Abstract
We have used an approach using 2-dimensional gel electrophoresis with mass spectrometry analysis combined with oligonucleotide chip hybridization for a comprehensive and quantitative study of the temporal patterns of protein and mRNA expression during myeloid development in the MPRO murine cell line. This global analysis detected 123 known proteins and 29 “new” proteins out of 220 protein spots identified by tandem mass spectroscopy, including proteins in 12 functional categories such as transcription factors and cytokines. Bioinformatic analysis of these proteins revealed clusters with functional importance to myeloid differentiation. Previous analyses have found that for a substantial number of genes the absolute amount of protein in the cell is not strongly correlated to the amount of mRNA. These conclusions were based on simultaneous measurement of mRNA and protein at just a single time point. Here, however, we are able to investigate the relationship between mRNA and protein in terms of simultaneous changes in their levels over multiple time points. This is the first time such a relationship has been studied, and we find that it gives a much stronger correlation, consistent with the hypothesis that a substantial proportion of protein change is a consequence of changed mRNA levels, rather than posttranscriptional effects. Cycloheximide inhibition also showed that most of the proteins detected by gel electrophoresis were relatively stable. Specific investigation of transcription factor mRNA representation showed considerable similarity to those of mature human neutrophils and highlighted several transcription factors and other functional nuclear proteins whose mRNA levels change prominently during MPRO differentiation but which have not been investigated previously in the context of myeloid development. Data are available online athttp://bioinfo.mbb.yale.edu/expression/myelopoiesis.
Introduction
The study of myeloid differentiation provides important insights both into normal developmental processes that generate peripheral blood leukocytes, as well as into abnormalities that lead to myeloid aplasia, dysplasia, and leukemia.1-9Access to normal myeloid precursors at homogenous stages of development and in quantities sufficient for biochemical analysis is not generally practicable so information about myeloid differentiation has generally been obtained by studies of leukemic cells arrested at various developmental stages.10 Informative results have also come from studies of humans with genetic abnormalities affecting neutrophil accumulation11-13 and gene targeting experiments, particularly of transcription factors.14 Overall, cell lines that can be induced to undergo myeloid differentiation in vitro continue to provide many of the most useful models for understanding of this process.15
Human and murine hematopoietic precursor lines have been developed that can be induced to mature to various degrees toward adult neutrophils.8,16 Several of these lines fail to form a full complement of proteins or to fully undergo morphologic changes characteristic of mature neutrophils, but the murine MPRO cell line provides a relatively favorable system for studying myeloid differentiation.8 The cells are arrested at the promyelocytic stage because of the presence of a dominant-negative retinoic acid receptor. Differentiation can be induced by adding appropriate concentrations of all-trans retinoic acid (ATRA). On differentiation, most cells mature to the level of band forms and mature polymorphonuclear neutrophils and express secondary granule mRNAs and proteins.8
Current methods that provide broad surveys of the patterns of mRNA expression include oligonucleotide chip hybridization17and 3′ end restriction fragment gel display analysis18; both have been used to study MPRO cell development. Although the chemical heterogeneity of proteins prevents similar global methods of protein abundance analysis, recent improvements in 2-dimensional gel electrophoresis, especially the development of immobilized pH gradient isoelectric focusing gels, have made it possible to semiquantitatively examine the levels of a substantial fraction of the proteins of a cell.19 This approach, termed proteome analysis, has provided important contributions to disease-related gene discovery, developmental program analysis, and drug discovery. Interest in this area has been spurred by recent studies indicating a modest to poor correlation between transcriptional profiles and actual protein levels in cells. These studies make it clear that cellular protein analysis is complementary to genomic analysis and that no biologic program can be successfully analyzed without the incorporation of a proteomics platform.
Previously, we used oligonucleotide chips and gel displays to study the patterns of mRNA expression during MPRO cell differentiation and compared these with a very limited set of protein analyses from wide pH range 2-dimensional gel electrophoresis.20 We have expanded these studies to a more global analysis of a much wider array of mRNA and protein species. The current studies use higher resolution narrow-range 2-dimensional gel systems and tandem mass spectrometry to identify a substantial portion of the more abundant proteins whose levels change during MPRO development. Bioinformatic and functional tools were then used to analyze the role of these proteins in myeloid differentiation. We have also used a new generation of oligonucleotide chips to compare mRNA levels in MPRO cells 0 hours and 72 hours after induction of differentiation. In particular, we have further examined the expression of transcription factor mRNAs in MPRO cells and compared this pattern with transcription factor expression in mature human neutrophils.
Materials and methods
Cell line growth and induction
Two-dimensional immobilized pH gradient gel electrophoresis
MPRO cells were disrupted in lysis buffer.20 We applied 50 to 100 μL of each MPRO cell lysate (1.25 × 106cells/100-2.5 × 106cells/100 μL, about 100-200 μg protein) at the cathodic end of the immobilized pH gradient gel (IPG) strips (pH 3-10 L, pH 4-7 and pH 6-11, Pharmacia Biotech, Uppsala, Sweden), and 2-dimensional IPG electrophoresis (2D-IPG) was conducted for 10 to16 hours (13 000 to 20 100 V-h) using Electrophoresis Power Supply ESP 3500 XL and Immobiline DryStrip Kit (Pharmacia Biotech). The electrophoresis in the second dimension was carried out in a 12% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) gel with the Laemmli-SDS-continuous system in a PROTEAN (II xi 2-D cell, Bio-Rad, Hercules, CA), run at 40 mA constant current for 5 hours.21 22
The 2-dimensional gels were stained with Coomassie brilliant blue G-colloidal following the vendor's recommendations.23Destaining was performed by soaking the gels in 10% acetic acid and 25% methanol solution for 60 seconds, then in 25% methanol solution for 24 hours at room temperature. Silver staining was performed according to the protocol of the manufacturer.24 25
The 2-dimensional maps of MPRO cells were compared by using the Adobe Photoshop 4.0 program Melanie III 2-D PAGE software (Genebio, Geneva, Switzerland) and checked manually. Proteins were recovered by punching out spots with a MultiFit Research Pipet Tips (Volume: 100-1000 μL; Dot Scientific, Burton, MI). More than 200 visible protein spots were punched for later mass spectrometry analysis.
Mass spectrometry analysis
The punched samples were washed at room temperature in the following solutions: in 50% acetonitrile for 5 minutes; in 50% CH3CN/50 Mm NH4HCO3 for 30 minutes; then in 50% CH3CN/10 Mm NH4HCO3for 30 minutes. After drying the sample gels in a SpeedVac Concentrator (Eppendorf, Hamburg, Germany), trypsin solution (0.05 μg trypsin/7 μL 10 Mm NH4HCO3) was added to the samples and they were incubated at 37°C for 24 hours. The supernatants of the trypsin digestion products were collected, 1 μL sample digest was mixed with 1.0 μL α-cyano-4-hydroxy cinnamic acid (CHCA; 4.5 mg/mL in 50% CH3CN, 0.05% trifluoroacetic acid [TFA]) matrix solution, and 1 μL calibrants (100 fmol each). The mixture was loaded on a target of the sample plate, then injected to the Perseptive Biosystem Voyager-DE STR instrument (Perseptive Biosystem, Boston, MA). The spectra of the peptides were acquired in reflector/delayed extraction mode. The standards used for calibration of peptide masses are bradykinin (average (M+H) is 1061.23) and ACTH Clip (average (M+H) is 2466.70). The criteria we are using currently to identify proteins are: (1) “Coverage,” ratio of the portion of protein sequence covered by matched peptides to the whole length of protein sequence, is 25% or more; (2) “Z score” is more than 1.5; (3) “Probability” is “1.0e + 000”; (4) “Coverage graphical” of the matched peptides from the protein candidate crosses the all length of the protein.
Peptide identification and database establishment
Peptides were identified using the ProFound-Peptide Mapping search engine (http://www.proteometrics.com/profound_bin/WebProFound.exe), and subsequently searched against the SWISS-PROT (http://www.expasy.ch/) or PIR (http://www-nbrf.georgetown.edu/) sites. The differential patterns of protein expression were analyzed with Melanie II 2-D Page Software (Bio-Rad) (http://www.expasy.ch/melanie/MelanieII/description.html).
The 2-dimensional reference maps and the identified protein information were collected in a database (dbMCp) that contained information for each protein including: GenBank matches, Locus Link or UniGene clusters, expression patterns, tissue distribution, synonym(s) protein name, gene name(s), notations of possible functions in myeloid cell biology and differentiation, and hyperlinks to the database searches, 2-dimensional images, and related references. These data were gathered as separate entries in a file. Supplementary information is available on our website (http://bioinfo.mbb.yale.edu/expression/blood).
The proteins identified from different sets of 2-dimensional gels were grouped into 12 categories according to their functions as documented in SWISS-PROT and National Center for Biotechnology Information (NCBI) databases. Furthermore, these proteins were classified into 5 expression patterns by their similarity to the ideal expression patterns.20 The correlations at various levels of proteins or RNA were compared using both visual estimates and Melanie software estimates of protein spot intensities and the average difference between match and mismatch probe sets for each gene on the oligonucleotide chips.
Protein synthesis inhibition by cycloheximide treatment
A pilot dose-response experiment determined the dose that produced 95% inhibition of MPRO cell protein synthesis, assayed by incorporation of radiolabeledl-[35S]-methionine. Based on dose-response experiment, MPRO cells (2 × 105 cells/mL) were treated with or without cycloheximide (final concentration 10 μL/mL) for 2 hours, then collected and sampled for proteomic analysis as described above.
mRNA isolation and analysis
The mRNA was isolated from MPRO cells at indicated time points during differentiation as previously described.20Oligonucleotide chip analysis was also performed as previously described,20 except for the use of the more advanced Affymetrix chip probes (Murine Genome U74Av2 array), interrogating approximately 36 000 full-length mouse genes and expressed sequence tag (EST) clusters from the UniGene database. The resulting data were compared with human neutrophil gene expression analysis using the Affymetrix U60 set of oligonucleotide chips. Human neutrophils were prepared according to the method described previously.18Criteria for considering cDNAs “present” and for selecting those with significant average differences, as well as rescaling, threshold, and normalization methods were applied as previously described.20
To study mRNA expression we first tested the incorporation of results from previous work20 using Affymetrix 11K chips along with the present work set of measurements using the newer generation Affymetrix murine genome U74Av2 array. Comparison of the differences in expression at times 0 and 72 hours between the 2 different chips requires preprocessing of the data, because the probe sets corresponding to any given gene in the old and new chips are different. Genes were identified by their Locus Link ID, by extracting the ID for each accession number in both the 11K and 36K chips using the Stanford Source database (http://genome-www5.stanford.edu/cgi-bin/SMD/source/sourceSearch). We filtered out probe sets that had missing values of expression levels at either 0 or 72 hours. The remaining probe sets of the 11K chip were linked with the remaining probe sets of the 36K chip through a common Locus Link ID. Most of the remaining distinct 1906 Locus Link IDs had a single probe set per Locus Link ID, both in the old and new chips. However, 63 probe sets from the 11K chip were linked with more than one probe set on the 36K chip and 400 probe sets from the 36K chip were linked with more than one probe set of the 11K chip. We chose not to average the RNA levels of probe sets that belong to the same gene, because it would not be appropriate when the expression levels of one probe set dominate the others. Therefore, we evaluated the correlation between mRNA from the 11K chip and the 36K chip using only the subset of genes that had single probe sets on both chips. Using this subset we found that the correlation between mRNA levels of the 11K chip and the 36K chip is 0.75 at 0 hours and 0.7 at 72 hours. These correlations were lower than the correlations between the mRNA levels at 0 and 72 hours using only the 11K chip (r = 0.89) or only the 36K chip (r = 0.84). Therefore, changes in RNA levels were not entirely reproducible using these 2 completely different chips. We compared the time course trends of 10 genes previously studied using Northern blots with the corresponding trends of the 11K and 36K chips. The trends of the 11K chip agreed with the Northern blots only in 6 of 10 instances, whereas the new 36K chip success rate was 9 of 10. The mRNA for several of the proteins we detected from 2-dimensional gels was reported as present on the new chip set but absent from the old chip set. We therefore used only data form the new chip set for comparisons with proteins and for further examination of changes in transcription factors. The use of only one replica of the new 36K chip, although not ideal, should be sufficient for exploring global relations between protein and mRNA.
Northern blot analysis was performed as described previously.20
Results
Proteomic analysis of MPRO differentiation
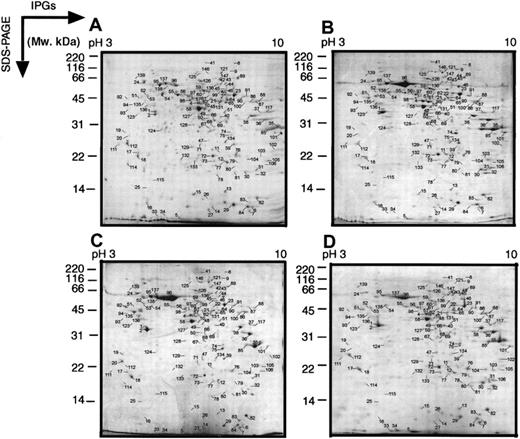
The MPRO cell model is particularly useful for studying aspects of myeloid differentiation because large numbers of cells can be obtained, arrested at the promyelocyte stage of development, and, most importantly, synchronous differentiation can be induced by adding ATRA. The fully differentiated cells resemble mature neutrophils both morphologically and in the expression of secondary granule proteins. For the purpose of initially scanning changes in protein levels during myeloid differentiation, we used 2D-IPG with wide-range, linear IPGs (pH 3-10) in the first dimension. Figure1 shows analytical colloidal blue-stained 2D-IPG standard maps of differentiated MPRO cells at 0, 24, 48, and 72 hours after the cells were induced with ATRA. The expression patterns of more than 300 protein spots were followed through the entire series of gels. The protein spots in different gels could easily be cross-matched to each other, using Melanie III software, indicating the reproducibility of the method. A large portion of these products changed their relative intensities among the 4 maps, suggesting extensive protein expression changes during the course of MRPO differentiation.
Two-dimensional electrophoretograms of wide pH range of MPRO cells.
MPRO cells differentiate to mature neutrophils in the presence of ATRA. Following exposure to 10 μM ATRA for 0, 24, 48, or 72 hours, MPRO cell lysate (2.5 × 106 cells/sample) was loaded for 2-dimensional electrophoretic (2DE) analysis. The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) MPRO cells induced with ATRA for 24 hours; (C) MPRO cells induced with ATRA for 48 hours; (D) matured MPRO cells induced with ATRA for 72 hours. The most visible protein spots in the maps were subjected to MS analysis. The marked 2 DE maps could be found in our website (http://bioinfo.mbb.yale.edu/expression/myelopoiesis). *2 DE maps of panels A and D were published in our previous paper.20
Two-dimensional electrophoretograms of wide pH range of MPRO cells.
MPRO cells differentiate to mature neutrophils in the presence of ATRA. Following exposure to 10 μM ATRA for 0, 24, 48, or 72 hours, MPRO cell lysate (2.5 × 106 cells/sample) was loaded for 2-dimensional electrophoretic (2DE) analysis. The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) MPRO cells induced with ATRA for 24 hours; (C) MPRO cells induced with ATRA for 48 hours; (D) matured MPRO cells induced with ATRA for 72 hours. The most visible protein spots in the maps were subjected to MS analysis. The marked 2 DE maps could be found in our website (http://bioinfo.mbb.yale.edu/expression/myelopoiesis). *2 DE maps of panels A and D were published in our previous paper.20
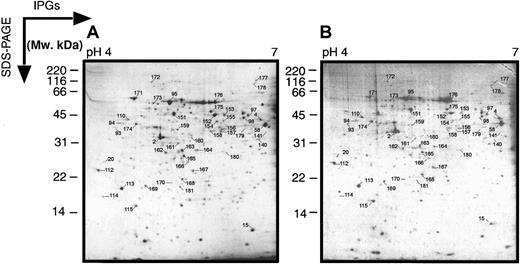
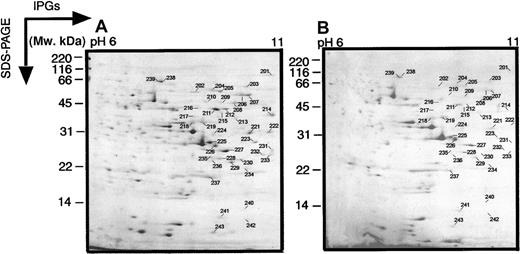
In these wide-range 2-dimensional maps, there is a loss of resolution in the region pH 4 to 7, most probably due to the fact that the pI values of many proteins occur in this range. Therefore, we also performed electrophoresis on pH 4 to 7 and pH 6 to 11 narrow-range IPGs to get better protein separation (Figures2 and 3). These narrower pH gels allowed a higher resolution and more protein spots in the relative pH zones. The abundant protein spots could also be cross-correlated between the wide and narrow gels.
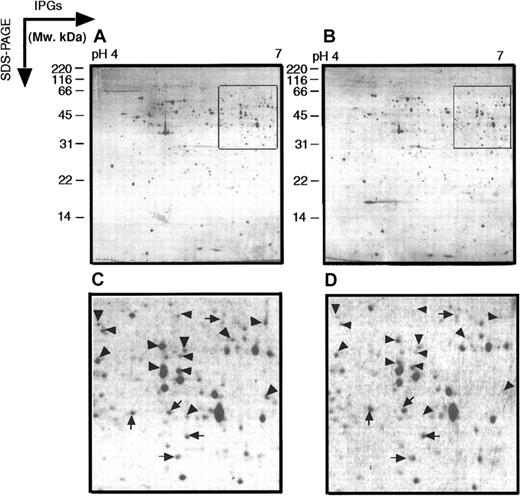
Two-dimensional electrophoretograms of MPRO cells in pH range 4 to 7.
MPRO cell lysate (1.5 × 106 cells/sample) was loaded for 2DE analysis (pH 4-7). The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) matured MPRO cells induced with ATRA for 72 hours. The other information is presented as in the legend to Figure 1.
Two-dimensional electrophoretograms of MPRO cells in pH range 4 to 7.
MPRO cell lysate (1.5 × 106 cells/sample) was loaded for 2DE analysis (pH 4-7). The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) matured MPRO cells induced with ATRA for 72 hours. The other information is presented as in the legend to Figure 1.
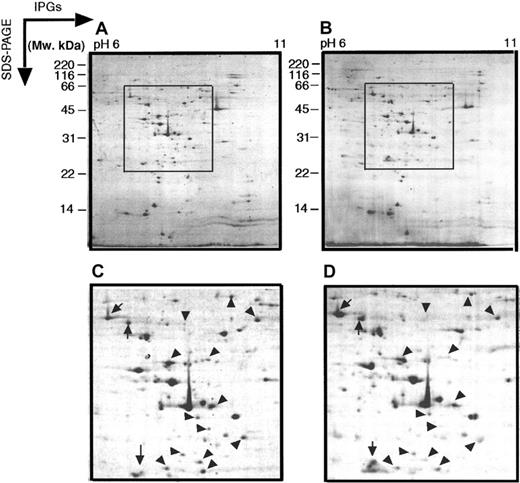
Two-dimensional electrophoretograms of MPRO cells in pH range 6 to 11.
MPRO cell lysate (1.5 × 106 cells/sample) was loaded for basic pH 2DE analysis (pH 6-11). The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) matured MPRO cells induced with ATRA for 72 hours. The other information is presented as in the legend to Figure 1.
Two-dimensional electrophoretograms of MPRO cells in pH range 6 to 11.
MPRO cell lysate (1.5 × 106 cells/sample) was loaded for basic pH 2DE analysis (pH 6-11). The gels were stained with brilliant blue G-colloidal dye. (A) Uninduced MPRO cell (0 hour); (B) matured MPRO cells induced with ATRA for 72 hours. The other information is presented as in the legend to Figure 1.
Protein identification
The protein spots in the different sets of the gels were identified by MALDI-MS on the basis of peptide mass matching with the theoretical peptide masses in tryptic digests of all known proteins from mouse and human species.26 Of 220 protein spots analyzed, 193 yielded high-quality spectral data. The experimental peptide masses were matched to a total of 143 spots corresponding to 123 different known proteins, as presented in Table1. The accession numbers, protein names, and theoretical pI and Mr values, as well as the number of peptide matches and probability of wrong assignment, are presented in the database dbMCp (http://bioinfo.mbb.yale.edu/expression/myelopoiesis). There were 29 spots with high-quality spectra but poor matches in public databases; another 21 spots with good mass spectra matched many different proteins in the mouse database. The latter finding was probably attributable to high sequence homology, but can also be the result of a mixture of proteins in a single spot.
Distribution of protein spots identified during myeloid differentiation
| Gel resource . | Spots analyzed . | Distinct proteins identified . | Multiple spots/protein (distinct proteins) . |
|---|---|---|---|
| Known proteins | |||
| pH 3–10 | 95 | 80 | 29 (14) |
| pH 4–7 or 6–11 | 48 | 43 | 9 (4) |
| Sum | 143 | 123 | 38 (18) |
| No hit | 29 | ||
| Multiple hits | 21 | ||
| Total | 193 |
| Gel resource . | Spots analyzed . | Distinct proteins identified . | Multiple spots/protein (distinct proteins) . |
|---|---|---|---|
| Known proteins | |||
| pH 3–10 | 95 | 80 | 29 (14) |
| pH 4–7 or 6–11 | 48 | 43 | 9 (4) |
| Sum | 143 | 123 | 38 (18) |
| No hit | 29 | ||
| Multiple hits | 21 | ||
| Total | 193 |
On the pH 3 to 10 maps, 14 protein species were represented by multiple spots (Table 2) that differed due to the pI or Mr. These differences might be the result of alternative splicing or posttranslational modifications, or of chemical modification by protease inhibitors during sample preparation. Interestingly, some of these proteins showed the same phenomena in Jurkat T-cell 2-dimensional protein maps.27Some spots with high-quality spectra, but shifted from their expected position in the gel, might also represent posttranslational modifications. Proteins with lower than expected molecular weights may be digestion fragments of larger proteins. Most proteins with low molecular weight (< 14 kDa) usually presenting multiply matches, could not be identified.
Protein species represented by multiple spots
| Symbol . | Accession . | Gi#* . | Protein ID . | Theoretical value . | % . | Practical value . | ||
|---|---|---|---|---|---|---|---|---|
| kDa . | pl . | kDa . | pl . | |||||
| Aldh2 | NP_033786 | 6753036 | MPRO-004 | 56.52 | 7.7 | 23 | 31∼50 | 6.4∼6.6 |
| MPRO-006 | 56.52 | 7.7 | 20 | 6∼14 | 7.3∼7.6 | |||
| Atp5a1 | NP_031531 | 6680748 | MPRO-087 | 59.73 | 9.3 | 24 | 45∼55 | 7.6∼7.8 |
| MPRO-088 | 59.73 | 9.3 | 24 | 45∼55 | 7.8∼8.0 | |||
| Ddx5 | NP_031866 | 6681157 | MPRO-206 | 69.3 | 9.3 | 22 | 45∼66 | 9.1∼9.3 |
| MPRO-207 | 69.3 | 9.3 | 26 | 45∼55 | 9.1∼9.4 | |||
| Gapd | NP_032110 | 6679937 | MPRO-035 | 35.79 | 8.7 | 39 | 25∼35 | 8.0∼8.2 |
| MPRO-085 | 35.79 | 8.7 | 34 | 28∼38 | 7.7∼7.9 | |||
| Hnrpa2b1 | NP_058086 | 7949053 | MPRO-223 | 35.98 | 8.7 | 55 | 25∼31 | 9.2∼9.3 |
| MPRO-227 | 35.98 | 8.7 | 55 | 21∼31 | 9.2∼9.3 | |||
| MPRO-229 | 35.98 | 8.7 | 55 | 21∼33 | 9.1∼9.2 | |||
| Hnrph1 | NP_067485 | 10946928 | MPRO-155 | 49.18 | 5.9 | 26 | 45∼66 | 5.9∼6.0 |
| MPRO-154 | 49.18 | 5.9 | 40 | 45∼66 | 5.8∼5.9 | |||
| Hmg2 | NP_032278 | 11527222 | MPRO-076 | 24.16 | 6.9 | 26 | 18∼28 | 7.2∼7.4 |
| 6680229 | MPRO-104 | 14.16 | 6.9 | 26 | 14∼21 | 7.6∼7.8 | ||
| Pk3 | NP_035229 | 6755074 | MPRO-023 | 57.9 | 7.2 | 48 | 45∼66 | 7.2∼7.4 |
| 2506796 | MPRO-008 | 57.87 | 7.2 | 42 | 150∼200 | 7.0∼7.5 | ||
| Rbm3 | NP_058089 | 7949121 | MPRO-014 | 16.59 | 6.8 | 25 | 7∼14 | 6.6∼6.8 |
| MPRO-015 | 16.59 | 6.8 | 25 | 12∼16 | 6.2∼6.4 | |||
| STEFIN 3 | P35175 | 461911 | MPRO-005 | 10.99 | 5.9 | 48 | 1∼6.5 | 6.2∼6.4 |
| MPRO-033 | 10.99 | 5.9 | 53 | 1∼6.5 | 5.8∼6.0 | |||
| Tpi | NP_033441 | 6678413 | MPRO-012 | 26.69 | 6.9 | 26 | 15∼25 | 6.9∼7.1 |
| MPRO-073 | 26.69 | 6.9 | 40 | 18∼28 | 6.7∼6.9 | |||
| Tpm5 | P21107 | 136097 | MPRO-083 | 29 | 4.7 | 46 | 6.5∼14 | 7.5∼7.7 |
| MPRO-112 | 29 | 4.7 | 27 | 21∼31 | 4.6∼4.8 | |||
| Vim | 2078001 | MPRO-093 | 51.55 | 4.9 | 25 | 31∼45 | 4.7∼4.9 | |
| MPRO-110 | 53.67 | 5.1 | 28 | 40∼50 | 4.9∼5.0 | |||
| Vdac1 | Q60932 | 10720404 | MPRO-228 | 32.33 | 8.7 | 49 | 21∼33 | 8.8∼9.0 |
| MPRO-235 | 32.33 | 8.7 | 35 | 21∼31 | 8.7∼8.9 | |||
| Symbol . | Accession . | Gi#* . | Protein ID . | Theoretical value . | % . | Practical value . | ||
|---|---|---|---|---|---|---|---|---|
| kDa . | pl . | kDa . | pl . | |||||
| Aldh2 | NP_033786 | 6753036 | MPRO-004 | 56.52 | 7.7 | 23 | 31∼50 | 6.4∼6.6 |
| MPRO-006 | 56.52 | 7.7 | 20 | 6∼14 | 7.3∼7.6 | |||
| Atp5a1 | NP_031531 | 6680748 | MPRO-087 | 59.73 | 9.3 | 24 | 45∼55 | 7.6∼7.8 |
| MPRO-088 | 59.73 | 9.3 | 24 | 45∼55 | 7.8∼8.0 | |||
| Ddx5 | NP_031866 | 6681157 | MPRO-206 | 69.3 | 9.3 | 22 | 45∼66 | 9.1∼9.3 |
| MPRO-207 | 69.3 | 9.3 | 26 | 45∼55 | 9.1∼9.4 | |||
| Gapd | NP_032110 | 6679937 | MPRO-035 | 35.79 | 8.7 | 39 | 25∼35 | 8.0∼8.2 |
| MPRO-085 | 35.79 | 8.7 | 34 | 28∼38 | 7.7∼7.9 | |||
| Hnrpa2b1 | NP_058086 | 7949053 | MPRO-223 | 35.98 | 8.7 | 55 | 25∼31 | 9.2∼9.3 |
| MPRO-227 | 35.98 | 8.7 | 55 | 21∼31 | 9.2∼9.3 | |||
| MPRO-229 | 35.98 | 8.7 | 55 | 21∼33 | 9.1∼9.2 | |||
| Hnrph1 | NP_067485 | 10946928 | MPRO-155 | 49.18 | 5.9 | 26 | 45∼66 | 5.9∼6.0 |
| MPRO-154 | 49.18 | 5.9 | 40 | 45∼66 | 5.8∼5.9 | |||
| Hmg2 | NP_032278 | 11527222 | MPRO-076 | 24.16 | 6.9 | 26 | 18∼28 | 7.2∼7.4 |
| 6680229 | MPRO-104 | 14.16 | 6.9 | 26 | 14∼21 | 7.6∼7.8 | ||
| Pk3 | NP_035229 | 6755074 | MPRO-023 | 57.9 | 7.2 | 48 | 45∼66 | 7.2∼7.4 |
| 2506796 | MPRO-008 | 57.87 | 7.2 | 42 | 150∼200 | 7.0∼7.5 | ||
| Rbm3 | NP_058089 | 7949121 | MPRO-014 | 16.59 | 6.8 | 25 | 7∼14 | 6.6∼6.8 |
| MPRO-015 | 16.59 | 6.8 | 25 | 12∼16 | 6.2∼6.4 | |||
| STEFIN 3 | P35175 | 461911 | MPRO-005 | 10.99 | 5.9 | 48 | 1∼6.5 | 6.2∼6.4 |
| MPRO-033 | 10.99 | 5.9 | 53 | 1∼6.5 | 5.8∼6.0 | |||
| Tpi | NP_033441 | 6678413 | MPRO-012 | 26.69 | 6.9 | 26 | 15∼25 | 6.9∼7.1 |
| MPRO-073 | 26.69 | 6.9 | 40 | 18∼28 | 6.7∼6.9 | |||
| Tpm5 | P21107 | 136097 | MPRO-083 | 29 | 4.7 | 46 | 6.5∼14 | 7.5∼7.7 |
| MPRO-112 | 29 | 4.7 | 27 | 21∼31 | 4.6∼4.8 | |||
| Vim | 2078001 | MPRO-093 | 51.55 | 4.9 | 25 | 31∼45 | 4.7∼4.9 | |
| MPRO-110 | 53.67 | 5.1 | 28 | 40∼50 | 4.9∼5.0 | |||
| Vdac1 | Q60932 | 10720404 | MPRO-228 | 32.33 | 8.7 | 49 | 21∼33 | 8.8∼9.0 |
| MPRO-235 | 32.33 | 8.7 | 35 | 21∼31 | 8.7∼8.9 | |||
Protein symbol, accession, and Gi# refer to NCBI UniGene database (if represented). Theoretical value refers from ProFound website (http://prowl.rockefeller.edu/cgi-bin/ProFound). Practical value is the observed value in 2 DE gels (see “”).
Protein expression patterns during MPRO development
The 123 “known” proteins identified here were classified into 12 categories on the basis of their function, including 18% categorized as cytoskeletal proteins, 15% metabolism-related molecules, and 10% signaling pathway-related proteins (Table3). These proteins were abundant in the cell and easily detected by 2-dimensional electrophoresis. Smaller sets of proteins included 7 possible transcription factors and 5 cytokines; other categories, such as kinases and chromatin remodeling factors, contain even fewer members.
Classification of known proteins
| Category . | Protein (gene) symbol . |
|---|---|
| Cytoskeleton | Actb, Actg, Anxa1, Anxa11, Anxa2, Anxa3, Arpc3, Cappa1, Coro1a, ECP, KER1, KER8, KER10, KER47, KER59, Krt2-6g, KT14, SAC, Tpm5, Tuba6, Tubb5, vim |
| Energy metabolism | Eno1, Gapd, Idh1, Idh2, Impdh2, Ldh1, Papss2/Atpsk2, Pygm, Taldo1, Tpi |
| Signaling pathway | Arhgdib, Arhn, Ephb2, G4-1-pending, Gnb2-rs1, Hcph, Nme1, Pgk1, Pk3, Ptpn1, Rac2, Ran, Rin, Vav2 |
| Cytokine | Hgf, IFI-205, IIIf5, Pbp, Spry1 |
| Transcription modulators | Hmgb1, Hmg2, KRZF80M, Rnf17, Stat5a, Taf2e, Zfp101, ZFP1A3, Zfp354a |
| Chaperone | Cab140, Cct2, Cct5, Cct6a, GROEL, Grp58, Hsc70, Hsp110, Hspa5/Grp78, Hspa8, P4hb, Ppia, Stip1 |
| Granule-related protein | Cas1, Es10, Psmc1, Psma7, Psmc2, Sod1, STEFIN3 |
| Mitochondrial | Got2, Aldh2, Atp5a1, Atp5b, Mor1 |
| RNA metabolism | Hnrpa1, Hnrpa2b1, Hnrph1, Nsap1-pending, Rbm3, RNPC |
| Transporter | Slc23a2, Vdac1 |
| Chromatin | Lmnb1, Pcna |
| Other categories | Abpa, Cftr, Crmp1, C4, Ddx5, Eef2, Eef1a1, Ehd1, Fut4, Gc, Gstm1, HPD76, IGVAP, Ltf, Tinag, Ube1x, H2-Ab1, Phb, Prdx1, Prdx2, Pdi4, Rag1, LOC56463, PRO2675, Tagln2, AA589396, Lgals3, Sfmbt |
| Category . | Protein (gene) symbol . |
|---|---|
| Cytoskeleton | Actb, Actg, Anxa1, Anxa11, Anxa2, Anxa3, Arpc3, Cappa1, Coro1a, ECP, KER1, KER8, KER10, KER47, KER59, Krt2-6g, KT14, SAC, Tpm5, Tuba6, Tubb5, vim |
| Energy metabolism | Eno1, Gapd, Idh1, Idh2, Impdh2, Ldh1, Papss2/Atpsk2, Pygm, Taldo1, Tpi |
| Signaling pathway | Arhgdib, Arhn, Ephb2, G4-1-pending, Gnb2-rs1, Hcph, Nme1, Pgk1, Pk3, Ptpn1, Rac2, Ran, Rin, Vav2 |
| Cytokine | Hgf, IFI-205, IIIf5, Pbp, Spry1 |
| Transcription modulators | Hmgb1, Hmg2, KRZF80M, Rnf17, Stat5a, Taf2e, Zfp101, ZFP1A3, Zfp354a |
| Chaperone | Cab140, Cct2, Cct5, Cct6a, GROEL, Grp58, Hsc70, Hsp110, Hspa5/Grp78, Hspa8, P4hb, Ppia, Stip1 |
| Granule-related protein | Cas1, Es10, Psmc1, Psma7, Psmc2, Sod1, STEFIN3 |
| Mitochondrial | Got2, Aldh2, Atp5a1, Atp5b, Mor1 |
| RNA metabolism | Hnrpa1, Hnrpa2b1, Hnrph1, Nsap1-pending, Rbm3, RNPC |
| Transporter | Slc23a2, Vdac1 |
| Chromatin | Lmnb1, Pcna |
| Other categories | Abpa, Cftr, Crmp1, C4, Ddx5, Eef2, Eef1a1, Ehd1, Fut4, Gc, Gstm1, HPD76, IGVAP, Ltf, Tinag, Ube1x, H2-Ab1, Phb, Prdx1, Prdx2, Pdi4, Rag1, LOC56463, PRO2675, Tagln2, AA589396, Lgals3, Sfmbt |
Protein symbols refer to NCBI databases (see “”).
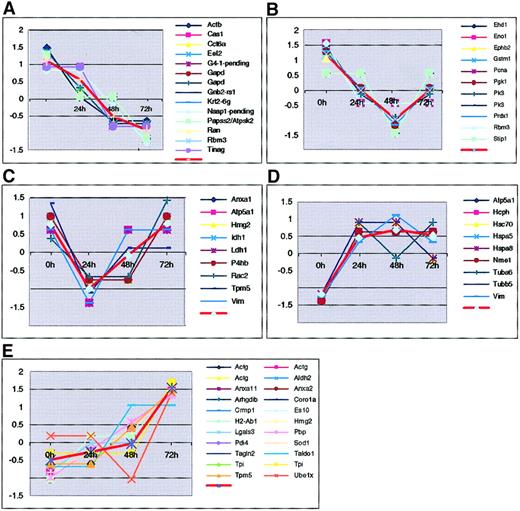
We also classified all known proteins according to their expression patterns during myeloid differentiation. We clustered the standardize protein expression level profiles (at 0, 24, 48 and 72 hours) using thegenecluster version of the self-organizing maps (SOMs) clustering algorithm,28 with a rectangular 3 × 2 grid as the input node geometry. The final position of the nodes in the 4-dimensional (time course) space, represents the centers of 6 clusters generated by the SOM algorithm. One of these clusters was empty. Figure4 shows the normalized expression profiles divided into the remaining 5 clusters representing trends such as down-regulation (Figure 4A) and up-regulation (Figure 4D,E) that occur during the cell maturation process. For example, the universal transcription factor Eef2 is down-regulated. This finding is consistent with the concurrent reduction of total RNA levels and cell size. Conversely, protein Es10 shows a pattern of up-regulation, as expected for a granule component. Thus, these profiles offer information about the roles of proteins in the different stages of the MPRO development.
Protein clusters according to their expression patterns.
The 72 protein spots were grouped into 6 clusters (1 empty cluster is not shown). Each cluster is represented by the centroid (average pattern represented by a thick red line) for genes in the cluster. Expression level of each gene was standardized to have zero mean and unit SD across the 4 time points. Standardized expression levels are shown on y-axis and time points on x-axis.
Protein clusters according to their expression patterns.
The 72 protein spots were grouped into 6 clusters (1 empty cluster is not shown). Each cluster is represented by the centroid (average pattern represented by a thick red line) for genes in the cluster. Expression level of each gene was standardized to have zero mean and unit SD across the 4 time points. Standardized expression levels are shown on y-axis and time points on x-axis.
Correlation of gene expression at the RNA and protein levels
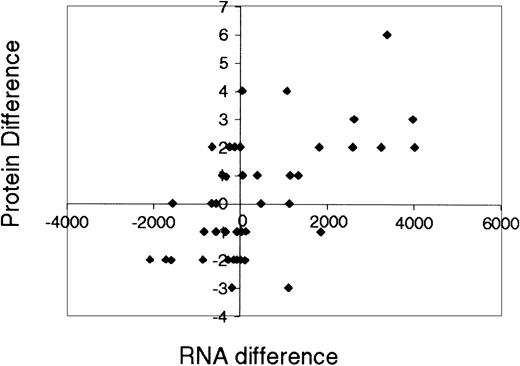
One of the goals of this work is to search for global relationships between mRNA and protein levels during MRPO cell maturation. Previous studies in yeast showed weak correlations between average mRNA levels and average protein levels.29-32 These studies focused on the relationship, at one instant, between absolute amounts of mRNA (measured from Affymetrix GeneChip experiments) and protein. Here we investigate another quantity: the correlation between changes over many time points, in mRNA levels and in protein levels. This is only possible because we have available experiments simultaneously done on protein and mRNA levels over an entire time course. In particular, we analyze the relationships between time course expression profiles of mRNA and proteins during a process of mammalian cellular development. This is the first time that the relationship between protein abundance and mRNA expression has been studied in terms of changes over time. To study mRNA expression, we used measurements taken at times 0 and 72 hours of the maturation process, using the Affymetrix 36K murine chip. To compare the mRNA changes with protein changes we first summed levels of proteins that are represented in more than one spot on the 2-dimensional gels. We retained only mRNAs with an Affymetrix oligonucleotide probe set with the suffix “_at” (representing a probe set corresponding to a single gene). This procedure removes the ambiguity of multiple probe sets per Locus Link. We then screened mRNA that had a “present” Affymetrix indicator and an amplitude more than 20 at 0 and 72 hours and found 51 different proteins that satisfied these conditions. The correlation between the mRNA difference at 0 and 72 hours with the corresponding protein difference is r = 0.58, as presented in Figure5 (the exact formula for the Pearson correlation coefficient r is given in the legend to Figure 5). Most proteins with increasing levels of mRNA also have increasing protein levels, with the exception of 2 outliers (enolase 1 and coronin). Overall, 11 of 51 proteins with upward/downward trends had an opposite mRNA trend.
The correlation between the mRNA difference at 0 and 72 hours and the corresponding protein difference.
Correlation between RNA expression level differences, ΔR ≡ RNA(t = 72) − RNA(t = 0), and protein level differences ΔP ≡ P(t = 72) − P(t = 0). Expression levels of proteins that have more than one conformation were summed. In this regression analysis we retained only RNA probe sets that correspond to single genes (the remaining probe sets lacked the ambiguity of multiple probe sets per Locus Link) and that had a “present” Affymetrix indicator and an amplitude more than 20 both at t = 0 and t = 72 hours. There were 51 different proteins that satisfy these conditions. The linear association r between changes in RNA levels (ΔR) and changes in protein levels (ΔP) of the remaining 51 genes is only 0.58, where r is the Pearson correlation coefficient defined as r(ΔP,ΔR) = ∑i (ΔPi −(ΔRi−/However, about 80% of the genes are located in the first and third quadrants, indicating a general trend that genes with increasing/decreasing levels of RNA also have increasing/decreasing protein levels.
The correlation between the mRNA difference at 0 and 72 hours and the corresponding protein difference.
Correlation between RNA expression level differences, ΔR ≡ RNA(t = 72) − RNA(t = 0), and protein level differences ΔP ≡ P(t = 72) − P(t = 0). Expression levels of proteins that have more than one conformation were summed. In this regression analysis we retained only RNA probe sets that correspond to single genes (the remaining probe sets lacked the ambiguity of multiple probe sets per Locus Link) and that had a “present” Affymetrix indicator and an amplitude more than 20 both at t = 0 and t = 72 hours. There were 51 different proteins that satisfy these conditions. The linear association r between changes in RNA levels (ΔR) and changes in protein levels (ΔP) of the remaining 51 genes is only 0.58, where r is the Pearson correlation coefficient defined as r(ΔP,ΔR) = ∑i (ΔPi −(ΔRi−/However, about 80% of the genes are located in the first and third quadrants, indicating a general trend that genes with increasing/decreasing levels of RNA also have increasing/decreasing protein levels.
The reproducibility of the protein results was studied by repeating the induction experiments of MPRO cells and also by repeated analyses of the same cell samples. The induction experiments were repeated 3 times. In each experiment, the MPRO cells from different time courses were analyzed by 2D-IPG 2 to 6 times. We found that the protein spot images were well reproduced, with only slight differences occurring at the far edges of gels. Quantitative analysis of 4 dilutions of the same samples showed that the intensity change of each protein was proportional. In comparisons of 2D-IPG of 0-hour and 72-hour cells between 2 different induction experiments, we found that among the 220 analyzed proteins, 199 (90%) were reproducibly observed, and 21 were not observed in all gel sets. The direction of expression changes of proteins in 72 hours against 0 hours was similar in both experiments, with a correlation coefficient of 0.88.
We measured protein abundance using both software and manual estimations of spot intensity. Using the Melanie III program from Genebio, we were able to compute the protein abundance of thousands of proteins across the gels and found a general consistency between measurements by eye and by software analysis (data not shown).
We did not expect to find a general correlation for the changes in levels of these proteins and their mRNAs; rather, as previously hypothesized,29 32 we sought correlations between smaller, better defined groups of proteins. Although the correlation over all proteins and mRNA hovered around 0.3 for each of the time points, we found that the median correlation for cytoskeletal proteins alone rose to approximately 0.65, highlighting the importance of analyzing mRNA expression and protein abundance using well-defined features and functions.
Protein stability
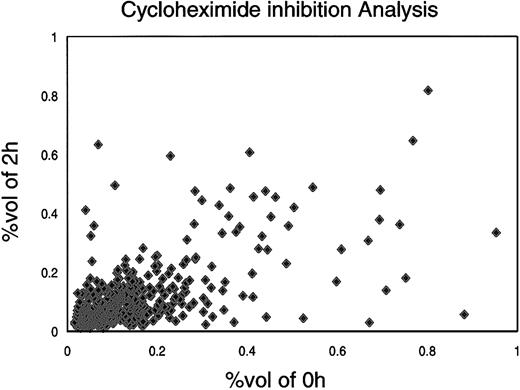
The level of any protein is theoretically determined by its cumulative rate of synthesis and by the rates of degradation or alteration (and an initial condition of protein level). For protein stability studies, MPRO cells (1.5 × 105 cells/mL) were treated for 2 hours with cycloheximide (final concentration 10 μL/mL, based on an initial dose-response experiment). The cycloheximide-treated and control cells were analyzed on 2 sets of IPGs (pH 4-7 and pH 6-11). As shown in Figures6 and 7, the relative expression of most proteins remained the same after 2 hours of treatment. Quantitative measurements showed that 27.5% proteins dropped off significantly (fold change > 2), whereas 63.7% of proteins were stable over this time period (Figure8). Nine proteins showed a relatively higher level of expression after cycloheximide treatment, indicating that posttranslation modifications of these proteins occurred less than 2 hours after their synthesis, or that their translation was relatively resistant to cycloheximide.
Two-dimensional electrophoretograms of cycloheximide inhibition of MPRO cells.
MPRO cells were treated with cycloheximide for 2 hours. MPRO cell lysate (1.5 × 106 cells/sample) was loaded for 2DE analysis (pH 4-7). (A) Control MPRO cells. (B) Cycloheximide-treated MPRO cells. The gels were stained with brilliant blue G-colloidal dye. (C,D) The magnified regions of 2 DE gels shown as inset in panels A and B. The arrowheads point to protein spots that decrease in intensity after cycloheximide treatment; the arrows point to spots whose intensity increases after cycloheximide treatment. The other information is presented as in the legend to Figure 1.
Two-dimensional electrophoretograms of cycloheximide inhibition of MPRO cells.
MPRO cells were treated with cycloheximide for 2 hours. MPRO cell lysate (1.5 × 106 cells/sample) was loaded for 2DE analysis (pH 4-7). (A) Control MPRO cells. (B) Cycloheximide-treated MPRO cells. The gels were stained with brilliant blue G-colloidal dye. (C,D) The magnified regions of 2 DE gels shown as inset in panels A and B. The arrowheads point to protein spots that decrease in intensity after cycloheximide treatment; the arrows point to spots whose intensity increases after cycloheximide treatment. The other information is presented as in the legend to Figure 1.
Two-dimensional electrophoretograms of cycloheximide inhibition of MPRO cells.
MPRO cells from cycloheximide inhibition experiment were also analyzed by basic pH range 2 DE. MPRO cell lysate (1.5 × 106 cells/sample) was loaded for IPGs-PAGE pH 6 to 11 and stained with brilliant blue G-colloidal dye. (A) Control MPRO cells. (B) Cycloheximide-treated MPRO cells. (C,D) The magnified regions of 2 DE gels shown as inset in panels A and B. The arrowheads point to protein spots that decrease in intensity after cycloheximide treatment; the arrows point to spots whose intensity increases after cycloheximide treatment. The other information is presented as in the legend to Figure 1.
Two-dimensional electrophoretograms of cycloheximide inhibition of MPRO cells.
MPRO cells from cycloheximide inhibition experiment were also analyzed by basic pH range 2 DE. MPRO cell lysate (1.5 × 106 cells/sample) was loaded for IPGs-PAGE pH 6 to 11 and stained with brilliant blue G-colloidal dye. (A) Control MPRO cells. (B) Cycloheximide-treated MPRO cells. (C,D) The magnified regions of 2 DE gels shown as inset in panels A and B. The arrowheads point to protein spots that decrease in intensity after cycloheximide treatment; the arrows point to spots whose intensity increases after cycloheximide treatment. The other information is presented as in the legend to Figure 1.
Distribution of protein spots from cycloheximide experiment.
In the cycloheximide experiment, MPRO cells were treated with cycloheximide for 2 hours; the untreated MPRO cells were used as a control. The protein inhibition patterns were compared with those of the control cells by Melanie-II software. For each protein, the x-axis value represents OD value of untreated with cycloheximide. The y-axis represents OD value after cycloheximide treatment. The information of proteins was collected in database dbMCp.
Distribution of protein spots from cycloheximide experiment.
In the cycloheximide experiment, MPRO cells were treated with cycloheximide for 2 hours; the untreated MPRO cells were used as a control. The protein inhibition patterns were compared with those of the control cells by Melanie-II software. For each protein, the x-axis value represents OD value of untreated with cycloheximide. The y-axis represents OD value after cycloheximide treatment. The information of proteins was collected in database dbMCp.
Comparison of differentiated MPRO cells with normal neutrophils
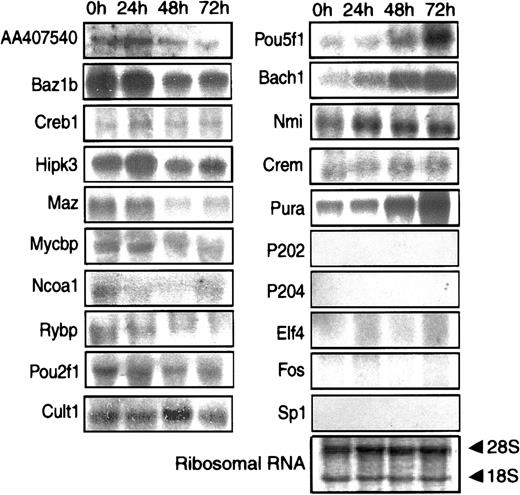
After 72 hours of ATRA treatment, the MPRO cells resembled mature neutrophils morphologically, including the presence of secondary granule proteins. To obtain a more complete picture of the differentiation state of the MPRO cells, we compared their RNA profiles with those of mature neutrophils. Human neutrophils were used rather than murine peripheral blood cells because the human cells are a more practical source of sufficient RNA for replicate analyses. In particular, we chose to focus on the levels of mRNA encoding transcription factors, because they control the differentiation process and determine the expression of the other genes. A total of 219 known or probable transcription factors were represented in mRNA isolated at some stage of MPRO cell development. Comparison of oligonucleotide chip analyses showed that there were 49 transcription factors whose mRNA was reported as present in resting human neutrophils but whose homologues were reported as absent in 72-hour MPRO cells. To obtain more precise data, we performed Northern blot analysis of 20 mRNAs encoding transcription modulators (Table 4). Of these, the oligonucleotide chips reported 12 as present in human neutrophils but absent in 72-hour MPRO cells. Eleven of these 12 were detected as present in 72-hour MPRO cells by Northern blot analysis (Figure 9). These includedBach1, not previously studied in myeloid cell differentiation, but markedly elevated in the mature cells. ConverselyRybp was markedly reduced as the cells matured. This finding is surprising because the protein is a presumptive transcriptional repressor and part of the mammalian homologue of theDrosophila polycomb complex.
Transcription factors analyzed by Northern blot assay
| Symbol . | 0 h . | 24 h . | 48 h . | 72 h . | MPRO 72 h4-150 . | Human 60K4-150 . |
|---|---|---|---|---|---|---|
| AA407540 | 2 | 3 | 1 | 1 | 169.81/P | N/A |
| Bach1 | 1 | 3 | 4 | 4 | 20/A | 679.32/P |
| Baz1b | 4 | 7 | 3 | 3 | 20/A | 679.32/P |
| Crem | 2 | 1 | 0 | 0 | 20/A | 821.08/P |
| Creb1 | 2 | 3 | 2 | 2 | 20/A | 233.22/P |
| Cutl1 | 2 | 1 | 3 | 3 | 20/A | 125.96/P |
| Hipk3 | 5 | 5 | 3 | 3 | 26.97/A | 41.14/P |
| Maz | 4 | 4 | 2 | 2 | 20/A | 1674.86/P |
| Mycbp | 1 | 2 | 2 | 2 | 20/A | 128.25/P |
| Nmi | 1 | 2 | 2 | 2 | 20/A | 407.37/P |
| pou2f1 | 3 | 2 | 1 | 1 | 20/A | 101.02/P |
| Pou5f1 | 1 | 2 | 2 | 3 | 20/A | 65.02/A |
| Pura | 3 | 3 | 5 | 8 | 81.62/P | 49.42/P |
| Rybp | 5 | 4 | 1 | 1 | 20/A | 592.25/P |
| Elf4 | 0 | 0 | 0 | 0 | 20/A | 729.51/P |
| Sp1 | 0 | 0 | 0 | 0 | 20/A | 20/A |
| Ncoa1 | 0 | 0 | 0 | 0 | 27.24/P | 392.04/P |
| Fos | 0 | 0 | 0 | 0 | 20/A | N/A |
| p202 | 0 | 0 | 0 | 0 | N/A | N/A |
| p204 | 0 | 0 | 0 | 0 | N/A | N/A |
| Symbol . | 0 h . | 24 h . | 48 h . | 72 h . | MPRO 72 h4-150 . | Human 60K4-150 . |
|---|---|---|---|---|---|---|
| AA407540 | 2 | 3 | 1 | 1 | 169.81/P | N/A |
| Bach1 | 1 | 3 | 4 | 4 | 20/A | 679.32/P |
| Baz1b | 4 | 7 | 3 | 3 | 20/A | 679.32/P |
| Crem | 2 | 1 | 0 | 0 | 20/A | 821.08/P |
| Creb1 | 2 | 3 | 2 | 2 | 20/A | 233.22/P |
| Cutl1 | 2 | 1 | 3 | 3 | 20/A | 125.96/P |
| Hipk3 | 5 | 5 | 3 | 3 | 26.97/A | 41.14/P |
| Maz | 4 | 4 | 2 | 2 | 20/A | 1674.86/P |
| Mycbp | 1 | 2 | 2 | 2 | 20/A | 128.25/P |
| Nmi | 1 | 2 | 2 | 2 | 20/A | 407.37/P |
| pou2f1 | 3 | 2 | 1 | 1 | 20/A | 101.02/P |
| Pou5f1 | 1 | 2 | 2 | 3 | 20/A | 65.02/A |
| Pura | 3 | 3 | 5 | 8 | 81.62/P | 49.42/P |
| Rybp | 5 | 4 | 1 | 1 | 20/A | 592.25/P |
| Elf4 | 0 | 0 | 0 | 0 | 20/A | 729.51/P |
| Sp1 | 0 | 0 | 0 | 0 | 20/A | 20/A |
| Ncoa1 | 0 | 0 | 0 | 0 | 27.24/P | 392.04/P |
| Fos | 0 | 0 | 0 | 0 | 20/A | N/A |
| p202 | 0 | 0 | 0 | 0 | N/A | N/A |
| p204 | 0 | 0 | 0 | 0 | N/A | N/A |
Band intensities at the different time courses from Northern blot assay were semiquantified on a scale from 1 (+) to 8 (++++++++).
The numbers in these columns are average differences in the value of hybridization intensity between the set of perfectly matched oligonucleotides and the set of mismatched oligonucleotides in the oligonucleotide array. “A” represents the genes that are absent, and “P” represents present in Affymetrix chip assay. The other information is presented as in the footnote to Table 2.
N/A indicates the gene is not presented in Affymetrix chips.
Northern blot analysis of selected mRNAs.
Equivalent amounts of RNA from MPRO cells induced by ATRA at different time points (0 hour, 24 hours, 48 hours, and 72 hours) were resolved by formaldehyde-agarose gel electrophoresis, stained to verify the amount of loading. Twenty transcription factor genes were separately probed on the RNA filters. The gene symbol of each probe was listed at the left of related Northern blot result. One of the RNA-blotted membrane photographs is shown with methylene blue-stained 28S and 18S RNA subunits demonstrating the quality and quantity of RNA loaded in individual lanes.
Northern blot analysis of selected mRNAs.
Equivalent amounts of RNA from MPRO cells induced by ATRA at different time points (0 hour, 24 hours, 48 hours, and 72 hours) were resolved by formaldehyde-agarose gel electrophoresis, stained to verify the amount of loading. Twenty transcription factor genes were separately probed on the RNA filters. The gene symbol of each probe was listed at the left of related Northern blot result. One of the RNA-blotted membrane photographs is shown with methylene blue-stained 28S and 18S RNA subunits demonstrating the quality and quantity of RNA loaded in individual lanes.
Discussion
We have used a 2-dimensional gel electrophoresis approach to explore the temporal patterns of protein expression during ATRA-induced myeloid development in the MPRO murine myeloid cell line.8This global analysis has detected 123 known proteins and 29 “new” proteins out of 220 protein spots identified by tandem mass spectroscopy, including proteins in 12 functional categories such as transcription factors, cytokines, and others. Bioinformatic analysis of these proteins has revealed clusters with functional importance to myeloid differentiation. Comparison of gene expression at the genomic and proteomic levels revealed some discrepancies between RNA and protein levels that indicate the importance of posttranscriptional and posttranslation processes during cell differentiation, although some differences undoubtedly arise at least in part from technical limitations of the current methods of measurement. These discrepancies may also be the result of varying translation and degradation efficiencies or might reflect posttranslation modifications. Nonetheless, overall there was a significant correlation between changes in mRNA and protein levels, consistent with the expectation that a substantial proportion of protein change is a consequence of changed mRNA levels, rather than posttranscriptional effects. Cycloheximide inhibition also showed that most of the proteins detected by gel electrophoresis were relatively stable, so that increased stability of proteins with maturation was not a likely explanation for the observed changes. We further examined the expression of transcription factor mRNA in MPRO cells and compared this with the expression pattern in mature human neutrophils. By combining oligonucleotide chip and Northern blot analysis, we observed that most of the transcription factor mRNAs detected in human neutrophils have homologues present in mature MPRO cells, although estimated relative RNA abundances could be quite different between species.
The first comparison of mRNA levels to the protein abundances of their gene products33 found a correlation coefficient of 0.48. These observations highlighted the limitations of functional studies performed only at mRNA level. Later, Anderson's group found a correlation coefficient of only 0.43 in a comparison of protein and mRNA abundances for a single gene product across 60 human cell lines by an immunoaffinity high-performance liquid chromatography method and quantitative Northern analysis.19 In 1999, Gygi et al30 quantitatively compared mRNA and protein expression levels for 128 different genes expressed in yeast, using serial analysis of gene expression (SAGE) and capillary liquid chromatography-tandem mass spectrometry methods. Their results showed a correlation coefficient of 0.935 for the most abundant proteins; but the coefficient was only 0.356 for the 69% of 106 genes34for which the transcript levels were less than 10 copies/cell.
These prior studies examined static expression levels without correlation of changes in protein and mRNA levels during cell development, as performed in the present study. In general, we found a moderately high correlation (coefficient 0.58) between estimated protein and RNA levels. There are multiple technical considerations, both in measuring RNA and protein levels that might affect the results, but the general conclusion supports previous contentions35that interpretations of changes in cell behavior based on changing mRNA levels is incomplete. Nevertheless, the correlation is sufficiently strong to indicate that the regulation of transcript levels is probably a major determinant of changes in protein levels during differentiation in this system. Because uninduced MPRO cells were in a steady state, one might expect to see better correlation at later time points, when changes in mRNA levels over time have been translated into protein levels.
Some loss of correlation could derive from unstable proteins that are differentially regulated during cellular maturation. Using cycloheximide to inhibit protein synthesis, we found that the large majority of the proteins in this system are relatively stable. However, protein stability is an important factor in posttranslational proteomic studies.
Much progress has been made in understanding transcriptional regulation of the myeloid differentiation program. Transcription factors such as PU.1 and members of the C/EBP family have been found to play important roles in the expression of a variety of myeloid genes, both by examination of individual gene regulatory regions and by gene knock-out studies in mice.36-39 Our previous work20initiated and the present study has established a database of transcription factors and target genes differentially regulated during myeloid differentiation. The results are limited by the sensitivity, accuracy, and comprehensiveness of the available oligonucleotide chips for mouse mRNAs.
Detection of transcription factor proteins is difficult because they are often present at low abundance, may have basic pIs, and may be present in various modified forms that alter their mobility on 2-dimensional gels. Encouragingly, the present study identified 7 proteins potentially important to transcriptional regulation, including RNA polymerase II, Stat5a, Aiolos, Hmg1 and 2, Kruppel-related zinc finger protein F80-m, and Zfp101. Previous studies have shown that all 7 members of the signal transducers and activator of transcription (STAT) family are involved in regulating expression of cytokine-induced and growth factor-induced genes.40 Among them, Stat5 appears to have an important role in myeloid cell development, primarily by mediating granulocyte-macrophage colony-stimulating factor (GM-CSF) signaling. At the mRNA level, several STAT proteins, including Stat1, 3, 5b, and 6, were moderately up-regulated in MPRO cells. Our data showed decreased expression of Stat5a protein at the late stage of MPRO differentiation, as reported in other systems.40Kruppel-related zinc finger protein F80-m and Aiolos are 2 newly identified transcription factors, with still unknown functions in myeloid cells, although Aiolos is known to interact with Rasto control cell death in T cells.41 In MPRO cells, we found that Aiolos is expressed at a fairly constant level throughout differentiation. In contrast, Kruppel-related zinc finger protein F80-m was strongly down-regulated and Zfp101 slightly up-regulated. The high mobility group (HMG) box domain defines a family of proteins, mostly transcription factors, that specifically interact with DNA on the minor groove.42,43 Surprisingly, recent studies suggest a second quite different function for Hmg1and 2 as cytokinelike factors.44 45 In this study, bothHmg1 and Hmg2 were detected by 2DE analysis.Hmg2 was significantly up-regulated indicating its possible important function in biologic processes in MPRO differentiation.
Oligonucleotide chip analyses showed the presence of mRNAs for about 123 transcription- or chromatin-modifying factors in differentiated MPRO cells and 147 factors in mature human neutrophils. Overall, 49 of these factors represented in neutrophil mRNA were not detected by chip analysis of MPRO cells, but 11 of 12 were detectable by Northern blot analysis. In some cases the failure to find an mRNA by chip analysis was probably because the amount of transcript was below the threshold for oligonucleotide chip detection,46 47 but in other cases relatively strong Northern signals were obtained.
Several subsets of transcription factor mRNAs had patterns of expression that could be interpreted in terms of known function of the products. Myc is a well-known transcription factor that promotes growth rather than differentiation,48 and in turn is regulated by interactions with a family of proteins includingMax, Mad, and Sin3B.49 In developing MPRO cells Myc is down-regulated andMad is up-regulated. The related protein Mad4 is slightly down-regulated and Mad5 is markedly down-regulated and apparently absent from the mature cells. Mad5 differs from other proteins of this group in that it may act to stimulate as well as repress transcription. In addition, Sin3b is one of the more markedly up-regulated transcription factor mRNAs. The combined changes in Mad, Myc, and Sin3b would be expected to synergistically prevent activation of Myc target genes.
PU.1 is a transcription factor implicated in the transcriptional control of neutrophil-specific genes and in neutrophil production, which is defective in PU.1 knockout mice.50Sp1, Purb, Klf9/Bteb1, and Maz are broadly expressed transcription factors that bind to purine-rich sites, including potentially some PU.1 sites. PU.1 is up-regulated almost 3-fold at the RNA level, whereas all 4 of the latter factors are down-regulated during MPRO development, as is the SP1-like factorKlfl3.
We have previously observed20 by Northern blot analysis that there is a shift in the balance of members of the C/EBP family of transcription factors at the mRNA level during MPRO differentiation, with some progressive down-regulation of C/EBP δ and up-regulation first of C/EBP α then C/EBP ε and β. These results are consistent with the role of these factors in neutrophil development, deduced from both transcriptional analysis of individual promoters and gene knockout effects on myelopoiesis. The present set of RNA analyses by oligonucleotide chip hybridization is more consistent with the Northern blot analyses than were the preliminary results,20although C/EBP ε is still not represented on the chip.
Overall, these coordinated changes in the expression of multiple transcription factors would serve to amplify differences in transcription and permit fine control of the timing and amplitude of regulation for multiple gene targets. As previously postulated,51 such reciprocal regulation of competing factors may be a common mechanism in differentiation. The changes in mRNA levels during maturation of myeloid cells include both the silencing of a number of genes and up-regulation of a number of other genes. The substantial changes in the level of some putative transcriptional repressors, both up (eg, Sin3b, Atf7ip) and down (eg, Rybp) during differentiation, suggest that specific repression of transcription provides an important and under-investigated means of regulating myeloid differentiation, in addition to more conventional mechanisms such as competition for binding sites and changes in activating factor levels.
The striking morphologic changes in the maturing nuclei of “polymorphonuclear leukocytes” remain mysterious both in terms of mechanism and teleology. Some possible clues may be observed in the current RNA expression data. For example, Ran is a small guanosine triphosphatase (GTPase) required for nuclear import and export, and mRNA levels for Ran and Ran binding proteins 1 and 2 decline as the cells mature. This change could either be a cause or consequence of decreased nuclear import of macromolecules coincident with nuclear condensation. Another protein, acinus, is implicated in causing chromatin condensation without DNA breakage during apoptosis52; its mRNA increases about 3-fold as MPRO cells mature and form highly condensed, multilobed nuclei.
In summary, we have comprehensively and quantitatively analyzed both RNA and protein expression patterns during myeloid differentiation. Changes in protein levels correlated moderately well with changes in mRNA expression. Investigation of transcription factor mRNA representation showed considerable similarity to those of mature human neutrophils and highlight several transcription factors and other functional nuclear proteins whose mRNA levels change prominently during MPRO differentiation but which have not been investigated previously in the context of myeloid development. The number of transcription factors expressed in these cells greatly exceeds those previously identified as important for the regulation of specific myeloid genes. Currently emerging techniques53-55 for genomic analysis of factor binding sites in mammalian DNA may help to elucidate their gene targets and potential roles in myeloid differentiation.
We express our gratitude to Dr S. Tsai (Program in Molecular Medicine, Fred Hutchinson Cancer Research Center, Seattle, WA) for his kind gift of MPRO cell line, and Mr Jeffrey J. Meyer (University of Chicago School of Medicine) for helpful advice.
This section contains the genes described in this paper, including figures, tables, and text. AA589396: dendritic cell protein; Abpa: androgen-binding protein: subunit alpha;Actb: put. Beta-actin; Actg: actin, gamma, cytoplasmic; Aldh2: aldehyde dehydrogenase 2, mitochondrial;Anxa1: lipocortin I protein annexin 1; Anxa11: annexin A11; Anxa2: annexin II calpactin I heavy chain;Anxa3: annexin A3; Arhgdib: RHO GDP-dissociation inhibitor 2(RHO GDI2); Arhn: rho7; Arpc3: actin-related protein 2/3 complex, subunit 3 (21 kDa); Arp2/3 complex subunit p21-Arc, Atp5a1: ATP synthase, H+transporting, mitochondrial F1 complex, alpha subunit, isoform 1;Atp5b: ATP synthase, H+ transporting mitochondrial F1 complex, alpha subunit; C4: MHC complement component C4; Cab140: 170 kDa glucose regulated protein GRP170 precursor; Cappa1: F-actin capping protein alpha-1 subunit; Cas1: catalase 1;Cct2: chaperonin containing TCP-1 beta subunit ;Cct5: chaperonin subunit 5 (epsilon); Cct6a: Chaperonin subunit 6a (zeta); Cftr: cystic fibrosis transmembrane conductance regulator homolog; Coro1a: coronin, actin-binding protein 1A; Crmp1: collapsin response mediator; Ddx5: DEAD (aspartate-glutamate-alanine-aspartate) box polypeptide 5; ECP: EndoA′ cytokeratin 5′ end put.); putative; Eef1a1: eukaryotic translation elongation factor 1 alpha 1; Eef2: elongation factor 2; Ehd1: “EH-domain containing 1, PAST, HPAST, H-PAST”; Eno1: alpha enolase; Ephb2: protein-tyrosine kinase (EC 2.7.1.112) sek-3, Eph receptor A4; Es10: sid478p/Esterase 10;Fut4: fucosyltransferase 4; G4–1-pending: phosphatase subunit gene g4–1; Gapd: glyceraldehyde-3-phosphate dehydrogenase; Gc: vitamin D-binding protein precursor;Gnb2-rs1: guanine nucleotide binding protein, beta-2, related sequence1, p205, Rack1, Gnb2l1, GB-like; Got2: glutamate oxaloacetate transaminase 2, mitochondrial; mitochondrial aspartate aminotransferase; GROEL: chaperonin groEL precursor; Grp58: glucose regulated protein, 58 kDa; endoplasmic reticulum protein; phospholipase C, alpha;Gstm1: glutathione-S-transferase, mu1; H2-Ab1: histocompatibility 2, class II antigen A, beta 1; Hcph: PTPN6 tyrosine phosphatase, me, hcp, PTPN6, Ptp1C, SHP-1;Hgf: hepatocyte growth factor precursor; Hmg2: high mobility group protein 2; Hmgb1: high mobility group protein 1; Hnrpa1: heterogeneous nuclear ribonucleoprotein A1; Hnrpa2b1: heterogeneous nuclear ribonucleoprotein A2; heterogenous nuclear ribonucleoprotein A2/B1; Hnrph1: heterogeneous nuclear ribonucleoprotein H1; HPD76: hypothetical protein DKFZp761C10121.1; Hsc70: dnaK-type molecular chaperone hsc73/Heat shock protein cognate 70;Hsp110: heat shock protein, 110 kDa; Hspa5: glucose-regulated protein, 78 kDa; Hspa5/Grp78: 78 kDa glucose-regulated protein precursor (GRP 78);Hspa8: dnaK-type molecular chaperone hsc70; Idh1: isocitrate dehydrogenase 1(NADP+), soluble;Idh2: NADP+-specific isocitrate dehydrogenase;IFI-205: interferon-activatable protein 205;IGVAP: Ig Vkappa, antiphenyloxazolone; Il1f5: interleukin 1 receptor antagonist homolog 1; Impdh2: inosine-5′-monophosphate dehydrogenase; KER1: keratin 1;KER8: keratin 8, type II cytoskeletal, embryonic;KER10: keratin 10, type I, cytoskeletal; KER14: keratin 8, type I cytoskeletal 14; KER47: 47 kDa keratin; KER59: keratin, 59K type I cytoskeletal;Krt2–6g: keratin, type II cytoskeletal 6;KRZF80M: Kruppel-related zinc finger protein F80-M;KT14: keratin, type I, cytoskeletal;Ldh1: lactate dehydrogenase 1, A chain;Lgals3: galectin-3; Lmnb1: lamin B1;LOC56463: p100coactivator; Ltf: lactotransferrin precursor; Mor1: malate dehydrogenase; Nme1: nucleoside diphosphate kinase A; Nsap1-pending: syncrip;P4hb: protein disulfide-isomerase, PDI;Papss2/Atpsk2: ATP sulfurylase/APS kinase, 2: PAPS synthetase; Pbp: hippocampal cholinergic neurostimulating peptide precursor protein, phosphatidylethanolamine-binding protein;Pcna: proliferating cell nuclear antigen; Pdi4: peptidyl arginine deiminase, type IV; PAD type IV; Pgk1: phosphoglycerate kinase 1; Phb: prohibitin; Pk3: pyruvate kinase 3; Ppia: peptidylprolyl isomerase A; cyclophilin A ; Prdx1: proliferation-associated gene A, osteoblast specific factor 3; Prdx2: Antioxidant protein 2; PRO2675: PRO2675; Psma7: proteasome (prosome, macropain) subunit, alpha type 7, Proteasome subunit RC6–1;Psmc1: protease (prosome, macropain) 26S subunit, ATPase 1;Psmc2: 26S protease regulatory subunit 7, MSS1 protein;Ptpn1: protein tyrosine phosphatase; Pygm: muscle glycogen phosphorylase; Rac2: RAS-related C3 botulinum substrate 2, p21-Rac2, EN-7 protein; Rag1: recombination activating gene 1; Ran: GTP-binding nuclear protein Ran (TC4); Rbm3: RNA binding motif protein 3; Rin: RAS-like protein expressed in neuro; Rnf17: RING finger protein Mmip-2; RNPC: RNP particle component;SAC: spectrin alpha chain; Sfmbt: Scm-related gene containing 4 mbt domains; Slc23a2: solute carrier family 23, (nucleobase transporters) member 1; Sod1: putative peroxisomal antioxidant enzyme, superoxide dismutase 1;Spry1: sprouty homolog 1 (Drosophila);Stat5a: signal transducer and activator of transcription 5A;STEFIN3: stefin 3; Stip1: extendin/Stress-induced phosphoprotein1; Taf2e: TATA box binding protein (Tbp)–associated factor, RNA polymerase II, E; Tagln2: transgelin 2; Taldo1: transaldolase; Tinag: tubulointerstitial nephritis antigen; Tpi: triosephosphate isomerase, TIM; Tpm5: tropomyosin 5, cytoskeletal type;Tuba6: tubulin alpha 6; Tubb5: tubulin, beta 5;Ube1x: ubiquitin-activating enzyme E1 X; Vav2: Vav2 oncogene; Vdac1: voltage-dependent anion-selective channel protein 1; vim: vimentin; Zfp101: zinc finger protein 101; ZFP1A3: Aiolos/zinc finger protein, subfamily 1A, 3; Zfp354a: transcription factor 17.
Prepublished online as Blood First Edition Paper, July 5, 2002; DOI 10.1182/blood-2002-03-0850.
Supported by National Institutes of Health (NIH) grants CA42556, AI43558, DK54369, and HL63357, and by Gene Logic (S.M.W.); NIH grant HL63357 (Z.L.); NIH grant DK 54369, grants from the Arthritis Foundation and Charles H. Hood Foundation, and the John H. Pierce Pediatric Oncology Research Fund (P.E.N.); and NIH grant P50 HG02357-01 (M.G.).
S.M.W. owns stock in and consults for Gene Logic Inc.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Peter E. Newburger, Department of Pediatrics, University of Massachusetts Medical School, 55 Lake Ave N, Worcester, MA 01655; e-mailpeter.newburger@umassmed.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal