSAs with nuclear extracts from different cell lines show different levels of Sp1 and Sp3 binding to the GC-/GA-rich oligonucleotide.
(A) Oligonucleotide sequence used for electrophoretic mobility shift assays. (B-E) HeLa and PC3 nuclear extracts, 10 μg each, were used in each lane with approximately 5 fmoles of labeled oligonucleotide. Competitions were done with 100-fold excess of cold oligonucleotides, and 2 μg of polyclonal antibodies were employed when required. On the left are indicated the Sp1 and Sp3 specific bands: mSp1 and mSp3 = multiple Sp1 and Sp3; lSp3 = long Sp3 isoforms; sSp3 = short Sp3 isoforms. Sp1/DNA complex is identified with asterisks. Panels C and E are simply overexposures of the autoradiograms shown in panels B and D. HeLa (B,C) and PC3 (D,E) nuclear extracts show that both Sp1 and Sp3 bind to the GC-/GA-rich oligonucleotide, albeit with different efficiencies.
EMSAs with nuclear extracts from different cell lines show different levels of Sp1 and Sp3 binding to the GC-/GA-rich oligonucleotide.
(A) Oligonucleotide sequence used for electrophoretic mobility shift assays. (B-E) HeLa and PC3 nuclear extracts, 10 μg each, were used in each lane with approximately 5 fmoles of labeled oligonucleotide. Competitions were done with 100-fold excess of cold oligonucleotides, and 2 μg of polyclonal antibodies were employed when required. On the left are indicated the Sp1 and Sp3 specific bands: mSp1 and mSp3 = multiple Sp1 and Sp3; lSp3 = long Sp3 isoforms; sSp3 = short Sp3 isoforms. Sp1/DNA complex is identified with asterisks. Panels C and E are simply overexposures of the autoradiograms shown in panels B and D. HeLa (B,C) and PC3 (D,E) nuclear extracts show that both Sp1 and Sp3 bind to the GC-/GA-rich oligonucleotide, albeit with different efficiencies.
With HeLa cells nuclear extract, the oligonucleotide forms a number of complexes (Figure 3B, HeLa, lane 1) that are specifically competed by a 100-fold higher concentration of the unlabeled oligonucleotide and by an oligonucleotide containing a canonical Sp1 binding site (Figure 3B, HeLa, lanes 2 and 4), but not by the same concentration of an unrelated oligonucleotide (Figure 3B, HeLa, lane 3).
Incubation of the nuclear extract with antibodies against Sp1 or Sp3 allows the depletion of specific complexes in the EMSA, and thus we could separate the complexes formed by Sp1 or Sp3 with the oligonucleotide. The most prominent band in the gel is made up of both Sp1 and Sp3 DNA complexes (Figure 3B, HeLa, lanes 5 and 6), as both antibodies partially deplete the band. The Sp1-containing complex migrates faster than the Sp3-containing complex, reflecting the difference in molecular weight of the 2 proteins (see above). An unrelated antibody (Figure 3B, HeLa, lane 7) does not affect the complexes. These results are compatible with the observation that Sp1 and Sp3 have the same affinity for GC- or GA-rich binding sites.
Sp3 also forms a rapidly migrating complex, specifically competed by the canonical Sp1 oligonuclotide and depleted by the anti-Sp3 antibody (more evident in Figure 3C, HeLa, lanes 11 and 13). This band is likely to be formed by the lower molecular weight splice forms of the protein binding to the −80/−30 oligonucleotide.
The Sp family polypeptides can also form slower migrating complexes with the oligonucleotide, possibly containing 2 molecules of the transcription factors (Figure 3C, HeLa, mSp1 and mSp3 bands). These bands are specifically competed by the cold oligonucleotide and the canonical Sp1 binding site (Figure 3C, HeLa, lanes 9 and 11) and the anti-Sp1 and anti-Sp3 antibodies specifically and selectively deplete them, indicating that the oligonucleotide is bound either by Sp1 or by Sp3, but not by a mixed population of these transcription factors (Figure 3C, HeLa, compare lanes 12 and 13).
This observation suggests that the binding of Sp1 or Sp3 molecules to the −80/−30 oligonucleotide, containing multiple binding sites, may occur as dimers or in a cooperative fashion, in line with the self-association of DNA-bound Sp1 molecules previously observed.37
Also, with PC3 cell nuclear extract, the labeled oligonucleotide specifically binds Sp1 and all the Sp3 isoforms as detected by the competitions with the cold oligonucleotide (Figure 3D, PC3, lane 2). However, when we specifically deplete the nuclear extract of Sp1 or Sp3 by using polyclonal antibodies, the band corresponding to the Sp1 DNA complex seems to be enriched when compared with the band of the Sp3 DNA complex (Figure 3D, PC3, lanes 4 and 5), differently from what observed with HeLa nuclear extract (compare with Figure 3B, HeLa, lanes 5 and 6). The behavior of the slowly migrating bands and the rapidly migrating bands (Figure 3E, PC3, lanes 10 and 11, mSp1, mSp3, and sSp3 complexes; compare with Figure 3C, HeLa, lanes 12 and 13) indicates the presence of specifically competed complexes, as observed with HeLa cell nuclear extracts.
We also performed EMSAs with oligonucleotides that were mutated either in the high- (GGGCGG) or low- (GGGAGG) affinity sites (Figure 1A), and the results (not shown) indicate that the binding of Sp1 and/or Sp3 to the GC-/GA-rich region in vitro largely depends on the high-affinity sites.
The overall results indicate that the GC-/GA-rich region upstream of the TATA box can bind both Sp1 and Sp3 in vitro and that multiple, adjacent sites bind more molecules of these polypeptides either as dimers or in a cooperative manner. They also indicate that the relative binding of these 2 molecules is different in HeLa and PC3 cells.
Endogenous Sp1 is phosphorylated in PC3 but not in HeLa cells
The results in Figure 3 suggest that the content of Sp1 and Sp3 may be different in the 2 cell lines. Thus we performed a Western analysis on nuclear extracts from HeLa and PC3 cells with the same anti-Sp1 and anti-Sp3 polyclonal antibodies used in the EMSAs.
The results in Figure 4 show that HeLa and PC3 cells apparently contain Sp1 and all 3 isoforms of Sp3 in different amounts. However, quantitation and normalization of the bands reveal a relative amount of protein that is comparable in the 2 cell lines (Table 1).
Detection of Sp1 and Sp3 in nuclear extracts from HeLa and PC3 cells by overlay with specific polyclonal antibodies of a Western blot analysis.
Proteins from HeLa and PC3 nuclear extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (10% [wt/vol] for 2 hours at 150 V) and transferred to a PVDF membrane. The Western blot analysis was sequentially overlaid with specific polyclonal antibodies against Sp1, Sp3, and HMGB-1, and protein detection was performed by chemoluminescence. Sp1 as well as all 3 isoforms of Sp3 are present in HeLa and PC3 cell nuclear extracts in comparable amounts (Table 1).
Detection of Sp1 and Sp3 in nuclear extracts from HeLa and PC3 cells by overlay with specific polyclonal antibodies of a Western blot analysis.
Proteins from HeLa and PC3 nuclear extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (10% [wt/vol] for 2 hours at 150 V) and transferred to a PVDF membrane. The Western blot analysis was sequentially overlaid with specific polyclonal antibodies against Sp1, Sp3, and HMGB-1, and protein detection was performed by chemoluminescence. Sp1 as well as all 3 isoforms of Sp3 are present in HeLa and PC3 cell nuclear extracts in comparable amounts (Table 1).
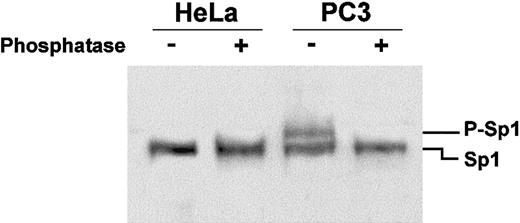
Since Sp1 from PC3 cell nuclear extracts seems to bind more efficiently to the labeled oligonucleotide than Sp1 from HeLa cell nuclear extracts, we asked if this was due to posttranslational modifications of the proteins in the 2 cell lines. Thus, nuclear extracts from HeLa and PC3 cells were treated with alkaline phosphatase, the proteins fractionated on an 8% polyacrylamide SDS gel, and analyzed by Western blotting technique, using the same polyclonal antibodies as in Figure3.
Figure 5 shows that while in HeLa cell nuclear extracts the antibodies detect only a single band corresponding to Sp1, in PC3 cells the antibodies reveal 2 bands corresponding to the unphosphorylated (lower band) and phosphorylated forms (upper band, Figure 5) of Sp1, in agreement with what was previously observed.38 Alkaline phosphatase treatment of the PC3 nuclear extracts abolishes the phosphorylated form of Sp1 (Figure 5). Thus, we can correlate the more efficient binding of Sp1 to its cognate sequence in PC3 extracts with its phosphorylation state.
Sp1 is phosphorylated in PC3 but not HeLa cells.
Nuclear extracts were treated with alkaline phosphatase for 45 minutes at 37°C prior to fractionation on an 8% SDS-polyacrylamide gel for 3 hours at 150 V. Transfer of proteins to PVDF membrane and overlay with anti-Sp1 polyclonal antibodies was as described in Figure 4. The upper band appearing in the PC3 nuclear extracts (P-Sp1), which disappears after alkaline phosphatase treatment, indicates that Sp1 is phosphorylated in this cell line.
Sp1 is phosphorylated in PC3 but not HeLa cells.
Nuclear extracts were treated with alkaline phosphatase for 45 minutes at 37°C prior to fractionation on an 8% SDS-polyacrylamide gel for 3 hours at 150 V. Transfer of proteins to PVDF membrane and overlay with anti-Sp1 polyclonal antibodies was as described in Figure 4. The upper band appearing in the PC3 nuclear extracts (P-Sp1), which disappears after alkaline phosphatase treatment, indicates that Sp1 is phosphorylated in this cell line.
We conclude that HeLa and PC3 cells do not differ in their relative content of Sp1 and Sp3 proteins, both quantitatively and qualitatively. Binding of the −80/−30 region by transcription factor Sp1 strongly depends on the posttranslational modifications (phosphorylation) of the protein molecule.
In vivo binding of Sp1 to the uPA GC-/GA-rich region
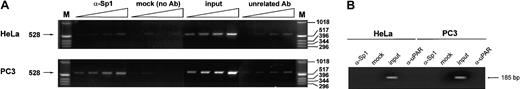
To directly assess the presence or absence of Sp1 on the proximal promoter of the endogenous uPA gene in HeLa and PC3 cells, we analyzed the occupancy of the GC-/GA-rich region in vivo by chromatin immunoprecipitation. Cross-linked, sonicated chromatin was immunoprecipitated with anti-Sp1 polyclonal antibodies, and purified DNA was analyzed by PCR with primers spanning the GC-/GA-rich region of the uPA gene. The PCR products were serially diluted and fractionated on an agarose gel.
The results in Figure 6A show that the immunoprecipitated DNA from PC3 cells was enriched in the 528-bp region spanning the uPA proximal promoter, as detected by conventional PCR, whereas no enrichment was detected in the immunoprecipitated DNA from HeLa cells. As a positive control, we amplified a genomic fragment containing an Sp1 site located 11 kb upstream from the transcription start site, which resulted enriched in the anti-Sp1 immunoprecipitated DNA from HeLa, but not from PC3 cells (data not shown). As a negative control, we also searched for immunoprecipitated exon XI of the uPA gene where no Sp1 sites are detectable by sequence analysis; in this case no signal was visible either in HeLa cells– or PC3 cells–derived genomic DNA (Figure 6B).
Detection of the presence of Sp1 and Sp3 on the uPA GC-/GA-rich region by chromatin immunoprecipitation assay.
Purified DNA from immunoprecipitated, cross-linked chromatin was amplified by conventional PCR and analyzed as described in “Materials and methods.” The PCR reaction produces a 528-bp band, as indicated on the left. For each sample (α-Sp1, mock, input, and α-uPAR = unrelated Ab), the stock DNA was amplified by PCR together with 3 decreasing dilutions as indicated by the triangles at the top and described in “Materials and methods.” (A) Sp1 is present in the region of the proximal promoter in PC3 cells but not in HeLa cells. (B) Amplification of an Sp1 unrelated region does not show a detectable signal either from HeLa cell– or PC3 cell–derived genomic DNA.
Detection of the presence of Sp1 and Sp3 on the uPA GC-/GA-rich region by chromatin immunoprecipitation assay.
Purified DNA from immunoprecipitated, cross-linked chromatin was amplified by conventional PCR and analyzed as described in “Materials and methods.” The PCR reaction produces a 528-bp band, as indicated on the left. For each sample (α-Sp1, mock, input, and α-uPAR = unrelated Ab), the stock DNA was amplified by PCR together with 3 decreasing dilutions as indicated by the triangles at the top and described in “Materials and methods.” (A) Sp1 is present in the region of the proximal promoter in PC3 cells but not in HeLa cells. (B) Amplification of an Sp1 unrelated region does not show a detectable signal either from HeLa cell– or PC3 cell–derived genomic DNA.
The results indicate that Sp1 is present on the GC-/GA-rich region of PC3 but not HeLa cells, thus indicating that regulation of transcription from the uPA proximal promoter may indeed depend on this transcription factor.
We also attempted in the same experiment with anti-Sp3 antibodies to detect the presence of the protein on the uPA proximal promoter in HeLa and PC3 cells, and with antibodies against DNA-dependent protein kinase (PK), shown to phosphorylate Sp1 in vitro, to check whether it was associated in vivo with this transcription factor. Unfortunately, in both cases, the commercially available antibodies were not sufficiently reliable to determine unequivocally the presence of the Sp3 and DNA-PK molecules on the uPA proximal promoter.
The GC-/GA-rich region is an independent transcriptional regulatory element of the uPA gene
The above in vitro results suggest that binding of Sp1 and/or Sp3 to the GC-/GA-rich region may play a role in the transcriptional regulation of the uPA gene. Thus we decided to analyze the contribution of this region to transcriptional activation. HeLa and PC3 cells were transiently transfected with constructs containing either the TATA box only (−30 construct) or the TATA box plus the GC-/GA-rich region (−86 construct) driving the luciferase gene. Figure7A shows the sequence of the −86 construct, where the Sp1/Sp3 binding sites and the position of the TATA box are indicated.
Transient transfections of HeLa and PC3 cells with luciferase reporter constructs driven by the −30 or −86 region of the human uPA promoter.
The sequences between −86 and +32 (−86 construct) and between −30 and +32 (−30 construct) driving transcription of the luciferase reporter gene are shown in Figure 1. (A) Transient transfections in HeLa and PC3 cells, showing the broad effect of the −86 construct in PC3 cells. (B) Transfection of HeLa and PC3 cells with constructs carrying the uPA enhancer (E −30 and E −86) cloned upstream of the −30 or −86 region. The contribution of the enhancer to transcriptional activation of the reporter construct in PC3 cells is marginal as compared to the effect of the proximal promoter alone (compare A and B).
Transient transfections of HeLa and PC3 cells with luciferase reporter constructs driven by the −30 or −86 region of the human uPA promoter.
The sequences between −86 and +32 (−86 construct) and between −30 and +32 (−30 construct) driving transcription of the luciferase reporter gene are shown in Figure 1. (A) Transient transfections in HeLa and PC3 cells, showing the broad effect of the −86 construct in PC3 cells. (B) Transfection of HeLa and PC3 cells with constructs carrying the uPA enhancer (E −30 and E −86) cloned upstream of the −30 or −86 region. The contribution of the enhancer to transcriptional activation of the reporter construct in PC3 cells is marginal as compared to the effect of the proximal promoter alone (compare A and B).
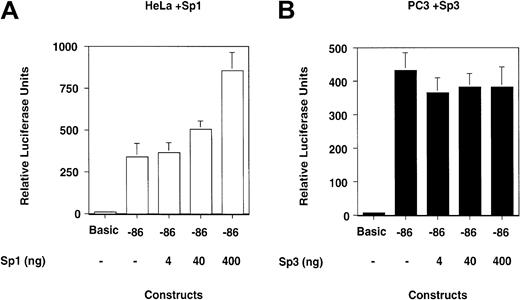
In HeLa cells, the −86 construct has a moderate effect on transcription, approximately 20-fold higher than the −30 construct (Figure 7B; Table 2). In PC3 cells, the difference between the levels of transcription of the −30 versus the −86 construct is approximately 100-fold (Figure 7B; Table 2). The results indicate that the contribution to transcription of the GC-/GA-rich region is largely different in HeLa and PC3 cells. Also, the contribution of the GC-/GA-rich element in the 2 cell lines correlates with the levels of binding of Sp1 and Sp3 proteins.
Fold increase in transcriptional activation of different constructs in HeLa and PC3 cells
| Plasmids . | Cells . | |
|---|---|---|
| HeLa . | PC3 . | |
| pGL2-Basic | 1 | 1 |
| pGL2-30 | 1.4 | 2.4 |
| pGL2-86 | 22.3 | 207.4 |
| pGL2 E-30 | 44.6 | 36 |
| pGL2 E-86 | 62.8 | 365.8 |
| Plasmids . | Cells . | |
|---|---|---|
| HeLa . | PC3 . | |
| pGL2-Basic | 1 | 1 |
| pGL2-30 | 1.4 | 2.4 |
| pGL2-86 | 22.3 | 207.4 |
| pGL2 E-30 | 44.6 | 36 |
| pGL2 E-86 | 62.8 | 365.8 |
Since the activity of the enhancer plays an important role in uPA gene transcription, we investigated the reciprocal role of this element and of the GC-/GA-rich region. The uPA enhancer was cloned in the −30 and −86 constructs, upstream of the TATA box and of the GC-/GA-rich region, respectively, and the resulting plasmids were used to transfect HeLa and PC3 cells.
In HeLa cells, the presence of the enhancer almost equally affects transcription from the −30 and the −86 constructs (Figure 7C; Table2), indicating that the GC-/GA-rich region plays a minor (if any) role. In PC3 cells, the effect of the enhancer on the −30 construct is similar to that observed in HeLa cells (Table 2). However, the presence of the GC-/GA-rich region downstream of the enhancer increases transcription 10-fold in this cell line, to a level slightly higher than that achieved with the −86 construct alone (Figure 7C; Table 2). Thus, in PC3 cells, it appears that the GC-/GA-rich region plays a more relevant role than the uPA enhancer in driving transcription from reporter constructs. Furthermore, the constructs containing either the enhancer cloned as a cassette immediately 5′ of the proximal promoter or containing the whole uPA regulatory region, in which enhancer and proximal promoter are approximately 2000 bp apart, yielded the same level of transcriptional activation, indicating that the genomic sequence separating the 2 elements had no effect on transcription in PC3 cells (W. Folk, personal written communication, February 2002).
We conclude that the GC-/GA-rich region upstream of the TATA box plays an important role in uPA gene transcription. This region can increase transcription from reporter constructs independently of the enhancer and in a manner correlated to the presence of Sp1 in vivo on the uPA proximal promoter and to its phosphorylation state.
Overexpression of Sp family proteins affects transcription of the proximal promoter reporter construct in HeLa but not PC3 cells
The above results suggest that the balance of Sp family proteins may be crucial for uPA gene expression in HeLa and PC3 cells and that posttranslational modifications may play a role in shifting the protein equilibrium. Thus, overexpression of Sp1 in HeLa cells may increase transcription from the reporter construct, whereas overexpression of Sp3 may decrease transcription from the reporter in PC3 cells. To test this hypothesis, HeLa cells were cotransfected with the proximal promoter reporter constructs and an expression vector for Sp1. Similarly, we cotransfected the reporter construct in PC3 cells together with an expression vector for Sp3.
The results shown in Figure 8 indicate that overexpression of Sp1 increases the transcriptional activity of the reporter construct in a fashion that parallels the amount of cotransfected Sp1 expression vector in HeLa cells (Figure 8A; Table3).
Overexpression of Sp1 increases transcription from the proximal promoter in HeLa cells, whereas overexpression of Sp3 does not reduce transcription in PC3 cells.
(A) HeLa and (B) PC3 cells were cotransfected with the reporter construct in which the uPA proximal promoter drives luciferase transcription and with expression vectors for Sp1 and Sp3, respectively. (A) Overexpression of Sp1 increases transcription of the luciferase construct in HeLa cells, whereas (B) overexpression of Sp3 does not affect transcription of this construct in PC3 cells.
Overexpression of Sp1 increases transcription from the proximal promoter in HeLa cells, whereas overexpression of Sp3 does not reduce transcription in PC3 cells.
(A) HeLa and (B) PC3 cells were cotransfected with the reporter construct in which the uPA proximal promoter drives luciferase transcription and with expression vectors for Sp1 and Sp3, respectively. (A) Overexpression of Sp1 increases transcription of the luciferase construct in HeLa cells, whereas (B) overexpression of Sp3 does not affect transcription of this construct in PC3 cells.
Fold increase in transcriptional activation of cotransfected HeLa and PC3 cells
| Plasmids . | Cells + overexpressed protein . | |
|---|---|---|
| HeLa + Sp1 . | PC3 + Sp3 . | |
| pGL2-Basic | 1 | 1 |
| pGL2-86 | 58.6 | 83.7 |
| pGL2-86 + 4 ng | 65.8 | 70.7 |
| pGL2-86 + 40 ng | 93.1 | 73.8 |
| pGL2-86 + 400 ng | 154.6 | 74.2 |
| Plasmids . | Cells + overexpressed protein . | |
|---|---|---|
| HeLa + Sp1 . | PC3 + Sp3 . | |
| pGL2-Basic | 1 | 1 |
| pGL2-86 | 58.6 | 83.7 |
| pGL2-86 + 4 ng | 65.8 | 70.7 |
| pGL2-86 + 40 ng | 93.1 | 73.8 |
| pGL2-86 + 400 ng | 154.6 | 74.2 |
Discussion
uPA is involved in extracellular matrix degradation and plays a role in normal and pathological processes that involve cell migration. In particular, tumor invasion and metastasis formation in a number of tissues correlate with the overexpression of the uPA gene.1 We have investigated the transcriptional role of the GC-/GA-rich region immediately upstream of the TATA box in the human uPA regulatory region in cell lines without (HeLa) and with invasive potential (PC3)27 28 that do not express or constitutively express the gene, respectively. We find that the sequence comprised between −80 and −30 is bound by the transcription factors Sp1 and Sp3 in vitro. Although the nuclear content of Sp1 and Sp3 of HeLa and PC3 cells is comparable, as detected by Western blot, Sp1 is phosphorylated in vivo only in PC3 cells. This observation correlates with the more efficient binding of this transcription factor to the GC-/GA-rich region of the uPA proximal promoter in EMSAs. This suggests that Sp1 phosphorylation plays an important role in the binding of this transcription factor to the GC-/GA-rich oligonucleotide.
In vivo experiments support the above results. Transient transfections in which reporter gene expression is driven by the uPA proximal promoter indicate that this region plays a major role in transcription in PC3 cells and has no effect in HeLa cells. It has been previously shown17 39 that the presence of multiple binding sites favors the binding of Sp3 molecules in a cooperative manner over that of Sp1. Our EMSA results are in agreement with these observations and, thus, Sp3 would act as a transcriptional repressor also in HeLa cells, preventing transcriptional activation of the reporter construct in this cell line.
On the other hand, phosphorylated Sp1 would bind more efficiently to the uPA proximal promoter in vivo in PC3 cells and substantially increase transcription from the reporter construct. In line with these observations, we were able to detect the presence of Sp1 in the proximal promoter region of the uPA gene in PC3 but not HeLa cells by chromatin immunoprecipitation experiments. Since chromatin immunoprecipitation could not be carried out with the available Sp3 antibodies, we resolved to overexpress Sp1 and Sp3 in HeLa and PC3 cells, respectively, and perform transient cotransfections with the proximal promoter constructs. We reasoned that if Sp3 was bound to the uPA proximal promoter, thus inhibiting transcriptional activation of the reporter construct in HeLa cells, an increase in the amount of Sp1 might shift the equilibrium and would result in transcriptional activation of the luciferase plasmid. Conversely, if Sp1 was bound to the GC-/GA-rich region of the uPA proximal promoter in the reporter construct in PC3 cells, overexpression of Sp3 might reduce the observed transcriptional activation by competing away Sp1 from this region. The results of these cotransfections in HeLa cells confirm the expectations. The absence of DNase I hypersensitive sites in HeLa cells on the proximal promoter and the lack of Sp1 in vivo on this element, as judged by chromatin immunoprecipitation, are in agreement with this hypothesis. The alternative, that binding of Sp3 to the uPA proximal promoter region of the uPA gene does not permit transcriptional activation to take place, is also a possibility, but not in line with the absence of a hypersensitive site, however faint. Conversely, in PC3 cells we could not detect any substantial reduction in the transcriptional activation of the reporter construct, suggesting that overexpression of Sp3 is unable to compete the binding of Sp1 to the proximal promoter. Possibly the phosphorylation of proximal promoter-bound Sp135 dramatically alters the functional Sp1/Sp3 balance of PC3 cells, rendering them constitutive for uPA expression. In this cell context, the contribution of the uPA enhancer to transcriptional activation of the reporter construct is only marginal and confirms that the GC-/GA-rich region of the proximal promoter is an independent transactivating cis-element of uPA transcription in vivo. Finally, the genomic sequence located between enhancer and proximal promoter had no effect on either elements in PC3 cells (W. Folk, personal written communication, February 2002).
The human uPA regulatory region, thus, contains another transactivating element, besides the enhancer, the “identity” and transcriptional role of which has been elucidated in this work. The genomic sequence located between the enhancer and the proximal promoter contains elements that affect uPA transcription (see, for instance, Cannio et al40). However, the role of the various elements in the context of the entire uPA regulatory region in different cells or tissues will be defined only once the basic elements have been individually characterized. The present study is another step in this direction.
It has been widely reported that proteases, in particular uPA, are expressed in normal tissue remodeling events41,42 and pathological phenomena such as tumor invasion and metastasis formation.43 44 The way in which the expression of proteases is controlled and the level at which they are expressed mark the difference between normal and pathological events.
Previous reports correlate the high and constitutive level of expression of the uPA gene in PC3 cells with their invasive phenotype (see, for instance, Angelucci et al45) and the lack of uPA gene expression in HeLa cells with the lack of invasive potential (see, for instance, Montero and Nagamine 46). Although far from explaining such a complex phenotype, our results suggest a molecular mechanism that may be involved in the onset of this phenotype and indicate the GC-/GA-rich region as one of the elements possibly involved.
We thank L. Guerrini, C. Kilstrup-Nielsen, S. Müller, V. Orlando, and V. Zappavigna for critically reading the manuscript, and Prof William Folk (Missouri University, Columbia) for sharing results prior to publication.
Supported by the Università Vita-Salute San Raffaele Excellence Center on Physiopathology of Cell Differentiation; the Italian Association for Cancer Research (AIRC); and the Italian Ministry of Education, University and Research grants (MIUR-PRIN 1999) and EU grants (QLG1-CT-2000-01131) to F.B.
I.I.-T., C.F., and E.L. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Massimo P. Crippa, Laboratory of Molecular Genetics, DIBIT-Ospedale S. Raffaele, Via Olgettina 58, 20132 Milano, Italy; e-mail: crippa.massimo@hsr.it.

![Fig. 4. Detection of Sp1 and Sp3 in nuclear extracts from HeLa and PC3 cells by overlay with specific polyclonal antibodies of a Western blot analysis. / Proteins from HeLa and PC3 nuclear extracts were fractionated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) (10% [wt/vol] for 2 hours at 150 V) and transferred to a PVDF membrane. The Western blot analysis was sequentially overlaid with specific polyclonal antibodies against Sp1, Sp3, and HMGB-1, and protein detection was performed by chemoluminescence. Sp1 as well as all 3 isoforms of Sp3 are present in HeLa and PC3 cell nuclear extracts in comparable amounts (Table 1).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/100/9/10.1182_blood.v100.9.3325/4/m_h82123326004.jpeg?Expires=1765889994&Signature=KcK2g~mjyb75eP7aJYSIkFMmn521JjhaNrHcRHa8ZN6-SFX9trPXDAV3q5W81yfoqw10c0dgzh37J7LkVFv4vj6p~-5pVlWvOkeSkyH1Iy0C7nxDqM--gFpICxdYgsIENd-uxPHsSp7e8M3LcciidCOCGlJ55b8Lx~4~QSIquGeheoj2w6YclR3Kia6XkQ0H4o5N1Wh8pf2YyGerimtu5j6xXDU0padP2BbpYV9qO-N~2U~2RJ-UVsaJtfhbgqkPaB0SyggxZicTA0cxg0N~Eu31DTHfS8Qmvg9BV4VRcyHhFLnB4jUCiqfqDeIYwS-hMtz7MYY-9HQdgvWoxdwX4Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal