The p47-phox gene, NCF-1, has 2 nearly identical pseudogenes (ψNCF-1) in proximity at chromosomal locus 7q11.23. A dinucleotide deletion (ΔGT) at the beginning of exon 2 that leads to a frameshift and premature stop codon is considered the signature sequence of the pseudogenes. It is also the most prevalent mutation in p47-phox–deficient (A47°) chronic granulomatous disease (CGD) as a result of the insertion of a ΔGT-containing fragment of pseudogene into NCF-1.Extending our study of the relationship between NCF-1 andψNCF-1 to 53 unaffected control individuals, we found that although in most (n = 44), the ratio of pseudogene (ΔGT) to functional gene (GTGT) sequence in amplicons spanning exon 2 was 2:1, as previously observed, surprisingly, in 7 persons the ratio was 1:1, and in 2 persons the ratio was 1:2. The lowered ratios are explained by the presence, in a heterozygous or homozygous state, respectively, of a pseudogene that contains GTGT rather than ΔGT. It is possible that this pseudogene has not undergone deletion of GT, but more likely, based on analysis of additional NCF-1/ψNCF-1 markers, it represents the previously unidentified product of the reciprocal crossover of DNA fragments between the functional gene and one of its pseudogenes. The mutated NCF-1 resulting from this event is the predominant A47°CGD allele. The existence of 2 extended haplotypes encompassing NCF-1/ψNCF-1 further complicates the detection of A47°CGD carriers. Although most have a ΔGT/GTGT ratio of 5:1, some have a ratio of 2:1 and are indistinguishable by this means from unaffected individuals.
Introduction
In stimulated normal phagocytes, reduced nicotinamide adenine dinucleotide phosphate (NADPH) oxidase catalyzes the reduction of oxygen to superoxide. Superoxide and its more potent microbicidal derivatives (eg, hydrogen peroxide, hypohalous acids) are important for killing invading microorganisms. In chronic granulomatous disease (CGD), an uncommon inherited disorder of the innate immune system, the primary defect occurs in any 1 of 4 genes encoding phox proteins of the phagocyte NADPH oxidase complex. Gp91-phox and p22-phox together form flavocytochrome b558, the catalytic core of the enzyme, whereas p47-phox and p67-phox are found in the soluble fraction of resting phagocytes. The association of p47-phox and p67-phox with the flavocytochrome at the plasma or phagosomal membrane is crucial for enzyme activation. In CGD patients, phagocyte NADPH oxidase activity is absent or occurs at very low levels. Consequently, patients are highly susceptible to severe, sometimes fatal, recurrent bacterial and fungal infections (reviewed by Roos and Curnutte1 and Segal et al2).
The p47-phox–deficient form of CGD (A47°CGD), which accounts for about 20% of all CGD cases, is inherited in an autosomal recessive manner and caused by mutations in the gene NCF-1.A47°CGD is unique among the 4 forms of the disease in that a common mutation has been identified in approximately 95% of affected alleles analyzed worldwide. Ninety-five of 104 unrelated patients reported to date were homozygous and a further 7 were heterozygous for a dinucleotide deletion (ΔGT) in a GTGT sequence at the beginning of exon 2 of NCF-1.3-9 The deletion predicts a frameshift and a premature stop codon at residue 51 and leads to complete absence of p47-phox protein from the patients' phagocytes (A47°CGD).3 Only 8 other mutations have been identified in NCF-1.4,8,10,11 In contrast, the mutations that cause the other forms of CGD are highly heterogeneous, with many of them being specific to each affected family (mutations and primary references are tabulated by Cross et al11 and Heyworth et al12). A47°CGD is also unusual in that it is at least 4 times more common than the other autosomal recessive forms of the disease. Defects in the genes for p22-phox (CYBA) and p67-phox (NCF-2) each account for approximately 5% or less of all cases. The remaining 70% of cases are inherited in an X-chromosome–linked manner and are caused by mutations in the gp91-phox gene, CYBB.1 13
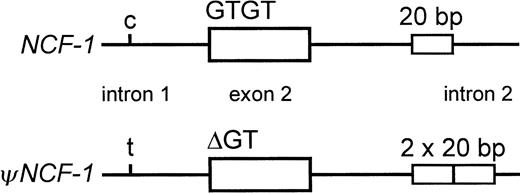
NCF-1 has at least 2 pseudogenes, each of which is highly homologous (approximately 98% identical) to the functional gene and colocalizes with it to chromosome 7q11.23.7,14-18 TheNCF-1 pseudogenes (ψNCF-1) are distinguished from the functional gene by 3 well-characterized differences (Figure1). One of them, considered the signature sequence of the pseudogenes, is the GT deletion at the beginning of exon 2 that causes CGD when it occurs in the functional gene. The others are a C>T transition in intron 1, 122–base pair (bp) upstream of the 5′ end of exon 2, and a 20-bp duplication 176 bp downstream from the 5′ end of intron 2. It is now apparent that the relatively high incidence of A47°CGD and the predominance of the ΔGT mutation are due to recombination events between NCF-1 and its pseudogenes resulting from their proximity, their high degree of similarity, and the presence within each gene of multiple recombination hot spots.7,9,15,19 Interestingly, these genes occur in a region of chromosome 7 that contains large (approximately 200-400 kilobase [kb]) duplicated segments (duplicons) of DNA, in each of which lies a single copy of NCF-1 or ψNCF-1, in addition to other gene/pseudogene sequences. The complex 7q11.23 region, which has been difficult to map because of the high level of duplication, has been intensively studied because it also contains the locus of Williams-Beuren syndrome.16-18 20
Differences between NCF-1 and its pseudogenes.
Within a small segment of the gene, the most well-characterized differences between NCF-1 and its pseudogenes(ψNCF-1) are shown. These are a C/T transition in intron 1 at −122 bp from the start of exon 2, GTGT or ΔGT at the beginning of exon 2, and the single or duplicated 20-bp stretch in intron 2 at +176 bp from the end of exon 2.
Differences between NCF-1 and its pseudogenes.
Within a small segment of the gene, the most well-characterized differences between NCF-1 and its pseudogenes(ψNCF-1) are shown. These are a C/T transition in intron 1 at −122 bp from the start of exon 2, GTGT or ΔGT at the beginning of exon 2, and the single or duplicated 20-bp stretch in intron 2 at +176 bp from the end of exon 2.
The minimum incidence of CGD was recently estimated at between 1 per 200 000 and 1 per 250 000 live births.13 Therefore, the estimated incidence of A47°CGD is approximately 1 per million births, from which the carrier frequency can be calculated as 1 per 500 individuals. The high degree of homology between NCF-1 andψNCF-1, which results in the coamplification of DNA strands from the functional gene and its pseudogenes with most oligonucleotide primers, complicates the molecular analysis of families affected by A47°CGD as well as the detection of the carrier state in other individuals. The majority of patients (ie, with the common ΔGT/ΔGT genotype) can be detected with relative ease because the GTGT-containing sequence is absent from polymerase chain reaction (PCR) products encompassing exon 2. Recently, we also described an allele-specific strategy to simplify the detection of rare non-ΔGT mutations in NCF-1.8 The main difficulty lies in confidently diagnosing the common carrier state (GTGT/ΔGT) because the genomes of all unaffected individuals also include DNA sequence with the GT deletion within the p47-phox pseudogenes. To determine whether the ratio of pseudogene (ΔGT) to functional gene (GTGT) sequence8 can be used reliably to identify carriers of A47°CGD,21 we studied in more detail the relationship between NCF-1 and ψNCF-1 in unaffected control individuals and obligate carriers of the disease. In so doing, we identified a novel form of ψNCF-1 that does not contain ΔGT. The presence of this pseudogene sheds new light on the recombination events that cause A47°CGD but further complicates the detection of the ΔGT carrier state.
Materials and methods
Collection of blood samples and preparation of DNA
Protocols and consent forms for the collection of blood samples were approved by the Human Subjects Committee of the Scripps Office for the Protection of Research Subjects. Informed consent was obtained according to the Declaration of Helsinki. Whole blood was collected with EDTA (ethylenediaminetetraacetic acid) as an anticoagulant, and genomic DNA was isolated using the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN). Custom-synthesized oligonucleotide primers were purchased from Sigma-Genosys (The Woodlands, TX).
Determination of the ratio of ΔGT- to GTGT-containing sequence
Two independent methods were used to estimate the ratio of ΔGT- to GTGT-containing sequence in genomic DNA samples. In the first method, exon 2 of NCF-1/ψNCF-1 was amplified using intronic primers 2LB2 (GTGCACACAGCAAAGCCTCT) and 2RB2 (CTAAGGTCCTTCCCAAAGGGT). Reaction conditions have been described previously.8 PCR-amplified fragments were purified using a QIAquick PCR purification kit (Qiagen, Valencia, CA) and sequenced using the ABI Prism BigDye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, CA) and an ABI Prism 310 Genetic Analyzer. Nucleotide peak heights were measured directly using the instrument software over a 27-bp stretch of sequence, corresponding to nucleotides 81 to 107 of the p47-phox cDNA. At each position, the ratio of nucleotide peak heights was calculated and a mean ratio was determined for the entire section. Peak heights were not measured at the 3 positions within this stretch at which nucleotides in the pseudogene and functional gene coincided (Figure2). When peak heights were measured over the entire exon, the calculated ratios were very similar to those obtained by measurements over the 27-bp stretch.
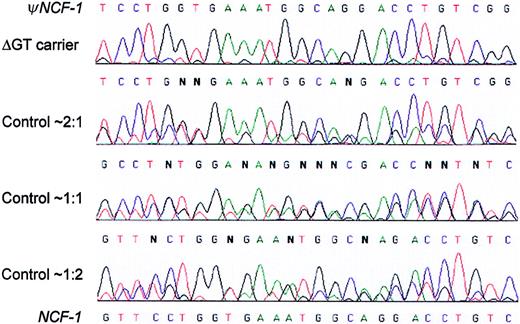
Sequencing electropherograms distinguish among 3 differentψNCF-1/NCF-1 genotypes in the normal population, as well as carriers of the common NCF-1 ΔGT mutation.
Genomic DNA from control individuals unaffected by CGD and obligate carriers of A47°CGD was amplified using primers 2LB2 and 2RB2, which do not distinguish between NCF-1 and ψNCF-1.The figure shows representative electropherograms that cover a 27-bp stretch starting 9 nucleotides downstream from the start of exon 2 (ie, nucleotides 81-107 of the NCF-1 cDNA). In each case, double sequence is observed because it diverges after the initial GT at the start of the exon (not shown). The nucleotide sequences of the pseudogenes and functional gene are shown at the top and bottom of the figure, respectively. Unaffected control individuals fell into 3 distinct groups with approximate ΔGT/GTGT sequence ratios of 2:1, 1:1, and 1:2, as shown, based on peak height measurements. This is well illustrated, for example, by the G (ψNCF-1) and C(NCF-1) at the fifth position. Most obligate carriers of the ΔGT mutation had a much higher ratio of ΔGT (pseudogene) sequence, as seen in the top electropherogram.
Sequencing electropherograms distinguish among 3 differentψNCF-1/NCF-1 genotypes in the normal population, as well as carriers of the common NCF-1 ΔGT mutation.
Genomic DNA from control individuals unaffected by CGD and obligate carriers of A47°CGD was amplified using primers 2LB2 and 2RB2, which do not distinguish between NCF-1 and ψNCF-1.The figure shows representative electropherograms that cover a 27-bp stretch starting 9 nucleotides downstream from the start of exon 2 (ie, nucleotides 81-107 of the NCF-1 cDNA). In each case, double sequence is observed because it diverges after the initial GT at the start of the exon (not shown). The nucleotide sequences of the pseudogenes and functional gene are shown at the top and bottom of the figure, respectively. Unaffected control individuals fell into 3 distinct groups with approximate ΔGT/GTGT sequence ratios of 2:1, 1:1, and 1:2, as shown, based on peak height measurements. This is well illustrated, for example, by the G (ψNCF-1) and C(NCF-1) at the fifth position. Most obligate carriers of the ΔGT mutation had a much higher ratio of ΔGT (pseudogene) sequence, as seen in the top electropherogram.
In the second method, radiolabeled PCR products encompassing the start of exon 2 were separated on denaturing acrylamide gels, and quantified. DNA from 62 bp 5′ from the start of exon 2 through 38 bp into exon 2 was amplified using 32P end-labeled 2LB2 and unlabeled cDNA2R (TCCGACAGGTCCTGCCA). Primer 2LB2 was end-labeled using 90 ng 2LB2, 180 μCi (6.66 MBq) γ-32P adenosine triphosphate, 0.5 μL T4 10× buffer, and 2.5 U T4 polynucleotide kinase in a final volume of 5 μL. The radiolabeling mixture was incubated at 37°C for 30 minutes, after which the kinase was inactivated by heating to 65°C for 20 minutes. The amplification reaction was performed in the following buffer: 33.5 mM Tris-HCl (pH 8.8), 8.3 mM (NH4)2SO4, 3.35 mM MgCl2, 85 μg/mL bovine serum albumin, 5% dimethylsulfoxide, 0.125 mM each deoxyribonucleoside triphosphate, 90 ng cDNA2R, 90 ng 32P-labeled 2LB2, 2.5 U AmpliTaq polymerase, and 100 ng DNA. An initial denaturation for 3 minutes at 94°C was followed by 40 cycles at 94°C for 5 seconds and 70°C for 1 minute, with a final extension for 15 minutes at 72°C. This PCR resulted in a 98-bp or 100-bp fragment, depending on whether the sequence contained ΔGT or GTGT. For separation of the fragments, 5 μL PCR product was mixed with 4 μL denaturing dye solution, denatured for 5 minutes at 95°C, chilled on ice, and then run on a 6% acrylamide sequencing gel (SequaGel 6; National Diagnostics, Atlanta, GA) for 3 hours at 50 to 55 W. The radiolabeled bands were quantified either using a Cyclone Storage Phosphor System and OptiQuant image analysis software (Packard Instrument, Meriden, CT) or by Cerenkov counting following autoradiography and their excision.
Allele-specific PCR
To avoid coamplification of DNA with the GT deletion, we used an allele-specific PCR strategy to amplify the entire functional NCF-1 and, where present, the GTGT-containingψNCF-1. The amplification reactions, which are described fully by Noack et al,8 used either forward or reverse primers including the GTGT at the beginning of exon 2. Allele specificity of the reactions was checked by sequencing the reaction products to ensure that only GTGT-containing sequence was present.7 8 Purification and sequencing of amplified fragments were performed as described above. Sequence numbering in this report is based on the convention that +1 is the A of the ATG initiator codon. This is 12 nucleotides less than the numbering of the cDNA sequence for NCF-1 in GenBank (accession numbers M25665and M26193).
Allele-specific RT-PCR
Total RNA was isolated from whole blood using the RNeasy Blood Mini Kit (Qiagen). Reverse transcription (RT)–PCR was performed exactly as described previously using the SuperScript Preamplification System for first-strand cDNA synthesis.8 Briefly, 2 allele-specific PCRs were performed. In the first, a fragment from the beginning of exon 1 to the beginning of exon 2 was amplified with primers cDNA1F and GTGT-R. In the second, primers cDNAGTGT and cDNA11R were used to amplify from the start of exon 2 to the end of exon 11. Primer sequences are given in Table 1 of the previous publication.8 The allele specificity of the RT-PCR products was checked by sequencing into the region of the allele-specific primer to ensure that only GTGT sequence was present.
The ratio of ΔGT- to GTGT-containing sequence in unaffected individuals and carriers of A47°CGD
| Population group . | Method 1: electropherogram . | Method 2: PCR product size* . | Average‡ . | ||
|---|---|---|---|---|---|
| No. . | Ratio† . | No. . | Ratio† . | ||
| Unaffected controls | 44 | 2.25 ± 0.14 | 8 | 1.66 ± 0.12 | 2.16 (2:1) |
| 7 | 1.02 ± 0.05 | 4 | 0.86 ± 0.07 | 0.96 (1:1) | |
| 2 | 0.46 ± 0.03 | 2 | 0.46 ± 0.02 | 0.46 (1:2) | |
| A47°CGD carriers | 7 | 5.97 ± 0.64 | 7 | 3.69 ± 0.23 | 4.83 (5:1) |
| 2 | 2.27 ± 0.01 | — | nd | 2.27 (2:1) | |
| Population group . | Method 1: electropherogram . | Method 2: PCR product size* . | Average‡ . | ||
|---|---|---|---|---|---|
| No. . | Ratio† . | No. . | Ratio† . | ||
| Unaffected controls | 44 | 2.25 ± 0.14 | 8 | 1.66 ± 0.12 | 2.16 (2:1) |
| 7 | 1.02 ± 0.05 | 4 | 0.86 ± 0.07 | 0.96 (1:1) | |
| 2 | 0.46 ± 0.03 | 2 | 0.46 ± 0.02 | 0.46 (1:2) | |
| A47°CGD carriers | 7 | 5.97 ± 0.64 | 7 | 3.69 ± 0.23 | 4.83 (5:1) |
| 2 | 2.27 ± 0.01 | — | nd | 2.27 (2:1) | |
nd indicates not determined.
All samples included under method 2 were also analyzed using method 1.
Mean ± SD.
Average of the 2 methods with predicted ratio in parentheses.
Results and discussion
We have shown previously that examination of the electropherogram peak heights for the p47-phox pseudogene and functional gene sequence in exon 2 can be a useful, preliminary guide for distinguishing between patients with the common ΔGT/ΔGT genotype and those with rare non-ΔGT mutations.8 In the relatively small number of control individuals (unaffected by CGD) included in that previous study, the ratio of ΔGT-containing sequence to GTGT-containing sequence was approximately 2:1, consistent with the presence of 2 copies of ψNCF-1 and a single copy of the functional gene.15-17 In obligate heterozygous carriers (ΔGT/GTGT) of the predominant form (ΔGT/ΔGT) of A47°CGD, the peak height ratios were predictably much higher (approximately 5:1).8
Three different ΔGT/GTGT ratios in the general population
Our earlier findings therefore raised the possibility that a simple measurement of the ratio of ΔGT to GTGT sequence could be used to reliably identify carriers of the ΔGT mutation. Indeed, a method using this general principle has recently been published.21 To study further the relationship between theNCF-1 and ψNCF-1 genes and to assess the reliability of this technique, we analyzed DNA from a group of 53 unaffected and unrelated control individuals and additional obligate carriers of the disease. Using oligonucleotide primers that do not distinguish between the gene and its pseudogenes, we amplified and sequenced fragments spanning exon 2. We initially compared electropherogram peak heights over a 27-bp stretch of double sequence because this method had yielded results in our previous study consistent with published data. As shown in Figures 2 and3, analysis of genomic DNA from this large group of control individuals revealed that 3 distinct electropherogram patterns with different ΔGT/GTGT sequence ratios were reproducibly obtained. In 44 of the 53 control individuals (Figure3), the ratio of ΔGT sequence to GTGT sequence was approximately 2:1 (2.25 ± 0.14; mean ± SD), as we had previously observed and consistent with data from human genome sequencing. In 7 persons, the amount of GTGT sequence was approximately equal to that of the ΔGT sequence, giving a ratio of 1:1 (1.02 ± 0.05). Surprisingly, in DNA from 2 control subjects, the ratio of ΔGT to GTGT sequence was completely reversed at approximately 1:2 (0.46 ± 0.03). As previously observed,8 21 most obligate carriers of the ΔGT mutation had a much higher ratio of ΔGT to GTGT sequence (Figure 2, top).
Estimated ratios of ΔGT/GTGT sequence in the normal population and in carriers of A47°CGD.
The ratios of ΔGT- to GTGT-containing sequence in 53 unrelated control individuals unaffected by CGD were determined from electropherogram peak height measurements as described in “Materials and methods.” Within this group, separation of symbols on the horizontal axis is for purposes of clarity only. DNA samples from 9 carrier parents of A47°CGD patients homozygous for ΔGT (ΔGT/ΔGT) were analyzed in the same way. For the experiment shown in the final column, DNA from each of 3 controls (2:1 ratio) was mixed with an equal amount of DNA from a ΔGT/ΔGT patient prior to PCR amplification and estimation of the ΔGT/GTGT ratio. Mean values are presented in Table 1, together with confirmatory data using a second, independent method.
Estimated ratios of ΔGT/GTGT sequence in the normal population and in carriers of A47°CGD.
The ratios of ΔGT- to GTGT-containing sequence in 53 unrelated control individuals unaffected by CGD were determined from electropherogram peak height measurements as described in “Materials and methods.” Within this group, separation of symbols on the horizontal axis is for purposes of clarity only. DNA samples from 9 carrier parents of A47°CGD patients homozygous for ΔGT (ΔGT/ΔGT) were analyzed in the same way. For the experiment shown in the final column, DNA from each of 3 controls (2:1 ratio) was mixed with an equal amount of DNA from a ΔGT/ΔGT patient prior to PCR amplification and estimation of the ΔGT/GTGT ratio. Mean values are presented in Table 1, together with confirmatory data using a second, independent method.
Measuring electropherogram peak height ratios is not necessarily a reliable way to estimate relative numbers of each gene because it requires not only equal efficiency of the NCF-1 andψNCF-1 DNA amplification reactions, but also quantitative data from the sequencing reaction. To verify our results, we used a second independent method, using the same upstream primer (2LB2) end-labeled with 32P and a different downstream primer (cDNA2R). Representative results using this method to analyze the ΔGT/GTGT ratios in control individuals and A47°CGD carriers are shown in Figure 4. The pooled data in Table 1 show that the results from the 2 methods were generally in good agreement. The largest difference occurred in the group of 7 A47°CGD carriers, where the frequency of the functional gene appeared to be consistently underestimated based on peak height measurements and overestimated using the gel-based technique, assuming a theoretical value of 5 (see below). However, the mean value with the 2 methods (4.83) was very close to the theoretical value and in good agreement with the results of Dekker et al,21 who used a similar technique with a fluorochrome-labeled forward primer.
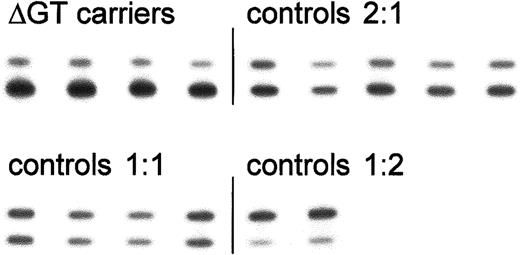
Analysis of the ratio of ΔGT- to GTGT-containing sequence based on PCR product size.
32P-labeled PCR products encompassing the start of exon 2 from control individuals and carriers of the ΔGT mutation were separated on denaturing acrylamide sequencing gels. Bands were detected by autoradiography (as shown here) or in a Cyclone storage phosphor system. The upper (100 bp) and lower (98 bp) bands represent fragments containing the functional GTGT or the ΔGT mutation, respectively. Results from a single representative experiment are shown; mean values for each group are presented in Table 1.
Analysis of the ratio of ΔGT- to GTGT-containing sequence based on PCR product size.
32P-labeled PCR products encompassing the start of exon 2 from control individuals and carriers of the ΔGT mutation were separated on denaturing acrylamide sequencing gels. Bands were detected by autoradiography (as shown here) or in a Cyclone storage phosphor system. The upper (100 bp) and lower (98 bp) bands represent fragments containing the functional GTGT or the ΔGT mutation, respectively. Results from a single representative experiment are shown; mean values for each group are presented in Table 1.
ψNCF-1 containing GTGT at the start of exon 2
The ΔGT/GTGT ratios of approximately 2, 1, and 0.5 that we have determined experimentally in unaffected controls are most easily explained by the presence in the general population of a second, previously unidentified type of NCF-1 pseudogene (or duplicate copy of the gene) containing GTGT at the start of exon 2 rather than the signature ΔGT. We refer to this novel pseudogene as Type II and the more common ΔGT-containing pseudogenes as Type I, but for this classification we do not take into account possible single-nucleotide differences between pseudogenes.15 The presence of 2 types of pseudogenes in the general population would generate at least 2 extended NCF-1 gene/pseudogene haplotypes. The more common haplotype would have 2 copies of Type IψNCF-1 and one copy of the functional NCF-1.The less common haplotype would have one copy each of Type I and Type II ψNCF-1 and one copy of the functional gene. The 3 genotypes that result from different combinations of these extended haplotypes are shown in Figure 5, together with the theoretical ΔGT/GTGT ratios. Our observed ratios (Table 1) match these values very closely. The data in the table indicate that 11 of 106 (about 10%) of the chromosomes 7 analyzed (in genomic DNA from 53 control individuals) contained the Type II pseudogene, as 7 persons had ΔGT/GTGT ratios of approximately 1.0 (heterozygous for Type II ψNCF-1) and 2 had ratios of approximately 0.5 (homozygous for Type II ψNCF-1). Dekker et al21 also identified one individual, out of a control group of 16, who had equal amounts of ΔGT- and GTGT-containing sequence.
Predicted
ψNCF-1/NCF-1 genotypes based on observed ΔGT/GTGT ratios in unaffected individuals and in carriers and patients with A47°CGD. We have identified unaffected control individuals with 3 different ratios of ΔGT/GTGT sequence in exon 2 of theψNCF-1/NCF-1 genes: 2:1, 1:1, or 1:2. The most likely explanation for these ratios is the existence in the population of 2 main extended haplotypes (see “Results and discussion”), giving rise to the ψNCF-1/NCF-1 genotypes shown. This figure reflects only the nucleotide sequence at the start of exon 2; it does not take into consideration other known differences between the gene and its pseudogenes. The solid and open bars represent GTGT-containing and ΔGT-containing (nonfunctional) genes, respectively. As depicted, the central gene on each chromosome is NCF-1. It is flanked by its 2 pseudogenes,16 18 which are either of Type I (ΔGT) or Type II (GTGT). In the majority of A47°CGD patients, both copies of NCF-1 contain the ΔGT mutation. Their carrier parents are heterozygous for the mutation, and although most have a ΔGT/GTGT sequence ratio of 5:1, as shown, approximately 10% would be predicted to show a ratio of 2:1 (see “Results and discussion”).
Predicted
ψNCF-1/NCF-1 genotypes based on observed ΔGT/GTGT ratios in unaffected individuals and in carriers and patients with A47°CGD. We have identified unaffected control individuals with 3 different ratios of ΔGT/GTGT sequence in exon 2 of theψNCF-1/NCF-1 genes: 2:1, 1:1, or 1:2. The most likely explanation for these ratios is the existence in the population of 2 main extended haplotypes (see “Results and discussion”), giving rise to the ψNCF-1/NCF-1 genotypes shown. This figure reflects only the nucleotide sequence at the start of exon 2; it does not take into consideration other known differences between the gene and its pseudogenes. The solid and open bars represent GTGT-containing and ΔGT-containing (nonfunctional) genes, respectively. As depicted, the central gene on each chromosome is NCF-1. It is flanked by its 2 pseudogenes,16 18 which are either of Type I (ΔGT) or Type II (GTGT). In the majority of A47°CGD patients, both copies of NCF-1 contain the ΔGT mutation. Their carrier parents are heterozygous for the mutation, and although most have a ΔGT/GTGT sequence ratio of 5:1, as shown, approximately 10% would be predicted to show a ratio of 2:1 (see “Results and discussion”).
Evidence to support the existence of the genotypes presented in Figure5 was provided by analysis of genomic DNA from the parents and siblings of 1 of the 2 control individuals identified as having a ΔGT/GTGT ratio of approximately 0.5 (1:2) in Figure 3 (referred to here as subject P). For our model to be correct, both his parents would have to have at least one copy of a Type II NCF-1 pseudogene within their genomes. As shown in the pedigree of this family (Figure6), the mother and father of subject P both had ΔGT/GTGT ratios of 1:1. This is internally consistent with their having copies of each of the 2 extended haplotypes, allowing subject P (indicated by the asterisk) to have inherited a Type IIψNCF-1 from each of his parents. Both siblings of subject P had ΔGT/GTGT ratios of 1:1. Additional support for our model came from experiments in which we mixed equal amounts of DNA (measured spectrophotometrically) from 3 different 2:1 control individuals with DNA from a single A47°CGD patient homozygous for ΔGT. These samples represent the first and final genotypes in Figure 5 and, as would be predicted, when combined gave ΔGT/GTGT ratios very similar to those of most A47° carriers—close to the theoretical value of 5:1 (Figure3).
The parents of control subject P, who has the uncommon ΔGT/GTGT ratio of 1:2, both have a ratio of 1:1, suggesting that they carry one copy of each of the extended haplotypes.
Exon 2 of NCF-1/ψNCF-1 in genomic DNA of the parents and siblings of control subject P (indicated by an asterisk in the pedigree) was amplified, and the ratio of ΔGT to GTGT sequence was estimated using the sequence- and gel-based methods. Both parents were found to have a ratio of 1:1, which is consistent with our hypothesis that subject P is homozygous for the extended haplotype that contains a single copy each of the Type I and Type II pseudogenes. The siblings both had ΔGT/GTGT ratios of 1:1. The family is unaffected by CGD. All other details are the same as in Figure 5.
The parents of control subject P, who has the uncommon ΔGT/GTGT ratio of 1:2, both have a ratio of 1:1, suggesting that they carry one copy of each of the extended haplotypes.
Exon 2 of NCF-1/ψNCF-1 in genomic DNA of the parents and siblings of control subject P (indicated by an asterisk in the pedigree) was amplified, and the ratio of ΔGT to GTGT sequence was estimated using the sequence- and gel-based methods. Both parents were found to have a ratio of 1:1, which is consistent with our hypothesis that subject P is homozygous for the extended haplotype that contains a single copy each of the Type I and Type II pseudogenes. The siblings both had ΔGT/GTGT ratios of 1:1. The family is unaffected by CGD. All other details are the same as in Figure 5.
Obligate carriers of ΔGT can have a ΔGT/GTGT ratio of 2:1
As indicated in Figure 5, the existence of 2ψNCF-1/NCF-1 extended haplotypes in the population predicts that although most carriers of the common A47°CGD allele would have a ΔGT/GTGT sequence ratio of 5:1, a minority would have a ratio of 2:1. Based on the prevalence calculated above for the Type IINCF-1 pseudogene, approximately 1 in 10 ΔGT carriers would be expected to have the Type I ψNCF-1/Type IIψNCF-1/NCF-1 (1:2) extended haplotype on their second (functional) copy of chromosome 7. In our analysis of 9 parents of homozygous ΔGT A47° patients, we identified 2 such carriers (Figure3). In one case, we could not categorically rule out the possibility that a de novo ΔGT mutation had occurred in a germline cell, raising the possibility that the parent with a 2:1 ratio was unaffected and not a carrier. However, in the second case the evidence was much stronger that the mother, whose ΔGT/GTGT ratio was 2:1, was indeed a carrier. As shown in this family's pedigree (Figure7), 2 daughters had A47°CGD and were homozygous for ΔGT, making the possibility of a de novo mutation very unlikely. In addition, and more conclusively, the third sibling, who was unaffected by CGD, had a ΔGT/GTGT ratio of 1:1. She must therefore have acquired the more common (2:1) haplotype from her father (who had a ratio of 5:1) and the less common (1:2) haplotype from her mother. Two parents of homozygous ΔGT patients with ΔGT/GTGT ratios of 2:1 were also identified in a previous study,21 but de novo ΔGT mutations were not excluded.
NCF-1/ψNCF-1 genotypes in a family affected by A47°CGD.
Exon 2 of ψNCF-1/NCF-1 in genomic DNA from the members of a family affected by A47°CGD was amplified, and the ratio of ΔGT to GTGT sequence was estimated. Two sisters (●) have A47°CGD and are homozygous for the ΔGT mutation. Both parents were considered obligate carriers of the disease, but the ΔGT/GTGT ratio of the mother (⊙) was 2:1, identical to that of an unaffected individual. A third sister (○) was found to have a ΔGT/GTGT ratio of 1:1. These data indicate that the mother carries the ΔGT mutation inNCF-1 on one copy of chromosome 7 and one each of the type I and type II pseudogenes on the other copy. All other details are the same as in Figure 5.
NCF-1/ψNCF-1 genotypes in a family affected by A47°CGD.
Exon 2 of ψNCF-1/NCF-1 in genomic DNA from the members of a family affected by A47°CGD was amplified, and the ratio of ΔGT to GTGT sequence was estimated. Two sisters (●) have A47°CGD and are homozygous for the ΔGT mutation. Both parents were considered obligate carriers of the disease, but the ΔGT/GTGT ratio of the mother (⊙) was 2:1, identical to that of an unaffected individual. A third sister (○) was found to have a ΔGT/GTGT ratio of 1:1. These data indicate that the mother carries the ΔGT mutation inNCF-1 on one copy of chromosome 7 and one each of the type I and type II pseudogenes on the other copy. All other details are the same as in Figure 5.
Origin of the GTGT-containing pseudogene
There appear to be 2 main possibilities for the origin of the GTGT-containing (Type II) pseudogene. One is that it represents a duplication of the functional gene in which the deletion of GT has not (yet) occurred, perhaps because this duplication is more recent from an evolutionary standpoint. A second possibility is that it represents the previously unidentified product of the reciprocal crossover of a DNA fragment between the functional NCF-1 gene and one of its pseudogenes. In an attempt to distinguish between these events, we analyzed additional markers previously shown to discriminate between the gene and its pseudogenes. These included the C/T transition in intron 1; the 20-bp stretch in intron 2 that is duplicated in the pseudogenes; and the single-nucleotide differences 269G>A, 496A>G, 558A>G, and 849A>G.9 15 We used an allele-specific strategy that amplifies only GTGT-containing DNA (ie, NCF-1and the Type II pseudogene) and sequenced the appropriate regions of introns 1 and 2; we also sequenced exons 2 (to confirm the absence of ΔGT), 4, 6, and 9 in their entirety. As expected, in 9 unaffected controls with ΔGT/GTGT ratios of 2:1, onlyNCF-1 sequence was detected at all these positions. In contrast, in the 2 control individuals with ΔGT/GTGT ratios of 1:2, both functional gene and pseudogene sequences were observed at each of the markers except the C/T transition in intron 1 (where only C was detected). These results demonstrate that the Type II pseudogene has undergone multiple mutations identical to those seen in Type I and suggest that it is unlikely to represent a newer duplication ofNCF-1.
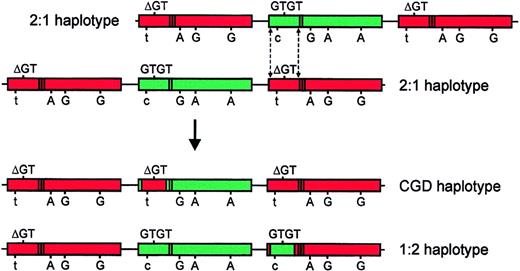
Although it is not possible to conclusively distinguish between the mechanisms by which the Type II pseudogene has arisen, based on our analysis of these gene/pseudogene markers, we believe that it most likely represents the product of a reciprocal crossover of a DNA fragment between NCF-1 and one copy of ψNCF-1.Figure 8 illustrates one possible crossover mechanism. As a result of this crossover event, one pseudogene acquires a GTGT-containing fragment from NCF-1(shown in green) to generate a Type II pseudogene. Concurrently, the previously functional NCF-1 acquires the ΔGT mutation originating in the corresponding fragment of pseudogene (shown in red), thereby generating the common A47°CGD allele. In the 2 1:2 control subjects studied, only functional gene sequence was observed at the C/T transition in intron 1, suggesting that the 5′ crossover site is upstream of this position. This is consistent with previous reports regarding the most common crossover sites withinNCF-1/ψNCF-1.7,9 Given the reported variation in size of recombination fragments in A47° patients,7 9it is likely that a similar heterogeneity will be seen in the size of the NCF-1 fragment inserted to form the Type II pseudogene.
Model of a possible crossover mechanism giving rise to the GTGT-containing ψNCF-1 and the prevalent A47°CGD allele.
In the top part of the figure, 2 chromosomes, each of the more common 2:1 extended haplotype, are misaligned. This event is probably most likely to occur between chromatids during meiosis. Dashed lines indicate possible sites of a double, reciprocal crossover event.NCF-1 is shown in green and its pseudogenes in red. The sequence at the start of exon 2 (GTGT or ΔGT) is shown above each gene, and the informative sequence differences (C/T in intron 1; 269G>A; 496A>G; 849A>G) are shown below (558A>G is omitted for the sake of clarity). The presence of the single (NCF-1) or duplicate (ψNCF-1) 20-bp stretch in intron 2 is indicated by bands in the body of the gene. The products of the crossover are shown in the bottom part of the figure, below the arrow. In the uppermost of these chromosomes, NCF-1 has acquired a pseudogene fragment containing ΔGT, resulting in the prevalent A47°CGD haplotype. One copy of ψNCF-1 on the lower chromosome has acquired a GTGT-containing fragment fromNCF-1 to form the Type II pseudogene (1:2 haplotype). In this model, the 3′ crossover site is based on data from subject P, but published data suggest that it is likely to vary. The exact position of the 5′ site is unknown, except that it is upstream of the C/T transition in intron 1 (see “Results and discussion”).
Model of a possible crossover mechanism giving rise to the GTGT-containing ψNCF-1 and the prevalent A47°CGD allele.
In the top part of the figure, 2 chromosomes, each of the more common 2:1 extended haplotype, are misaligned. This event is probably most likely to occur between chromatids during meiosis. Dashed lines indicate possible sites of a double, reciprocal crossover event.NCF-1 is shown in green and its pseudogenes in red. The sequence at the start of exon 2 (GTGT or ΔGT) is shown above each gene, and the informative sequence differences (C/T in intron 1; 269G>A; 496A>G; 849A>G) are shown below (558A>G is omitted for the sake of clarity). The presence of the single (NCF-1) or duplicate (ψNCF-1) 20-bp stretch in intron 2 is indicated by bands in the body of the gene. The products of the crossover are shown in the bottom part of the figure, below the arrow. In the uppermost of these chromosomes, NCF-1 has acquired a pseudogene fragment containing ΔGT, resulting in the prevalent A47°CGD haplotype. One copy of ψNCF-1 on the lower chromosome has acquired a GTGT-containing fragment fromNCF-1 to form the Type II pseudogene (1:2 haplotype). In this model, the 3′ crossover site is based on data from subject P, but published data suggest that it is likely to vary. The exact position of the 5′ site is unknown, except that it is upstream of the C/T transition in intron 1 (see “Results and discussion”).
Besides the C/T transition, only one additional marker has been described that is 5′ of the start of exon 2 and distinguishes betweenNCF-1 and its pseudogenes. Görlach et al15 reported that all pseudogene clones analyzed contained a single 30-bp stretch of sequence in intron 1 that was present as a tandem duplication in the functional gene clones. We analyzed this region in genomic DNA to determine whether it was a potentially useful marker to identify the upstream site of recombination. Using allele-specific PCR, we amplified and sequenced the fragment from exon 1 to the start of exon 2 in 10 control individuals (all with ΔGT/GTGT ratios of 2:1) and confirmed in each case that only GTGT-containing sequence was present. DNA from 5 persons contained only the 30-bp duplication as expected, but 3 individuals were heterozygous at this position and in 2 persons, both alleles contained only a single copy of the 30-bp stretch. Therefore, it does not reliably distinguish between gene and pseudogene and is not likely to be a good marker to map crossover sites.
One advantage of our model is that it explains the intriguing anomaly that although there is a pool within the population of additional DNA fragments containing ΔGT (in ΔGT/ΔGT patients and carriers of the mutation), no corresponding fragment had been identified that contains GTGT. Because crossover events betweenNCF-1 and ψNCF-1 most commonly account for the insertion of ΔGT in NCF-17 9 and crossovers are reciprocal in nature, it was puzzling that a GTGT-bearing fragment had not been located in the genome. The Type II pseudogene identified here appears to carry this missing fragment. Although the crossover mechanism shown in Figure 8 would generate an equal number of CGD and 1:2 extended haplotypes, negative selection pressure would tend to remove the CGD haplotype from the population. This has apparently resulted in approximately a 50-fold excess within the population of the 1:2 haplotype (with an incidence of 1 in 10) compared with the CGD haplotype (1 in 500).
Type II “pseudogene” may be functional
The 5′ regulatory regions of the p47-phoxpseudogenes are almost identical to the equivalent region of the functional gene, and the pseudogenes are known to be transcriptionally active.15 19 With GT deleted at the start of exon 2, the predicted translation product of the Type I pseudogene is 50 amino acids in length, with only the first 25 being identical to the corresponding region of p47-phox. This altered, truncated polypeptide is unlikely to be functional and, to our knowledge, it has never been detected. Of the other well-characterized differences that distinguish between the gene and its pseudogenes, 2 are intronic and unlikely to affect transcription and splicing, and those that are exonic are single-nucleotide changes that are mostly silent. Only 2 predict amino acid substitutions, 269G>A and 496A>G predicting Arg90His and Asn166Asp, respectively, and these changes may or may not lead to a loss of function. Based on our relatively small sample, 17% (9 of 53) of the healthy population has at least one copy of Type IIψNCF-1. Sequencing of allele-specific RT-PCR products from subject P revealed heterozygosity at the same exonic gene/pseudogene markers as found in GTGT-containing genomic DNA, confirming that Type II ψNCF-1 is transcribed. With a paucity of mutations within the coding region, it is feasible that it produces intact protein, but the analysis required to show this conclusively is beyond the scope of this study.
In conclusion, we have provided evidence for the presence in the general population of a previously undetected NCF-1pseudogene/gene chimera. In contrast to the predominant A47°CGD allele, which arises from the insertion of ΔGT into NCF-1,rendering it nonfunctional, this new fusion allele appears to represent the insertion of a GTGT-bearing fragment into a nonfunctional pseudogene. Whether the resulting gene is functional remains to be determined, but in either case, it is likely to be innocuous because, to date, we have found it only in combination with functionalNCF-1. However, the presence of at least 2NCF-1/ψNCF-1 extended haplotypes in the population further complicates the detection of ΔGT carriers because approximately 1 in 10 will have ΔGT/GTGT ratios of 2:1 and be indistinguishable from the majority of unaffected individuals.
We are grateful to all family members who provided blood samples for this study. This is manuscript 14865-MEM of The Scripps Research Institute.
Prepublished online as Blood First Edition Paper, May 13, 2002; DOI 10.1182/blood-2002-03-0861.
Supported by grants CA68276 (P.G.H.), AI24838 (A.R.C.), and RR00833 (to the General Clinical Research Center at The Scripps Research Institute) from the National Institutes of Health; and by the Stein Endowment Fund.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Paul G. Heyworth, MEM-241, Department of Molecular and Experimental Medicine, The Scripps Research Institute, 10550 North Torrey Pines Rd, La Jolla, CA 92037; e-mail: heyworth@scripps.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal