The C-terminal region of erythroid cytoskeletal protein 4.1R, encoded by exons 20 and 21, contains a binding site for nuclear mitotic apparatus protein (NuMA), a protein needed for the formation and stabilization of the mitotic spindle. We have previously described a splicing mutation of 4.1R that yields 2 isoforms: One, CO.1, lacks most of exon 20–encoded peptide and carries a missense C-terminal sequence. The other, CO.2, lacks exon 20–encoded C-terminal sequence, but retains the normal exon 21–encoded C-terminal sequence. Knowing that both shortened proteins are expressed in red cells and assemble to the membrane skeleton, we asked whether they would ensure 4.1R mitotic function in dividing cells. We show here that CO.2, but not CO.1, assembles to spindle poles, and colocalizes with NuMA in erythroid and lymphoid mutated cells, but none of these isoforms interact with NuMA in vitro. In microtubule-destabilizing conditions, again only CO.2 localizes to the centrosomes. These data suggest that the stability of 4.1R association with centrosomes requires an intact C-terminal end, either for a proper conformation of the protein, for a direct binding to an unknown centrosome-cytoskeletal network, or for both. We also found that 4.1G, a ubiquitous homolog of 4.1R, is present in mutated as well as control cells and that its C-terminal region binds efficiently to NuMA, suggesting that in fact mitotic spindles host a mixture of the two 4.1 family members. These findings led to the postulate that the coexpression at the spindle poles of 2 related proteins, 4.1R and 4.1G, might reflect a functional redundancy in mitotic cells.
Introduction
Protein 4.1R (4.1R) is a major component of the red cell membrane skeleton, which stabilizes the spectrin-actin lattice, and interacts with various skeletal and transmembrane proteins.1 Human 4.1R is encoded by the EPB41gene1 (p33-p34.2), which encompasses about 200 kb and contains over 25 exons, 22 of which are expressed in erythroid cells.2 The prototypical 80-kDa erythroid isoform of 4.1R is one of multiple protein isoforms, most of which are generated by pre-mRNA alternative splicing, in a tissue- and developmental-specific manner.2-4 In addition to this protein diversity generated from 4.1R gene, recent studies have identified 3 new proteins closely related to 4.1R but encoded by 3 different genes. Among the new members of the 4.1 family of genes, 4.1G is the most widely distributed in tissues.5 In fact, 4.1R and 4.1G show relatively opposite patterns of expression: 4.1R is most abundant in hematopoietic cells and tissues, whereas 4.1G is expressed at higher levels in a wide range of tissues, but is relatively less abundant in hematopoietic cells.5
Several studies have recently documented the expression of 4.1R-related isoforms in both the nuclear and the cytoplasmic compartments, as well as in perinuclear structures: 4.1R associates with the nuclear matrix,6,7 splicing factors,8 and centrosomes,9 where it binds to centrosomal P4.1-associated protein (CPAP), and might therefore act as an adapter to anchor the CPAP/γ-tubulin complex to the centrosome.10 On the other hand, in vitro studies have shown that 4.1R interacts specifically with tubulin11 and erythroid and skeletal muscle myosins.12,13 It has also been shown that one isoform, probably a 135-kDa–related isoform, binds with the nuclear mitotic apparatus protein (NuMA) at the spindle poles of dividing cells.14 NuMA is a nuclear protein that relocalizes to the spindle poles during mitosis and meiosis. Its presence is required to focus microtubules into spindle poles and to stabilize the mitotic spindle and control its size until anaphase.15 16
The erythrocyte membrane 4.1R protein isoform is composed of 4 domains, referred to as 30, 16, 10, and 22/24 kDa, from N- to C-terminus.1 The 30-kDa domain contains binding sites for band 3, glycophorin C/D, p55, and calmodulin. The 10-kDa domain ensures the binding of 4.1R to actin and spectrin β-chain. However, little is known about the function of the 22/24-kDa domain in mature red cells.
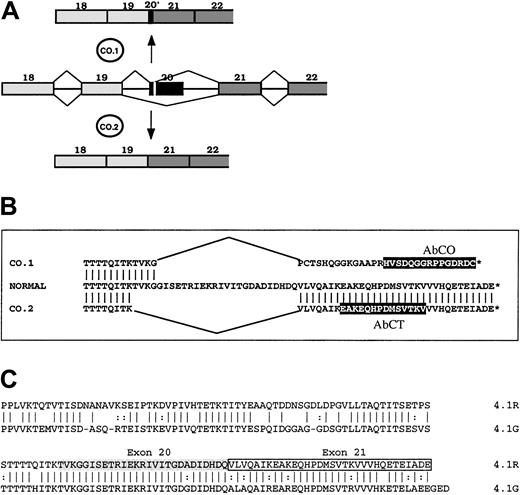
To gain new insights into the functional importance of the 22/24-kDa domain, we analyzed the expression and the functional properties of 2 4.1R variants resulting from a mutation in the gene sequence encoding the 22/24-kDa C-terminal domain (CTD).17 One of these 2 variants, named 4.1R Coimbra, derived from a splicing mutation, a G→A substitution at position 2720, which is the last position of exon 20 (AA/gt→AG/gt).17 The mutation yielded 2 abnormal spliceoforms (Figure 1). One spliceoform, named CO.1, resulted from the activation of a cryptic donor site in the 5′ region of exon 20, and left only the first 10 nucleotides of this exon. The out-of-frame sequence downstream met a stop codon only 7 nucleotides ahead of the physiological stop codon. All together, it allowed a C-terminal tail (downstream of the exon 19–encoded segment) of 33 amino acids, among which the last 29 amino acids accounted for a missense sequence. The other spliceoform (CO.2) arose from the skipping of exon 20, leaving a C-terminal sequence of 32 amino acids identical to the wild-type sequence, encoded by exon 21. The 2 4.1R isoforms were truncated by about the same length. They both appeared in nearly equal quantities, yet the total amount of both was 30% of normal in the homozygous patient. That patient presented with an unusually mild picture, intermediate between the common heterozygous and homozygous 4.1R hereditary elliptocytosis. We concluded that the CTD must have no major functional role in mature red cell skeleton, and that the mild functional loss is correlated with the total amount of 4.1R translatable mRNA, rather than with the primary structure of the mutated proteins.17
Schematic representation of 4.1R, CO.1, CO.2, and 4.1G in their C-terminal regions.
(A) Abnormal splicing events generated at 4.1R pre-mRNA from exon 20 mutation, and leading to the spliceoforms encoding CO.1 and CO.2. (B) Amino acid sequence alignment of normal 4.1R, CO.1, and CO.2. Peptide epitopes for specific antibodies AbCO and AbCT are highlighted in black boxes. (C) Sequence alignment of 4.1R and 4.1G CTDs. Note the high degree of amino acid identity between the 2 proteins at 4.1R exons 20–encoded (shaded) and 21–encoded (boxed) sequence.
Schematic representation of 4.1R, CO.1, CO.2, and 4.1G in their C-terminal regions.
(A) Abnormal splicing events generated at 4.1R pre-mRNA from exon 20 mutation, and leading to the spliceoforms encoding CO.1 and CO.2. (B) Amino acid sequence alignment of normal 4.1R, CO.1, and CO.2. Peptide epitopes for specific antibodies AbCO and AbCT are highlighted in black boxes. (C) Sequence alignment of 4.1R and 4.1G CTDs. Note the high degree of amino acid identity between the 2 proteins at 4.1R exons 20–encoded (shaded) and 21–encoded (boxed) sequence.
The question arose as to whether in the homozygote any of the 2 abnormal 4.1R protein isoforms would bind NuMA in nucleated cells, and whether NuMA could assemble to the spindle poles in the absence of intact 4.1R. To address these issues, we analyzed 4.1 expression in purified erythroblasts and immortalized lymphoblasts obtained from peripheral blood in 4.1R Coimbra carriers. Messenger RNA analysis showed that the splicing site mutation at the end of exon 20 generates in both erythroid and lymphoid cells the same splicing alterations described in reticulocyte mRNA.17 Cultured erythroid precursors displayed normal morphological features through day 14 of culture. Using confocal microscopy, we observed that the CO.2, but not the CO.1, isoform colocalizes with NuMA in the nuclei of erythroid precursors and lymphoid cells, whereas in vitro binding assays showed no interaction between NuMA and either of the shortened protein isoforms. In nocodazole-treated cells, the CO.2 shortened form of 4.1R appears to colocalize with centrosomal γ-tubulin, whereas CO.1 fails to assemble to centrosomes. These data suggest that an intact CTD is required for 4.1R assembly to the centrosome and to the mitotic spindle poles; this CTD would provide either a proper conformation for the whole 4.1R protein, and/or a bona fide protein-binding site for a centrosomal component. On the other hand, we showed that 4.1G is also targeted to spindle poles in mitotic cells and colocalizes with NuMA. In vitro binding assays further supported a direct interaction between NuMA and 4.1G. These results suggest a possible functional redundancy between 4.1R and 4.1G.
Patients, materials, and methods
Case reports
Most of the experiments were carried out on blood samples obtained from a patient homozygous for the 4.1R Coimbra mutation and her heterozygous grandson after informed consent. Further details regarding this family were presented previously.17
Cell culture
After Ficoll hypaque separation of mononuclear cells, erythroid-progenitor enrichment was performed by negative selection with the use of carbonyl iron (Sigma, St Louis, MO) and an immunomagnetic bead procedure with the use of Dynabeads anti-CD2 (Dynal, Oslo, Norway). Erythroid progenitors were grown for 7 or 14 days in standard methylcellulose assays with the use of 50 ng/mL recombinant stem cell factor, 3000 IU/L recombinant erythropoietin, and 30 IU/mL recombinant interleukin-3. Colonies derived from erythroid burst-forming units (BFU-Es) were enumerated and plucked at days 7 and 14 and then processed for May-Grünwald-Giemsa staining, immunofluorescence microscopy, and RNA extraction.
Epstein-Barr virus (EBV)–infected lymphoid cell lines were kindly established by Dr A. Calender (Laboratoire de Cytogénétique, Hôpital Edouard Herriot, Lyons, France). The cells were grown in RPMI 1640 with Glutamax, HEPES (Life Technologies, Paisley, United Kingdom), and 10% fetal bovine serum. They were harvested during the exponential growth phase.
To test the microtubule dependence on 4.1R association with centrosomes, cells were exposed to nocodazole at 2 μg/mL in culture medium for 2 hours, then washed with phosphate-buffered saline (PBS), and stained for tubulin and 4.1R immunofluorescence (see below).
Reverse-transcriptase polymerase chain reaction
Total RNA was isolated from erythroid and lymphoid cells by means of RNAble (Eurobio, Les Ulis, France). Following a reverse transcription step using Superscript II and random hexamers (Life Technologies), cDNA was amplified by polymerase chain reaction (PCR) as previously described,17 with the use of a sense primer (5′TGAGACCAAGACCATCACTT3′) within exon 18 and a reverse primer (5′TCAGCAATCTCGGTCTCCTG3′) within exon 21.
Antibodies
The antibodies used were as follows: (1) A polyclonal antibody against the N-terminal sequence of 4.1R (headpiece antibody [AbHP]), was kindly provided by Dr T. K. Tang (Institute of Biomedical Sciences, Academia Sinica, Taipei, Taiwan). The corresponding epitope encompassed mostly exon 2–encoding sequence. (2) Affinity-purified polyclonal antibodies against the C-terminal peptides of normal 4.1R (AbCT) and CO.1 (AbCO) were previously described (Figure 1).17 Note that AbCT also reacted with CO.2 isoform.17 (3) A mouse monoclonal antibody against NuMA (AbNuMA) was used as described previously.14(4) An affinity-purified polyclonal antibody specific for human 4.1G, AbHG13, was raised in chicken by Agro-Bio (La Ferté-Saint-Aubin, France). It was directed against the peptide NH2-CDGDGRREVRSPTKAP-CONH2 prepared by Synt:em (Nı̂mes, France; the underlined C residue was added for coupling purposes). This peptide is present within a 4.1G sequence, which appears as one of the most divergent peptide sequences among 4.1 protein members.5 (5) Goat antibodies against glutathione S-transferase (GST; AbGST) and mouse anti–γ-tubulin (AbγT) were purchased from Amersham Pharmacia Biotech Europe (Freiburg, Germany) and Sigma-Aldrich (St Louis, MO), respectively.
Immunofluorescence and confocal microscopy
Erythroid or lymphoid cells were washed in PBS and cytocentrifuged onto poly-L-lysine–coated slides. Cells were then fixed and permeabilized by immersion in PBS containing 2% paraformaldehyde and 0.2% Triton X-100 for 25 minutes at room temperature. After 3 washes in PBS, preparations were blocked for 2 hours with PBS containing 10% bovine serum albumin (BSA) at room temperature. The slides were incubated with primary antibodies with the use of the following dilutions in PBS/3% BSA/0.1% Tween 20 (AbCT, 1/40; AbCO, 1/40; AbHP, 1/1000; and AbHG13, 1/50) for 1 hour at room temperature. After 3 washes in PBS, cells were incubated with AbNuMA or AbγT diluted at 1/50 or 1/500, respectively, in the same conditions. Following thorough washing with PBS, cells were incubated with the secondary antibodies: goat antirabbit antibodies conjugated with Alexa 488 (Molecular Probes, Eugene, OR), goat antimouse antibodies conjugated with Alexa 546 (Molecular Probes), or donkey antichicken immunoglobulin Y (IgY) conjugated to fluorescein isothiocyanate (Interchim, Montluçon, France), diluted according to the supplier's recommendations for 1 hour at room temperature. Negative controls were prepared by replacing primary antibodies by nonimmune rabbit serum, mouse serum, or chicken IgY. All the incubations were performed at room temperature in a humidified chamber. Finally, preparations were mounted in 150 mM Tris-HCl (tris(hydroxymethyl)aminomethane–HCl), glycerol 25%, pH 8.60. The samples were observed with a confocal Zeiss Axiovert 135 microscope (Göttingen, Germany) through a × 40 oil immersion objective. Images were processed by means of Photoshop software (Adobe Systems, San Jose, CA).
Recombinant plasmid constructs
A fragment encompassing the CTD of 4.1R was obtained by reverse-transcriptase PCR (RT-PCR) amplification of a control RNA template, with the use of the following primers designed within exons 17 and 21: sense, 5′AAGCGGCCGCCCCGGGGGATCCCGAACTCTTAACATCAATGGG3′; antisense, 5′AACCCGGGGCGGCCGCTCTAGATCACTCATCAGCAATCTCGGT3′. The truncated CTDs of CO.1 and CO.2 isoforms were also obtained by RT-PCR from a homozygous 4.1R Coimbra RNA sample, with the use of the same set of primers.
A cDNA fragment encoding amino acids 856 through 1005 at the 3′ region of 4.1G mRNA5 was amplified by PCR with the following primers: sense, 5′AAGCGGCCGCCCCGGGGGATCCGTTGACATTGATGTTTTGCCAC3′; antisense, 5′AACCCGGGGCGGCCGCTCTAGATTAATCTTCCCCTTCCTCAGC3′. Following agarose gel electrophoresis, the bands were cut out, digested withBamHI and NotI, repurified by agarose gel electrophoresis, and ligated in frame with GST into pGEX-4T-1 plasmid (Amersham Pharmacia Biotech Europe). The 4 recombinant plasmids obtained are referred to as pGEX/R, pGEX/CO.1, pGEX/CO.2, and pGEX/G, respectively.
All the constructs were further sequenced to ascertain the inframe insertion of the DNA fragments and the absence of nucleotide mismatches. Finally the NuMA1/TOPO construct containing amino acids 1697 through 2101–encoding region of NuMA was previously described and used for in vitro translation experiments.14
In vitro binding assays
The pGEX constructs described above were expressed in bacteria, and GST-fusion proteins, designated GST/R (for normal), GST/CO.1, GST/CO.2, and GST/G, were purified basically according to the manufacturer's recommendations (Amersham Pharmacia Biotech Europe): The expression of the GST-fusion proteins was carried out in the presence of 0.1 mM isopropylthiogalactopyranoside (IPTG) at 37°C. After cell harvest and centrifugation, the cell pellet was resuspended in a PBS (150 mM NaCl, 10 mM Na2HPO4, pH 7.50) containing 0.1 mM phenylmethyl sulfonyl fluoride or 1 mM Pefabloc, 5 mM dithiotreitol, and 1000 mg/L lysozyme. After 30 minutes of incubation on ice, the cells were disrupted in a Sonifier II disrupter (Branson Ultrasonic, Carouge-Geneva, Switzerland) for a maximum of 3 pulses of 10 seconds each, avoiding frothing. The postsonication solution was incubated in the presence of 1% Triton X-100 for 1 hour. The homogenate was then centrifuged at 13 000 rpm for 30 minutes at 4°C. The GST-fusion peptides present in the supernatant were immediately mixed with glutathione-Sepharose 4B beads for different times (20 minutes; 2, 20, and 43 hours). All the purification steps were carried out on ice. After incubation, the beads were washed by the addition of 10 bead volumes of PBS. Finally, the GST-fusion peptides were eluted from the beads by adding 1 vol glutathione elution buffer (10 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0) and analyzed by Western blot with the use of the above-described AbCT, AbCO, and AbGST antibodies.
A minor band of 24 kDa gradually developed in the course of incubation of the bacterial lysate with glutathione-Sepharose 4B beads (20 minutes; 2, 20, and 43 hours), with GST/CO.1, but not with GST/R or with GST/CO.2 (not shown). The 24-kDa band stemmed presumably from proteolysis, in spite of the presence of protease inhibitors. Upon Western blotting (not shown), GST/R GST/CO.2, and, notably, GST/G reacted with AbCT and AbGST. The cross-reaction of 4.1G C-terminal peptide with AbCT antibody was rather expected considering the high degree of homology observed between all the 4.1 family members at their CTDs.18 GST/CO.1 (38.5-kDa band) reacted with AbCO and AbGST. The 24-kDa band reacted with AbGST only. The GST/CO.1 high susceptibility to degradation was addressed by reducing the incubation time to 20 minutes. After the bacterial lysate was eliminated, GST/CO.1 bound to glutathione-Sepharose beads could be kept at 4°C for 10 hours without degradation.
The in vitro transcription/translation experiments were performed on NuMA1/TOPO,14 with the use of the TNT Quick coupled transcription/translation system (Promega, Madison, WI) in the presence of [35S]-methionine. Equivalent amounts of labeled NuMA1 peptides were incubated with each of the affinity-purified GST-fusion proteins, coupled to glutathione-Sepharose beads for 1 hour at 4°C in PBS binding buffer (150 mM NaCl, 10 mM Na2HPO4), containing leupeptine (4 μg/mL), antipain (4 μg/mL), and pepstatin (12 μg/mL). After washing the loaded column with PBS, the bound protein complex was eluted with glutathione elution buffer (30 mM reduced glutathione in 50 mM Tris-HCl, pH 8.0). The eluted protein complexes were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE), and the bound NuMA was visualized by autoradiography.
Results
Normal morphology but slight decrease in growth of 4.1R Coimbra cells
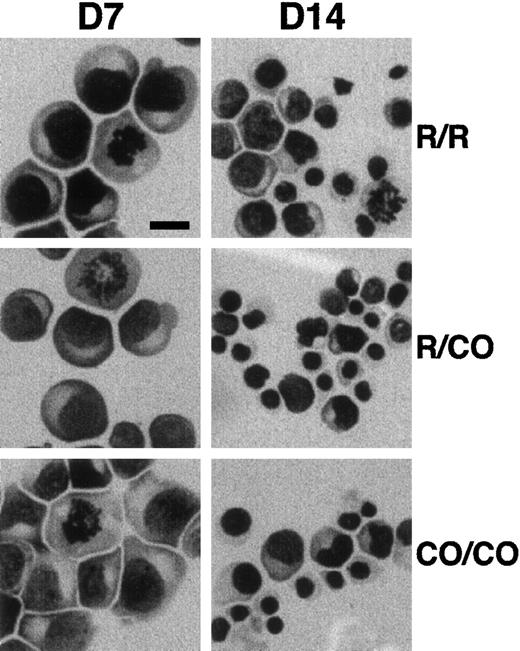
Erythroid progenitors were purified from peripheral blood and cultured for 7 and 14 days in methyl cellulose standard assay. The number of BFU-Es and of total cells were slightly, but consistently, decreased in 4.1R Coimbra cells (CO cells), in comparison with control cells (Table 1). Similarly, slight but not significant differences in the time of lymphoid cell doubling were repeatedly observed (3 sets of experiments; Table 1). On the other hand, May-Grünwald-Giemsa staining of cultured erythroblasts showed normal appearance of CO cells, as compared with control cells, with more than 90% of immature proerythroblasts at 7 days of culture (Figure 2).
Effect of 4.1R Coimbra mutation on cell proliferation
| Blood sample . | Erythroid cells* . | Lymphoid cells† . | |
|---|---|---|---|
| Day 7 . | Day 14 . | Population-doubling time . | |
| Control | 16.6 (10.4/22.8) | 19.8 (14.6/25.0) | 40.3 ± 8.0 |
| Heterozygote | 13.6 | 16.6 | 44 |
| Homozygote | 11.0 | 12.8 | 46.0 ± 8.0 |
| Blood sample . | Erythroid cells* . | Lymphoid cells† . | |
|---|---|---|---|
| Day 7 . | Day 14 . | Population-doubling time . | |
| Control | 16.6 (10.4/22.8) | 19.8 (14.6/25.0) | 40.3 ± 8.0 |
| Heterozygote | 13.6 | 16.6 | 44 |
| Homozygote | 11.0 | 12.8 | 46.0 ± 8.0 |
The number of BFU-Es obtained in standard methylcellulose assay after 7 and 14 days of culture is expressed as a mean of 4 experiments (minimum/ maximum values).
Population-doubling time of EBV-transformed cell lines calculated during exponential growth phase by the formula T = [(t1 − t0) × log 2]/(log n1 − log n0), with n1 and n0 being the numbers of cells at times t1 and t0.
Effect of 4.1R Coimbra mutation on cell morphology.
Erythroid cells from control (R/R), heterozygous (R/CO), and homozygous (CO/CO) persons after 7 (D7) and 14 (D14) days of culture. No obvious morphologic changes were noticed in CO cells in comparison with the control cells. Bar indicates 10 μm.
Effect of 4.1R Coimbra mutation on cell morphology.
Erythroid cells from control (R/R), heterozygous (R/CO), and homozygous (CO/CO) persons after 7 (D7) and 14 (D14) days of culture. No obvious morphologic changes were noticed in CO cells in comparison with the control cells. Bar indicates 10 μm.
Effect of 4.1R Coimbra mutation on exon 20 splicing
Exon 20 mutation generates a partial or total skipping of the exon in reticulocyte RNA.17 We asked whether this mutation acts in a similar fashion in dividing erythroid and lymphoid cells. RT-PCR amplification of the exon 19–exon 21 region yielded 2 shorter bands, corresponding to CO.1 and CO.2 (Figure 1). In heterozygous cells, the smaller bands were present along with the normal fragment, whereas they were the exclusive products in homozygous CO cells (not shown). These data suggested that unlike the tissue- and/or stage-specific behavior of 4.1R alternatively spliced motifs,2-4 the exon 20 mutation is associated with the same splicing alteration in different nucleated cells and during erythroid development.
Expression of 4.1R variant isoforms in mitotic erythroblasts and lymphoid cells
To assess the expression of CO.1 and CO.2 and their assembly at the spindle poles of dividing cells, a series of double labeling experiments using various anti-4.1R antibodies and anti-NuMA antibody were performed on erythroid and lymphoid cells.
The 4.1R displays a punctate distribution within the cytoplasm and the nucleus of interphasic normal cells,6,7 with no obvious colocalization with the nuclear protein NuMA, which appears uniformly distributed, as shown previously14 (Figure3). In mitotic cells, NuMA accumulates almost entirely at the poles of the mitotic spindle in a crescent-shaped distribution. The 4.1R colocalizes with NuMA at the spindle poles of mitotic cells. However, this colocalization only partly covers the NuMA labeling at the poles and seems in some cases to be concentrated at a single central spot, suggestive of the centrosome (Figure 3).
Immunolocalization of 4.1R and NuMA in erythroid and lymphoid cells.
Column A shows 4.1 isoform distribution; column B, NuMA distribution; column C, colocalization of various 4.1 isoforms and NuMA; column D, higher magnification of the mitotic cells shown in column C; column E, results obtained with the use of lymphoid cells. In all of these, only the high magnifications are shown. Bar indicates 5 μm. AbCT gave a positive reaction with 4.1R in the control and with CO.2 in the patient. AbCO recognized only the missense sequence of CO.1 in the patient (note the complete absence of fluorescence in the control, lane 3). Normal 4.1R (control), and CO.2 and CO.1 (homozygote) showed a punctate distribution within the cytoplasm and the nucleus of interphasic and dividing cells. The 4.1R partly concentrated at the center of each mitotic pole in dividing cells. AbNuMA gave a positive reaction in both the control and the homozygote. NuMA had a diffuse distribution in the nucleus in interphasic cells and concentrated almost entirely in the mitotic spindle poles in dividing cells. The 4.1R and CO.2, on the one hand, and AbNuMA, on the other, colocalized in control and homozygote mitotic cells, while CO.1 was missing from the spindle poles (AbCO plus AbNuMA; lane 4, columns C, D, and E).
Immunolocalization of 4.1R and NuMA in erythroid and lymphoid cells.
Column A shows 4.1 isoform distribution; column B, NuMA distribution; column C, colocalization of various 4.1 isoforms and NuMA; column D, higher magnification of the mitotic cells shown in column C; column E, results obtained with the use of lymphoid cells. In all of these, only the high magnifications are shown. Bar indicates 5 μm. AbCT gave a positive reaction with 4.1R in the control and with CO.2 in the patient. AbCO recognized only the missense sequence of CO.1 in the patient (note the complete absence of fluorescence in the control, lane 3). Normal 4.1R (control), and CO.2 and CO.1 (homozygote) showed a punctate distribution within the cytoplasm and the nucleus of interphasic and dividing cells. The 4.1R partly concentrated at the center of each mitotic pole in dividing cells. AbNuMA gave a positive reaction in both the control and the homozygote. NuMA had a diffuse distribution in the nucleus in interphasic cells and concentrated almost entirely in the mitotic spindle poles in dividing cells. The 4.1R and CO.2, on the one hand, and AbNuMA, on the other, colocalized in control and homozygote mitotic cells, while CO.1 was missing from the spindle poles (AbCO plus AbNuMA; lane 4, columns C, D, and E).
The homozygous CO cells contained no full-length 4.1R, but only CO.1 and CO.2 truncated forms. Therefore, AbCT- and AbCO-specific antibodies must necessarily recognize only CO.2 or CO.1, respectively (Figure1).17 AbCT produced the same result as in control cells, suggesting a colocalization of CO.2 isoform with NuMA at the spindle poles. AbCO showed a similar punctate immunofluorescence labeling in CO cells, but virtually no staining in control cells. However, this antibody failed to show a colocalization with NuMA at the mitotic spindle poles in CO cells. It therefore appears that (1) both CO.1 and CO.2 are expressed at the protein level in both cell types (Figure 3) and preferentially condense at the plasma membrane of maturing erythroblasts (not shown), confirming our previous data stating a proper assembly of these truncated forms to the red cell skeleton17; and (2) CO.2, but not CO.1, assembles to the spindle poles in CO cells and colocalizes with NuMA. Moreover, CO.1 and CO.2 distribution is very similar in mitotic erythroid and lymphoid cells, as shown in Figure 3.
CO.2, but not CO.1, assembles to the centrosome
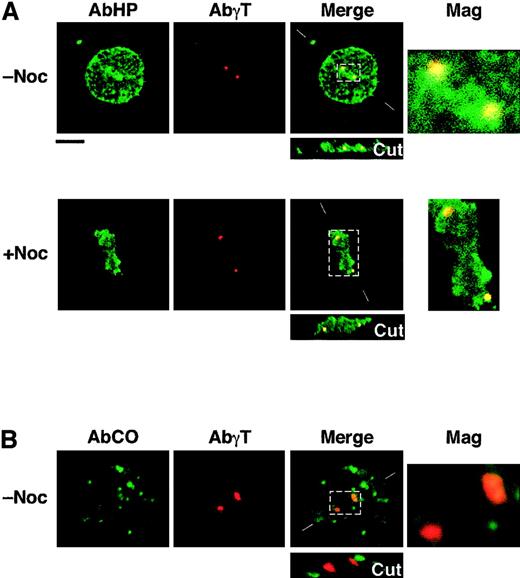
In light of previous in vitro binding assays,14 we hypothesized that none of the shortened 4.1R Coimbra isoforms would bind to NuMA. Therefore, it was rather unexpected to find CO.2 assembled at the spindle poles of CO cells. To test whether this assembly is dependent on NuMA, we performed a new set of immunofluorescence labeling in microtubule-destabilizing conditions (Figure 4). NuMA is known to be a pericentriolar protein; its assembly occurs at the minus ends of microtubules.16 Cells were treated with nocodazole prior to immunofluorescence labeling with AbHP. As shown in Figure 4, AbHP decorates 2 spots in mitotic cells and a single spot in some interphasic cells (not shown). This staining coincides with γ-tubulin, an inherent centrosomal component, in double-labeling experiments (Figure 4). Note that the colocalization of 4.1R and γ-tubulin is in most cases only partial and asymmetric; this observation is consistent with previous work by Krauss et al.9 Anti-NuMA antibody showed a diffused immunofluorescence in nocodazole-treated cells in mitosis, whereas DAPI (4,6-diamidino-2-phenylindole) staining ascertained the chromosome condensation at the median region between the centrosomes (not shown).
Immunolocalization of shortened CO.1 and CO.2 isoforms at the centrosome.
Lymphoid control (not shown) and CO cells (panels A and B) were cultured in the absence (−Noc) or presence (+Noc) of nocodazole. (A) Double staining with AbHP and AbγT revealed the presence of 4.1R epitopes at the centrosome, in a microtubule-independent manner. Further staining with specific antibodies ascertained the absence of NuMA at the spindle poles in nocodazole-treated cells and the chromosome condensation between the centrosomes. (B) Staining with AbCO antibody showed a punctate distribution of CO.1 isoform throughout the cell, but failed to show a specific localization at the centrosome, in both microtubule-stabilizing and microtubule-destabilizing (not shown) conditions. Bar indicates 10 μm.
Immunolocalization of shortened CO.1 and CO.2 isoforms at the centrosome.
Lymphoid control (not shown) and CO cells (panels A and B) were cultured in the absence (−Noc) or presence (+Noc) of nocodazole. (A) Double staining with AbHP and AbγT revealed the presence of 4.1R epitopes at the centrosome, in a microtubule-independent manner. Further staining with specific antibodies ascertained the absence of NuMA at the spindle poles in nocodazole-treated cells and the chromosome condensation between the centrosomes. (B) Staining with AbCO antibody showed a punctate distribution of CO.1 isoform throughout the cell, but failed to show a specific localization at the centrosome, in both microtubule-stabilizing and microtubule-destabilizing (not shown) conditions. Bar indicates 10 μm.
AbCO once again revealed CO.1 isoforms scattered within the cytoplasm and the nucleoplasm, but none was assembled at the centrosome (Figure 4).
All together, these results suggest that CO.2, but not CO.1, assembles to the centrosome in a NuMA- and microtubule-independent fashion.
Coexpression at the spindle poles of both 4.1R and 4.1G
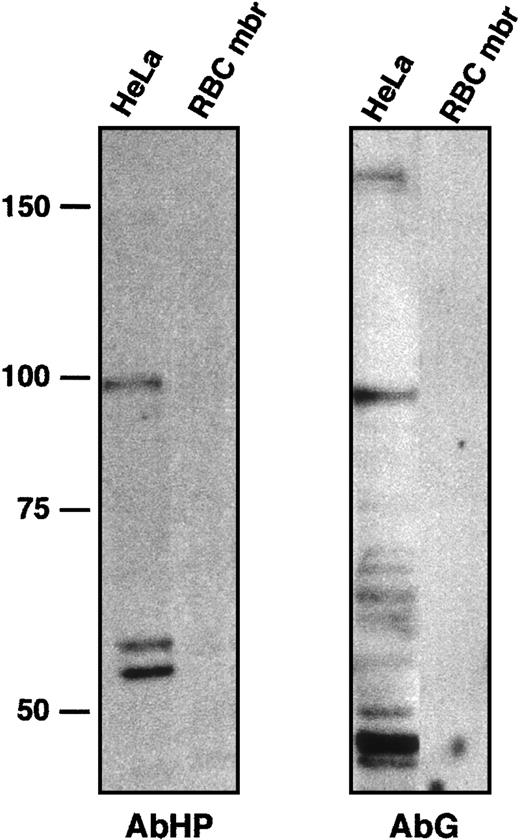
The anti-4.1R CTD antibody AbCT used here was directed against a peptide encoded by exon 21.17 However, this peptide showed only a 2–amino acid difference with its homolog in 4.1G (Figure 1C). In fact, AbCT does recognize a recombinant 4.1G C-terminal peptide upon Western blotting (GST/G; see “Patients, materials, and methods”). To ask whether the fluorescent labeling reflected a cross-reaction with 4.1G, and whether 4.1G compensated for 4.1R deficiency, we performed new sets of immunohistochemical observations using antibodies AbHG13 and AbHP. AbHG13 was raised against a 4.1G human sequence. This sequence is within the most divergent region among 4.1 gene family members2,5,18,19; it corresponds to exon 13–encoded peptide on 4.1R. AbHP was directed against the N-terminal extension of human 4.1R. This unique N-terminal region is not homologous among 4.1R family members.18 AbHG13 was tested for its specificity by immunoblotting, with the use of HeLa cell lysate and red blood cell membrane proteins (Figure 5). The same blot was reprobed with AbHP to confirm the absence of cross-reaction between these 2 antibodies (Figure 5).
Characterization of anti-4.1G antibody AbG.
To test a possible cross-hybridization between AbHP and AbG, HeLa whole-cell extract and red blood cell membrane (RBC mbr) samples were immunoblotted with AbHP (left panel). The blot was then stripped and probed with AbG (right panel). Red blood cell membrane proteins served as negative control for both antibodies. AbHP revealed bands around 98 and 54 to 59 kDa, comparable in size to 4.1R isoforms found in nuclear matrix.7 AbG revealed a high-molecular–weight isoform of approximately 160 kDa, similar to a full-length 4.1G isoform observed in transfected cells,5 or an endogenous 4.1G isoform found in PC12 cells.26 Other smaller and uncharacterized bands reacted with AbG, but not with AbHP. The band approximately 96 kDa in particular is slightly smaller than the 98-kDa isoform reacting with AbHP.
Characterization of anti-4.1G antibody AbG.
To test a possible cross-hybridization between AbHP and AbG, HeLa whole-cell extract and red blood cell membrane (RBC mbr) samples were immunoblotted with AbHP (left panel). The blot was then stripped and probed with AbG (right panel). Red blood cell membrane proteins served as negative control for both antibodies. AbHP revealed bands around 98 and 54 to 59 kDa, comparable in size to 4.1R isoforms found in nuclear matrix.7 AbG revealed a high-molecular–weight isoform of approximately 160 kDa, similar to a full-length 4.1G isoform observed in transfected cells,5 or an endogenous 4.1G isoform found in PC12 cells.26 Other smaller and uncharacterized bands reacted with AbG, but not with AbHP. The band approximately 96 kDa in particular is slightly smaller than the 98-kDa isoform reacting with AbHP.
Confocal microscopy using AbHG13 revealed a strong and homogeneous labeling mostly throughout the nucleus during interphase (Figure6). These findings contrast with the punctate aspect and mostly cytoplasmic distribution of 4.1R (Figures 3and 6). These differences are better evidenced in merged images in both Figures. Most remarkably, 4.1G was concentrated almost entirely in the mitotic spindle poles within dividing cells. Upon AbHG13 and AbNuMA double staining, 4.1G and NuMA appeared to colocalize at the spindle poles. Again, this colocalization is restricted to the central area of the crescent distribution of NuMA. Interestingly, similar results were obtained in normal and CO lymphoid and erythroid cells, suggesting that 4.1G is intrinsically a component of the mitotic spindle poles.
Immunolocalization of 4.1G and NuMA in the nuclei of erythroid and lymphoid cells.
Similar results were obtained in erythroid progenitors (columns A-D) and lymphoid cells (column E): Column A shows 4.1 isoform distribution; column B, NuMA distribution; column C, colocalization of various 4.1 isoforms and NuMA; column D, higher magnification of the mitotic cells shown in column C; column E, results obtained in lymphoid cells. In all of these columns, only the high magnifications are shown. AbHG13 (AbG) gave a positive reaction in both control and patient CO cells, and it colocalized with NuMA at the mitotic spindle poles. AbHP reacted with 4.1R (control) and indiscriminately with CO.2 and CO.1 (homozygote). This antibody reproduced the punctate distribution within the cytoplasm and the nucleus of interphasic cell seen in Figure 3. Even though AbHP recognized both isoforms, CO.1 and CO.2, which are shortened at their C-termini, colocalization with NuMA necessarily stems only from CO.2 (Figure 3). Bars indicate 5 μm.
Immunolocalization of 4.1G and NuMA in the nuclei of erythroid and lymphoid cells.
Similar results were obtained in erythroid progenitors (columns A-D) and lymphoid cells (column E): Column A shows 4.1 isoform distribution; column B, NuMA distribution; column C, colocalization of various 4.1 isoforms and NuMA; column D, higher magnification of the mitotic cells shown in column C; column E, results obtained in lymphoid cells. In all of these columns, only the high magnifications are shown. AbHG13 (AbG) gave a positive reaction in both control and patient CO cells, and it colocalized with NuMA at the mitotic spindle poles. AbHP reacted with 4.1R (control) and indiscriminately with CO.2 and CO.1 (homozygote). This antibody reproduced the punctate distribution within the cytoplasm and the nucleus of interphasic cell seen in Figure 3. Even though AbHP recognized both isoforms, CO.1 and CO.2, which are shortened at their C-termini, colocalization with NuMA necessarily stems only from CO.2 (Figure 3). Bars indicate 5 μm.
AbHP reproduced the punctate distribution within the cytoplasm and the nucleus of interphasic cells as AbCT and AbCO (Figure 4). Even though AbHP indiscriminately recognized CO.1 and CO.2, colocalization with NuMA is expected to stem only from CO.2 in homozygous CO cells, which again do not express any full-length 4.1R protein (compare patient rows in Figure 3 with AbCO and in Figure 6 with AbHP).
All together, these data suggest that both 4.1R and 4.1G are expressed at the mitotic spindle poles. Whether this coexpression reflects a functional redundancy between 4.1 protein members during cell division, or whether each component is endowed with a specific role, remains to be elucidated.
Altered CTDs of 4.1R Coimbra do not interact with NuMA in in vitro binding assays
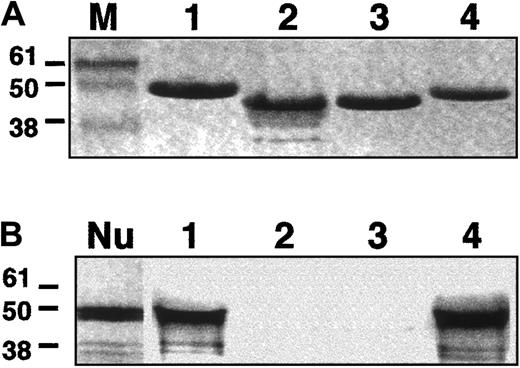
We next tested the ability of CO.1 and CO.2 to bind NuMA upon in vitro assays. Four different affinity-purified GST-fusion proteins were prepared. They included the CTDs of intact 4.1R, migrating at 41 kDa (GST/R); the altered CO.1 (GST/CO.1) and the altered CO.2 (GST/CO.2), both migrating at 38.5 kDa; and the intact 4.1G (GST/G) of 41 kDa apparent size.
Each of the fusion peptides was incubated with in vitro translated and radiolabeled NuMA C-terminal region (amino acids 1697 to 2102).14 The bound NuMA/4.1 protein complexes were then analyzed by electrophoresis. Coomassie blue staining and autoradiography were performed to detect protein 4.1 isoforms and NuMA, respectively. In agreement with previous observations,14the C-terminal region of NuMA interacted with the intact CTD of 4.1R (Figure 7), but not with 4.1R peptides lacking exon 20–encoding sequence (CO.2), or most of the exon 20– and 21–encoding sequence (CO.1). Most remarkably, NuMA also bound to 4.1G at least through its CTD, as shown in this assay. These findings, together with the immunochemical studies presented above, suggest that 4.1G is another 4.1 component of the mitotic spindle poles, where it potentially binds to NuMA.
In vitro binding assays.
A fragment (amino acids 1697-2102) of NuMA C-terminal region was tested in vitro for its binding to the CTDs of 4.1R, CO.1, CO.2, or 4.1G. (A) Coomassie blue detection of 4.1R (1), CO.1 (2), CO.2 (3), and 4.1G (4). M indicates molecular size markers (kDa). (B) Autoradiography of radiolabeled NuMA. NuMA bound to 4.1R and 4.1G (1 and 4), but not to either CO.1 or CO.2 (2 and 3). Nu: appearance of NuMA alone following the in vitro coupled transcription/translation reaction.
In vitro binding assays.
A fragment (amino acids 1697-2102) of NuMA C-terminal region was tested in vitro for its binding to the CTDs of 4.1R, CO.1, CO.2, or 4.1G. (A) Coomassie blue detection of 4.1R (1), CO.1 (2), CO.2 (3), and 4.1G (4). M indicates molecular size markers (kDa). (B) Autoradiography of radiolabeled NuMA. NuMA bound to 4.1R and 4.1G (1 and 4), but not to either CO.1 or CO.2 (2 and 3). Nu: appearance of NuMA alone following the in vitro coupled transcription/translation reaction.
Discussion
Recent studies gathered from different groups have emphasized the potential role of the CTD of 4.1R in nucleated cells. Hence, it appears to bear the 4.1R-binding site for NuMA at the mitotic spindle poles.14 It has also been shown to interact with ZO-2, a cell tight junction protein,20 and eIF3-p44, a subunit of the eukaryotic translation initiation factor 3.21 The interaction with NuMA involves the segment of protein 4.1R encoded by exons 20 and 21 (27 + 32 amino acids), that is its C-terminal region.14 The binding site for protein 4.1R lies within amino acids 1788 to 1810 on NuMA.
The 4.1R Coimbra mutation is associated with a mild elliptocytosis, which is expressed virtually only at the homozygous state.17 We showed that it stemmed from the fact that 4.1R was reduced by more than 50%, in keeping with an equivalent decrease in the corresponding mRNA, rather than from the qualitative changes in the CTD of the 2 isoforms of 4.1R generated. We assumed that protein 4.1R CTD probably had a limited role, if it had a role at all, in the membrane of the mature red cell. On the other hand, the patient failed to present any routine laboratory stigmata pointing to any abnormalities of erythropoiesis. Although preliminary, the slight decrease in growth of CO cells observed in this study appears to be genuine and would be worth further investigation using other cell-system models.
The molecular composition of the centrosome at interphase and during mitosis has been actively investigated.22 Recent works have described 2 new components of the centrosome: 4.1R and CPAP.9,10 A direct association between the HP of 4.1R 135-kDa isoform and CPAP has been demonstrated.10 CPAP localizes to the centrosome throughout the cell cycle and was defined as a bona fide core component of the centrosome. However, 4.1R was not detected in isolated centrosomes.10 Data presented here led to the observation that CO.1 isoform does not assemble to the spindle poles in intact cells. Nor does it assemble to the centrosomes in the presence of microtubule-disrupting drugs, whereas CO.2 assembles to the spindle poles and to the centrosomes, but does not bind to NuMA in vitro. These results collectively suggest that 4.1R might be located at the vicinity of the centrosome, within a cytoskeletal protein complex, as previously suggested.10 It would bind to CPAP through its N-terminal HP, but a stable assembly would require an association to a yet-to-be-defined cytoskeletal network. This uncharacterized association would involve a direct binding of the C-terminal exon 21–encoded peptide, or at least an intact C-terminal sequence needed for a specific conformation of the whole protein.
The finding that centrosomal distribution of CO.2 and CO.1 is independent of NuMA and microtubules, the apparent contradiction between CO.2 assembly at spindle poles in vivo, and the absence of interaction with NuMA in vitro all further support the view that the defined bindings of HP with CPAP on the one hand and the CTD with NuMA on the other, are probably not sufficient to stabilize the centrosome-cytoskeletal complex. In fact, it has been shown that NuMA is required for the mitotic spindle formation and stabilization. Its assembly to the spindle poles seems to occur through a direct interaction with dynein.15,23 The 4.1R also interacts with the dynein-dynactin complex, and it has been speculated that, at the spindle poles, 4.1R might stabilize a protein complex including NuMA, dynein, dynactin, and microtubule ends.14 Furthermore, analysis of potential 4.1R partners derived from immunoprecipitation experiments using anti-HP antibody showed that at least 15 components coimmunoprecipitate with 4.1R.14 The binding sites on 4.1R to one or the other of these proteins remain to be defined. It is however tempting to speculate that among these 4.1R partners, the dynein-dynactin complex appears to be a good candidate.
In the present study, we found that in addition to 4.1R, the mitotic spindle poles contain 4.1G, the 4.1R ubiquitous homolog. This observation was based on in vitro binding studies and immunocolocalization using anti-HP– and anti-4.1G–specific antibodies. Sequence alignment presented in Figure 1C shows that the CTDs of 4.1R and 4.1G are highly homologous. Most remarkably, the peptide sequences encoded by exons 20 and 21 in 4.1R are almost identical to their counterparts in 4.1G. The strong homology between these parts of 4.1R and 4.1G presumably accounted for the ability of 4.1G to bind NuMA. These findings raised the question as to whether the coexpression of these related 4.1 family members reflects a functional redundancy. The finding that 4.1R−/− knockout mice show no apparent abnormality of cell division and have instead a normal viability and fertility24 is very supportive of the idea that a 4.1 family member, probably 4.1G, can rescue for 4.1R function. Equally conceivable is the occurrence of phenotypic manifestations, associated with 4.1R deficiencies, only in mature red cells. The 4.1G is indeed virtually absent from red cell membrane (see Figure 5), and therefore could not compensate for 4.1R function at the red cell membrane skeleton.
It is not surprising that redundant proteins occur in relation with essential cellular functions, such as cell division. For instance, dynein and kinesin microtubule-based motor superfamilies act within the mitotic spindles to segregate replicated chromosomes to progeny cells. Seven of these motors have been identified in yeast, but owing to functional overlap or redundancy, none of them seems to be individually essential for cell division.25 It is therefore tempting to hypothesize that at least the 2 4.1 members, 4.1R and 4.1G, would play similar and perhaps complementary roles in stabilizing spindle pole protein complexes. It would be of interest to simultaneously alter the expression of both proteins in cells and to examine the effect on spindle formation and on cell division.
We are most grateful to Dr Shu-Ching Huang for critical discussion, A. Calender for establishing the lymphoblastoid cell lines, and Dr T. K. Tang for kindly providing us with anti-HP antibodies.
Supported by grants from the Institut National de la Santé et de la Recherche Médicale, the Fondation de France, the Association Française contre les Myopathies, the Assistance Publique-Hôpitaux de Paris, the Faculté de Médecine Paris Sud, the Association pour la Recherche sur le Cancer, and the Centre National de la Recherche Scientifique.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Faouzi Baklouti, Centre de Génétique Moléculaire et Cellulaire CNRS UMR 5534; UniversitéClaude-Bernard Lyon 1; Bât G Mendel; 16 rue Dubois; 69622 Villeurbanne Cedex, France; e-mail: baklouti@univ-lyon1.fr.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal