The CC chemokine macrophage inflammatory protein 1α (MIP1α) is a key regulator of the proliferation and differentiation of hematopoietic progenitor cells. The activity of MIP1α appears to be modulated by its binding to heparan sulfate (HS) proteoglycans, ubiquitous components of the mammalian cell surface and extracellular matrix. In this study we show that HS has highest affinity for the dimeric form of MIP1α. The predominantly dimeric BB10010 MIP1α interacts with an 8.3-kDa sequence in the HS polysaccharide chain, which it protects from degradation by heparinase enzymes. The major structural motif of this HS fragment appears to consist of 2 sulfate-rich S-domains separated by a short central N-acetylated region. The optimum lengths of these S-domains seem to be 12 to 14 saccharides. We propose that this binding fragment may wrap around the MIP1α dimer in a horseshoe shape, facilitating the interaction of the S-domains with the heparin-binding domains on each monomer. Molecular modeling suggests that these S-domains are likely to interact with basic residues Arg 17, Arg 45, and Arg 47 and possibly with Lys 44 on MIP1α and that the interconnecting N-acetylated region is of sufficient length to allow the 2 S-domains to bind to these sites on opposite faces of the dimer. Elucidation of the structure of the HS-binding site for MIP1α may enable us to devise ways of enhancing its myeloprotective or peripheral blood stem cell mobilization properties, which can be used to improve cancer chemotherapy treatments.
Introduction
Stem cells may exist in niches in the bone marrow environment where stromal cells, specific extracellular matrix components, and bound cytokines regulate proliferation, commitment, and terminal differentiation of undifferentiated hematopoietic progenitor cells (HPCs).1,2 A key regulatory cytokine of HPCs is the heparin-binding CC chemokine MIP1α, which is a reversible inhibitor of HPC proliferation.3 This property has been exploited in a number of experimental chemotherapy models and may provide a mechanism for protecting HPCs against the myelosuppressive effects of cancer chemotherapy.4-6
In the absence of stroma, specifically sulfated heparan sulfate (HS) glycosaminoglycans are required for maintenance of long-term culture-initiating cells (LTC-ICs) mediated by MIP1α and interleukin-3 (IL-3).7,8 The proteoglycan-binding site of MIP1α has been mapped to a highly conserved region between the third and fourth cysteine residues in a 3-dimensional cleft.9-11However, the role of proteoglycans in chemokine function is unclear. The interpretation of many MIP1α–proteoglycan studies is complicated by the use of heparin rather than the physiological ligand HS, a common pericellular constituent of mammalian cells. Furthermore, it has been demonstrated that in contrast to HS, unmodified heparin does not support LTC-IC maintenance.8
HS is a linear polysaccharide consisting of a series of hypervariable N- and O-sulfated domains (S-domains) connected by less sulfated, N-acetyl–rich regions. Spacing of these S-domains and their fine structure appear to be the major determinants of the specific binding of HS to protein ligands, including a number of cytokines.12 The mechanism of action of HS is controversial, but it is likely that in some cases it induces a conformational change in the bound ligand, which enables it to be recognized by cognate-signaling receptors. However, in the case of MIP1α, Graham et al9 demonstrated that binding affinity of MIP1α to at least one of its receptors, CCR1, is not reduced in CHO cells deficient in HS proteoglycan expression. Cell surface HS may be more important to sequester chemokines, such as MIP1α, to the correct location in the bone marrow, hence facilitating their binding to signaling receptors. Dimerization of MIP1α, shown to be induced by heparin,13 may also occur with HS to aid the concentration of MIP1α at appropriate sites of action.
In the present study we examined whether MIP1α aggregation is important for HS binding, investigated the prevalence of the binding site in bone marrow stromal HS, and determined the characteristics of the HS binding site, which may be important for MIP1α localization in a bone marrow niche. This is the first study in which the HS binding site for MIP1α has been isolated and analyzed.
Materials and methods
Materials
BB-10010 was kindly provided by British Biotechnology (Oxford, United Kingdom) as a genetically engineered variant of MIP1α equipotent to human MIP1α but with reduced multimerization abilities, giving it increased solubility.14 Monomeric, dimeric, and tetrameric aggregation variants of MIP1α were prepared as described in Graham et al.15 D-[6-3H] glucosamine hydrochloride (740-1665 GBq/mol [20-45 Ci/mol]) was obtained from Perkin Elmer Life Sciences (Cambridge, United Kingdom). Heparinase III (Flavobacterium heparinum, EC 4.2.3.8) was obtained from Grampian Enzymes (Orkney, United Kingdom). Chondroitinase ABC (Proteus vulgaris, EC 4.2.2.4) was obtained from Seikagaku Kogyo (Tokyo, Japan). Human platelet extract was generously provided by Paul Brenchley (University of Manchester, United Kingdom). Biogel P10 and Affi-Gel 10 were purchased from Bio-Rad Laboratories (Hemel Hempstead, United Kingdom). Sepharose CL6B, DEAE Sephacel and PD10 columns were purchased from Amersham Pharmacia Biotech (Buckinghamshire, United Kingdom).
Radiolabeling and preparation of intact HS chains
HS chains biosynthetically labeled with3H-glucosamine were prepared from subconfluent cultures of murine 3T3 fibroblasts, bovine aortic endothelial cells (isolated by Graham Rushton, Paterson), and human bone marrow stromal cells (generously provided by Erika de Wynter16), as described by Lyon et al.17 Residual amino acids from the core protein were removed by alkali borohydride treatment, and the size distribution of the material was analyzed by gel filtration chromatography.18
Specific degradation of HS
Nitrous acid hydrolysis was performed by the low pH method of Shively and Conrad.19 Heparinase III digestion and gel filtration column chromatography were performed as described in Stringer and Gallagher.18 Platelet heparinase digestion was performed by the addition of 4 μL platelet extract to 200 μL3H-radiolabeled HS in 50 mM NaAc buffer (pH 5.5, 6.5, or 7.4 to create a range of oligosaccharide sizes) and 0.01% bovine serum albumin for 16 hours at 37°C. Platelet proteins were then precipitated by the addition of ice-cold trichloroacetic acid (TCA) to 10% for 15 minutes at 0°C and pelleted by microcentrifugation, 12 000 rpm, for 15 minutes. The supernatant was neutralized by the addition of 5 M NaOH. Resultant HS fragments were then size separated on a Sepharose CL-6B column,18 and the required oligosaccharide sizes were pooled and freeze dried.
Preparation of MIP1α-protected domain of HS
Equimolar quantities of 3H-radiolabeled HS (500 000 cpm) and MIP1α (5 μM) were preincubated for 10 minutes at room temperature before digestion by heparinase III at 40 mIU/mL in 0.5 mM CaAc, 50 mM NaAc, and 0.1 mg/mL bovine serum albumin, pH 7.0, in 250 μm for approximately 24 hours at room temperature. An additional 10 mIU heparinase III (in 0.5 mM CaAc, 50 mM NaAc, and 0.1 mg/mL bovine serum albumin, pH 7.0) was added after 8 hours, and 10 mIU enzyme was added for 2 hours at 37°C. Resultant fragments were separated on a Biogel P10 column,18 and the void volume peak was pooled and freeze dried. Proteins were removed by ice-cold TCA treatment as above. The supernatant was desalted on a PD10 column eluted with double-distilled water.
Affinity chromatography
To prepare a MIP1α affinity gel column, 100 μg MIP1α was mixed with 100 μg heparin in 500 μL coupling buffer (0.1 M HEPES [N-2-hydroxyethylpiperazine-N'-2-ethanesulfonic acid], 80 mM NaCl, pH 7.0) and was incubated for 20 minutes at room temperature. MIP1α was then bound to Affi-Gel 10, and the column was prepared as previously described by Lyon et al17 alongside a control column in which the MIP1α was omitted. The final concentration of MIP1α was deduced to be 80 μg in 0.5 mL gel, by protein assay of the spent coupling buffer.
Affinity experiments were performed by application of radiolabeled HS samples in 20 mM sodium phosphate buffer of physiological ionic strength and pH level (0.15 M NaCl and pH 7.3). The sample of HS was recirculated through the column 5 times at room temperature to maximize its binding. The column was then washed with 2.5 mL of 0.15 M NaCl, 20 mM sodium phosphate, pH 7.3, followed by 2.5 mL each of a range of NaCl concentrations, as described in the figure legends. Because of interbatch variation of MIP1α, results for the various treated HS fractions or sources were always compared to those of intact 3T3 fibroblast HS eluted from the same column.
Protein model building
Protein models for a MIP-1α dimer were built using the program Modeler20 interfaced to the insightII protein modeling package (Accelrys, San Diego, CA). Two sets of protein models for MIP-1α were created. The first set used the known protein structure of platelet factor 4 (PF4; pdb code 1RHP chains A, B) as a template structure, and the second set used the known structure of IL-8 (pdb code 1ICW). Typically 4 protein models were constructed for each case. Protein models were evaluated using a scoring function21 based on nonlocal atomic interactions. This scoring scheme uses a database of known crystal structures to characterize favorable residue environments in terms of atom–atom interaction parameters. These parameters can then be applied to model structures to find potential misfolded regions or to objectively select the most physically realistic model (http://guitar.rockefeller.edu/∼fmelo/anolea/anolea.html).
Heparin protein docking
Docking of heparin pentasaccharide model ligands to protein model structures was performed with Autodock version 2.4.22 This program allows for flexibility in the ligand structure but uses a rigid body protein approximation to speed up the calculation. The pentasaccharides consisted of 3 GlcN2S6S residues separated by IdoA2S residues. Because the IdoA2S residue can adopt different ring conformations, 2 model ligands were used. In one, all IdoA2S residues were in the 1C4 ring form; in another, the 2S0 ring form was adopted. The model ligands had fixed glycosidic torsion angles taken from a reported heparin nuclear magnetic resonance (NMR) structure (PDB code1HPN). Flexibility was allowed for all exocyclic torsion angles. Docking was also performed using a heparin endecasaccharide with IdoA2S residues in the 1C4 conformation, but in this case no flexibility was allowed. Partial atomic charges required for the docking calculation were obtained by ab initio quantum chemistry calculations (optimization and charge fitting using an HF/6-31G* basis set) on 1-OMe 4-OMe substituted monosaccharides. This was performed using the Jaguar program (Schrodinger, Portland, OR). Typically the 10 lowest energy (most favorable) coordinate sets were extracted for each ligand type and were used for visualization in insightII.
Results
Interaction of aggregation variants of MIP1α with HS
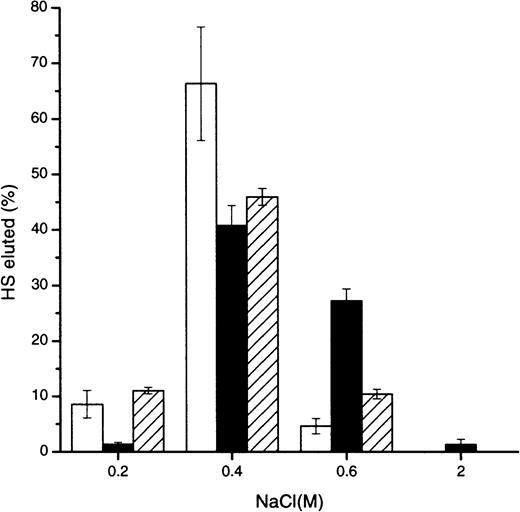
To determine whether HS interacted with greater affinity to a particular aggregation state of MIP1α, Affi-Gel 10 columns were prepared from MIP1α variants engineered to control the degree of multimerization.15 Most bound intact 3T3 fibroblast HS eluted from the monomeric, dimeric, and tetrameric MIP1α at approximately 0.4 M NaCl, but there was significantly more HS binding to the dimeric MIP1α at 0.6 M and greater than to the other aggregation variants (Figure 1). A control column did not exhibit any binding of HS greater than 0.15 M NaCl (not shown). Subsequent experiments were carried out using BB-10010 MIP1α, which appears to exist predominantly in a dimeric state in concentrations up to 1 mg/mL in phosphate-buffered saline and across a wide pH range.14 23
Comparison of the relative affinity of HS for monomeric, dimeric, and tetrameric MIP1α aggregation mutants.
Intact murine fibroblast 3H-radiolabeled HS was applied to an Affi-Gel column, prepared with the monomeric (■), dimeric (▪), or tetrameric (▨) MIP1α aggregation mutants, in 0.15 M NaCl. Bound material was eluted with the NaCl concentrations indicated. Results show the averages from 6 experiments. Error bars of the percentage HS eluted are depicted.
Comparison of the relative affinity of HS for monomeric, dimeric, and tetrameric MIP1α aggregation mutants.
Intact murine fibroblast 3H-radiolabeled HS was applied to an Affi-Gel column, prepared with the monomeric (■), dimeric (▪), or tetrameric (▨) MIP1α aggregation mutants, in 0.15 M NaCl. Bound material was eluted with the NaCl concentrations indicated. Results show the averages from 6 experiments. Error bars of the percentage HS eluted are depicted.
Comparison of the affinity of different HS types to MIP1α
Elution profiles of 3H-radiolabeled HS, prepared from human bone marrow stromal cultures, bovine aortic endothelial cells, and 3T3 fibroblast cells from a (BB-10010) MIP1α Affi-Gel 10 column were compared (Figure 2). Thirty-six percent of the bone marrow stromal HS, 53% of the fibroblast HS, and 24% of the endothelial HS bound above physiological NaCl concentrations. Most of the bound material had eluted by 0.8 M NaCl, but there was significantly more HS eluting above this NaCl concentration for both the bone marrow stromal HS (8%, A) and the fibroblast HS (6%, C) than the endothelial cell HS (3%, B). Therefore, fibroblast HS appeared to have a similar percentage of these higher ionic strength binding sites for MIP1α than bone marrow stroma HS and to have the most material binding overall, and it was used in the following experiments to isolate and characterize the binding site.
Determination of the affinity of MIP1α (BB10010) for HS from different cell types.
Intact 3H-labeled HS from human bone marrow stroma (A), bovine aortic endothelial cells (B), and murine fibroblast cells (C) were applied to an MIP1α (BB10010) Affi-Gel column (•) in 0.15 M NaCl. The same types of HS were also applied to a control column (●). Bound material was eluted with a stepwise NaCl gradient (. . . . . . .). Graphs are representative of duplicate experiments.
Determination of the affinity of MIP1α (BB10010) for HS from different cell types.
Intact 3H-labeled HS from human bone marrow stroma (A), bovine aortic endothelial cells (B), and murine fibroblast cells (C) were applied to an MIP1α (BB10010) Affi-Gel column (•) in 0.15 M NaCl. The same types of HS were also applied to a control column (●). Bound material was eluted with a stepwise NaCl gradient (. . . . . . .). Graphs are representative of duplicate experiments.
Effects of specific enzyme scission on MIP1α binding to HS
To identify HS domains important for MIP1α binding, HS digested by the enzymes heparinase I or III, which cleave in regions of high and low sulfation, respectively, was bound to MIP1α Affi-Gel 10. Heparinase I acts in the N-sulfated regions and specifically cleaves disaccharides that contain 2-O-sulfated iduronate—that is, GlcNS(± 6S)α–1,4-IdoA2S (where GlcNS indicates α-D-N-sulfated glucosamine; 6S, 6-O-sulfated; IdoA, α-L-iduronic acid; 2S, 2-O-sulfated).24 In contrast, heparinase III cleaves GlcA-containing disaccharides,24 principally GlcNAc/GlcNSα–1,4-GlcA (where GlcA indicates β-D-glucuronic acid; GlcNAc, α-D-N-acetylglucosamine), present in regions of low sulfation, and not contiguous N-sulfated sequences enriched in IdoA. Binding to MIP1α of HS fragments produced by either of the enzymes was significantly decreased in comparison with native HS (Figure3, compare panels B and C to A). This was most noticeable with heparinase III digestion (Figure 3C), through which less than 10% of the HS fragments remained bound to the column after elution with 0.15 M NaCl and bound material eluted at 0.2 M (most) and 0.4 M NaCl compared to 56% of the intact HS binding and elution at up to 0.7 M (Figure 3A). In contrast to heparinase III treatment, almost 3-fold more heparinase I fragments (28%) eluted from MIP1α between 0.2 to 0.4 M NaCl and another 6% bound greater than 0.4 M. These results indicate that both N-sulfated (S-domains) and N-acetylated regions of the HS chain are important for binding to MIP1α.
Investigation of the effect of specific enzyme scission on MIP1α binding to HS.
3H-labeled murine fibroblast HS chains were treated with either heparinase I or heparinase III enzymes as described in “Materials and methods.” Intact (A), heparinase I-digested, (B) and heparinase III–digested (C) HS were applied to an MIP1α (BB10010) Affi-Gel column (▪) in 0.15 M NaCl. Identically prepared HS was also applied to a control column (●). Bound material was eluted with a stepwise NaCl gradient (. . . . . . .). (D) Sulfated domains excised from HS by heparinase III were separated by Biogel P10 chromatography, then applied to the Affi-Gel MIP1α (BB10010) column. All the material was eluted with 0.2 M and 0.4 M NaCl steps. Most of the disaccharides and tetrasaccharides were eluted in the 0.15 M NaCl wash; hence, the bars at 0.2 M and 0.4 M are undetectable. ■ indicates hexasaccharides; ▪, octasaccharides; ▨, decasaccharides; ▩, dodecasaccharides; and ▤, tetradecasaccharides. Each graph is representative of 2 or more experiments.
Investigation of the effect of specific enzyme scission on MIP1α binding to HS.
3H-labeled murine fibroblast HS chains were treated with either heparinase I or heparinase III enzymes as described in “Materials and methods.” Intact (A), heparinase I-digested, (B) and heparinase III–digested (C) HS were applied to an MIP1α (BB10010) Affi-Gel column (▪) in 0.15 M NaCl. Identically prepared HS was also applied to a control column (●). Bound material was eluted with a stepwise NaCl gradient (. . . . . . .). (D) Sulfated domains excised from HS by heparinase III were separated by Biogel P10 chromatography, then applied to the Affi-Gel MIP1α (BB10010) column. All the material was eluted with 0.2 M and 0.4 M NaCl steps. Most of the disaccharides and tetrasaccharides were eluted in the 0.15 M NaCl wash; hence, the bars at 0.2 M and 0.4 M are undetectable. ■ indicates hexasaccharides; ▪, octasaccharides; ▨, decasaccharides; ▩, dodecasaccharides; and ▤, tetradecasaccharides. Each graph is representative of 2 or more experiments.
To elucidate the nature of the S-domains that constitute part of the MIP1α binding site, we isolated individual HS S-domains from fibroblast HS by heparinase III scission and Biogel P10 chromatography (Figures 4). Hexasaccharides (open bars) and octasaccharides (solid bars) were the smallest S-domains to show any binding to MIP1α, but only 18% and 13% of these bound and eluted at 0.2 M NaCl, respectively (Figure 3D). More than 3 times (53%) the decasaccharides and larger S-domains eluted at 0.2 M NaCl, with some dodecasaccharides (2%) and tetradecasaccharides (6%) eluted at 0.4 M NaCl, suggesting that one or more of these larger S-domains may make up part of the binding domain.
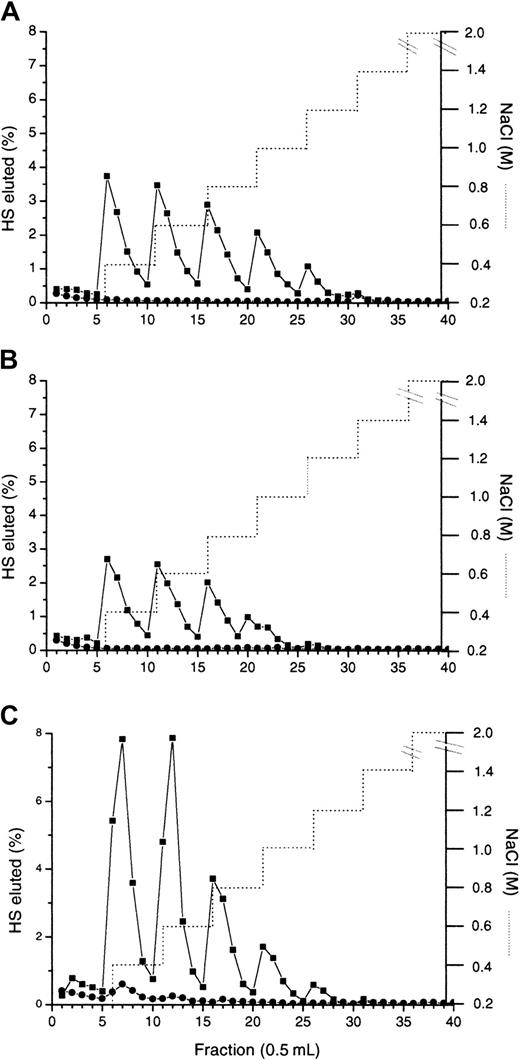
Heparinase III digestion in the presence or absence of MIP1α: isolation of the MPD.
3H-labeled HS chains were treated with heparinase III in the absence (A) or presence (B) of equimolar quantities of MIP1α (BB10010) as described in “Materials and methods.” Digests were analyzed by chromatography on a Biogel P10 column. The void (Vo) in panel B represents the MPD. (C) Molecular sizes of native HS chains (▪) and MPD (●) were compared by gel filtration on Sepharose Cl-6B. Vo indicates void volume; Vt, total volume. Distinct oligosaccharide peaks are labeled according to the number of monosaccharide units. (D) MPD (●) or intact HS (▪) were applied to an Affi-Gel MIP1α (BB10010) column in 0.15 M NaCl and eluted with a stepwise NaCl gradient (. . . . . . .). Each graph is representative of 2 or more experiments.
Heparinase III digestion in the presence or absence of MIP1α: isolation of the MPD.
3H-labeled HS chains were treated with heparinase III in the absence (A) or presence (B) of equimolar quantities of MIP1α (BB10010) as described in “Materials and methods.” Digests were analyzed by chromatography on a Biogel P10 column. The void (Vo) in panel B represents the MPD. (C) Molecular sizes of native HS chains (▪) and MPD (●) were compared by gel filtration on Sepharose Cl-6B. Vo indicates void volume; Vt, total volume. Distinct oligosaccharide peaks are labeled according to the number of monosaccharide units. (D) MPD (●) or intact HS (▪) were applied to an Affi-Gel MIP1α (BB10010) column in 0.15 M NaCl and eluted with a stepwise NaCl gradient (. . . . . . .). Each graph is representative of 2 or more experiments.
Isolation of the MIP1α binding site on HS (protection assay)
Because the MIP1α binding site appeared to overlap N-acetylated and N-sulfated regions of HS, the binding site could not be isolated from fragments produced by scission with either heparinase I or heparinase III. Therefore, a protection assay18 was used in which MIP1α was included in a heparinase III digest to prevent cleavage of the binding site. A prolonged digest was carried out to ensure that only the HS fragments with strong affinity for MIP1α were left undigested. When heparinase III breakdown of HS in the absence of MIP1α was compared by P10 gel filtration chromatography with profiles for MIP1α-protected HS (Figure 4A-B), the most striking difference was the presence of a large void peak in the MIP1α-protected digest (Figure 4B), surmised to be the MIP1α-protected domain (MPD). There was also a marked absence of the tetradecasaccharide and larger S-domains in the digest of the MIP1α-protected HS, presumably as these are incorporated into the MPD. Gel-filtration chromatography of MPD on Sepharose CL6B revealed a radioactive fraction, with a mean Kav (from several experiments) of 0.68 (Figure 4C), equal to a mass of 8.3 kDa by reference to the published calibration of Wasteson.25 This is equivalent to approximately 18 disaccharides, assuming 443 Da/disaccharide (molecular weight of a nonsulfated disaccharide). By comparison the intact fibroblast HS had a Kav of 0.38, equivalent to approximately 40 kDa (Figure 4C).
It was important to establish that MPD was indeed an MIP1α-binding fragment. Elution of intact HS and MPD from MIP1α Affi-Gel 10 demonstrated that the binding profiles were similar, with both binding up to at least 1 M NaCl (Figure 4D). Forty-six percent of MPD bound above the 0.15 M NaCl concentration level compared to 53% of intact fibroblast HS, and almost identical levels of each (approximately 15%) bound at the higher NaCl concentrations, above 0.6 M.
Effect of platelet heparinase on MIP1α-binding properties of HS
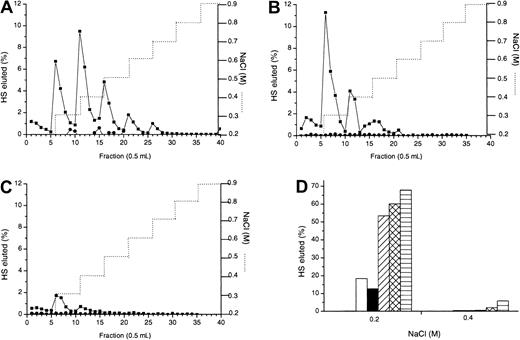
Platelet heparinase brings about a limited cleavage of HS by acting at glucuronate residues in mixed N-acetylated/N-sulfated regions adjacent to S-domains. Hence, we used platelet heparinase to create large fragments of HS to confirm the size range of the MIP1α-binding site. Figure 5A shows the size distributions on a Sepharose CL-6B column of oligosaccharides of mean sizes 5 kDa, 10 kDa, and 14 kDa pooled from platelet heparinase digests carried out to completion at pH 5.2, 6.5, or 7.4. The 10-kDa (Figure 5D), 14-kDa (Figure 5C), and 20-kDa (not shown) oligosaccharides showed similar ranges of binding to MIP1α Affi-Gel 10 as intact HS (Figure 5B), mostly eluting by 0.5 M NaCl. In contrast, the apparent affinity of the 5-kDa oligosaccharides for MIP1α were markedly reduced (Figure 5E), with most of the material eluting at 0.2 M. This suggests that the size of the MIP1α-binding site is between 5 and 10 kDa, in agreement with the 8.3-kDa protected fragment (MPD) as the binding domain.
Confirmation of the size of HS required for optimum binding to MIP1α.
3H-labeled HS chains were digested with platelet heparinase under different pH conditions, as described in “Materials and methods.” Resultant HS fragments were size fractionated on a Cl-6B Sepharose gel filtration column. (A) Pooled platelet heparinase fractions of average sizes 14 kDa (▴), 10 kDa (●), and 5 kDa (▪) were separately rerun on the Cl-6B Sepharose column to verify their size distribution. Intact (B), 14 kDa (C), 10 kDa (D), and 5 kDa (E) HS fragments were applied to an MIP1α (BB10010) (▪) and to a control Affi-Gel column (●) in 0.15 M NaCl and were eluted with a stepwise NaCl gradient (. . . . . . .). Graphs are representative of 2 experiments.
Confirmation of the size of HS required for optimum binding to MIP1α.
3H-labeled HS chains were digested with platelet heparinase under different pH conditions, as described in “Materials and methods.” Resultant HS fragments were size fractionated on a Cl-6B Sepharose gel filtration column. (A) Pooled platelet heparinase fractions of average sizes 14 kDa (▴), 10 kDa (●), and 5 kDa (▪) were separately rerun on the Cl-6B Sepharose column to verify their size distribution. Intact (B), 14 kDa (C), 10 kDa (D), and 5 kDa (E) HS fragments were applied to an MIP1α (BB10010) (▪) and to a control Affi-Gel column (●) in 0.15 M NaCl and were eluted with a stepwise NaCl gradient (. . . . . . .). Graphs are representative of 2 experiments.
Analysis of MPD compared with native HS
MPD was depolymerized by nitrous acid degradation and heparinase enzymes and then was analyzed on a P10 column to compare its structure with that of its parent fibroblast HS (Figure 6).
Analysis of specific degradation of HS and MPD by gel filtration chromatography.
3H-labeled HS (A, C) and MPD (B, D) were degraded by exhaustive treatment with low pH nitrous acid (A, B) and heparinase III (C, D), as described in “Materials and methods.” Digests were analyzed by chromatography on a Biogel P10 column. Distinct oligosaccharide peaks are labeled according to the number of monosaccharide units.
Analysis of specific degradation of HS and MPD by gel filtration chromatography.
3H-labeled HS (A, C) and MPD (B, D) were degraded by exhaustive treatment with low pH nitrous acid (A, B) and heparinase III (C, D), as described in “Materials and methods.” Digests were analyzed by chromatography on a Biogel P10 column. Distinct oligosaccharide peaks are labeled according to the number of monosaccharide units.
Treatment with low pH nitrous acid, which cleaves at GlcNS residues, yielded a distinct pattern of scission of MPD and HS, with an almost 2-fold enrichment of disaccharides in MPD and a 3-fold reduction of tetrasaccharides (Figure 6A-B). This indicates a significant increase in contiguous N-sulfated disaccharides in comparison with the original HS, consistent with MPD encompassing 2 S-domains and one GlcNAc-rich interconnecting region. The octasaccharides appeared to be the most predominant contiguous GlcNAc containing oligosaccharides (Figure 6B), with a 2-fold enrichment in MPD compared to intact HS.
Heparinase III scission provides data on the size range of the S-domains in HS, elucidating in part the arrangement of the contiguous N-sulfated disaccharides identified by nitrous acid hydrolysis. There was a 5-fold enrichment of dodecasaccharide and tetradecasaccharide S-domains released from MPD by heparinase III compared to intact HS (Figure 6C-D). These oligosaccharides were confirmed to be S-domains, as opposed to incompletely digested material, by their susceptibility to low pH nitrous acid degradation (not shown).
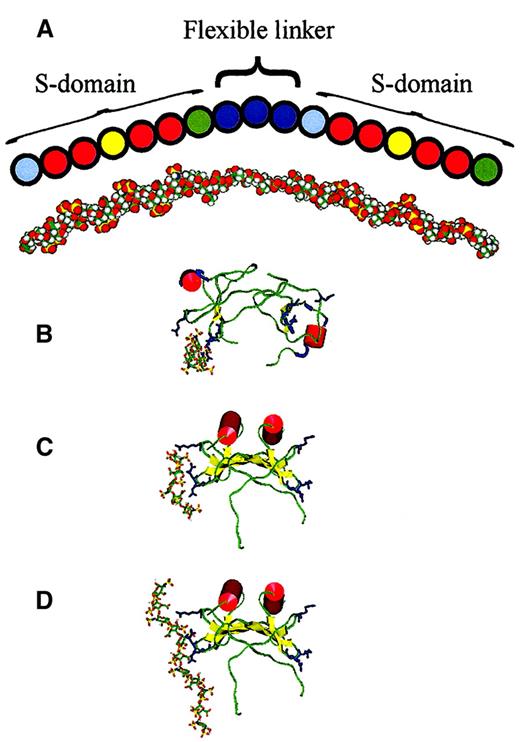
The prevalence in MPD of particular-sized fragments excised by heparinase III and low pH nitrous acid can be rationalized in the form of a major structural motif, with some permissive variations, which represents this enzyme-protected binding region for MPD in HS. Such a structure is depicted in Figure 7. Although only 17 disaccharides in length, the molecular weight of this N- and O-sulfated oligosaccharide is 8259 Da, the average size for (TCA-treated) MPD.
Diagram and model of predicted MPD structure and models of MIP1α–heparin complexes.
(A) An empirically determined structure for MPD is depicted based on the prevalence of particular-sized nitrous acid and heparinase-resistant fragments and frequencies of cleavage sites (Figure 6) and from information on fibroblast HS S-domain composition from sequencing studies.32 Pale blue spheres, GlcA-GlcNS; red spheres, IdoA(2S)-GlcNS; yellow spheres, IdoA(2S)-GlcNS(6S); green spheres, IdoA(± 2S)-GlcNAc; and deep blue spheres, GlcA-GlcNAc (see “Results” for abbreviations). This is roughly aligned with a model of MPD presented in an extended conformation. (B-D) Heparin ligand is in stick representation and MIP1α is shown rendered to display secondary structure; red α-helix, yellow β-strands, green random coil, and pale blue turns. Basic residues predicted to bind with heparin (Arg 17 on one loop, Arg 45 and Arg 47 on an adjacent loop) are shown in deep blue. (B) Dimeric MIP1α (pdb code 1B53) and (C) dimeric MIP-1α (model based on relative orientation of monomer subunits in PF4) docked with the energetically most favorably positioned heparin pentasaccharide ligand in each case. (D) Dimeric MIP-1α, as in panel C, docked with the energetically most favorable endecasaccharide.
Diagram and model of predicted MPD structure and models of MIP1α–heparin complexes.
(A) An empirically determined structure for MPD is depicted based on the prevalence of particular-sized nitrous acid and heparinase-resistant fragments and frequencies of cleavage sites (Figure 6) and from information on fibroblast HS S-domain composition from sequencing studies.32 Pale blue spheres, GlcA-GlcNS; red spheres, IdoA(2S)-GlcNS; yellow spheres, IdoA(2S)-GlcNS(6S); green spheres, IdoA(± 2S)-GlcNAc; and deep blue spheres, GlcA-GlcNAc (see “Results” for abbreviations). This is roughly aligned with a model of MPD presented in an extended conformation. (B-D) Heparin ligand is in stick representation and MIP1α is shown rendered to display secondary structure; red α-helix, yellow β-strands, green random coil, and pale blue turns. Basic residues predicted to bind with heparin (Arg 17 on one loop, Arg 45 and Arg 47 on an adjacent loop) are shown in deep blue. (B) Dimeric MIP1α (pdb code 1B53) and (C) dimeric MIP-1α (model based on relative orientation of monomer subunits in PF4) docked with the energetically most favorably positioned heparin pentasaccharide ligand in each case. (D) Dimeric MIP-1α, as in panel C, docked with the energetically most favorable endecasaccharide.
Protein-docking results
Two different models of the MIP-1α dimer, both based on experimental evidence, have been proposed—the published dimeric NMR structure pdb code 1B53 and a more compact dimer with a different interface.23 We have constructed protein dimer models, similar to those reported by Ashfield et al,23 using monomer–monomer orientations adopted by IL-8 (pdb code1QE6) that have a distinct quaternary structure but retain a tertiary structure similar to that of 1B53. We have also created novel alternative dimers using PF4 (pdb code 1RHP). Both IL-8 and PF4-based dimers adopt a compact structure in contrast to the extended structure reported on the basis of NMR data26 (pdb code 1B53). Systematic evaluation of protein structure quality according to the method of Melo and Feytmans21 indicates that the compact dimers predicted on the basis of IL-823 or PF4 (Figure 7C) are more likely to be physically realistic than the extended dimer proposed on the basis of NMR data.26 The 2 compact models for the dimer are similar in quaternary structure, with the PF4-based model having a marginally better score than the IL-8 model. In both compact dimer models, Lys 60 is less solvent-exposed than Lys 36 or Lys 44, whereas in the published PDB structure 1B53, Lys 60 is the most exposed. Ashfield et al23 have shown that of these lysine residues Lys 60 is the most protected from methylation, indicating that Lys 60 is less exposed to solvent. In both dimer models, 1 of the 2 possible heparin-binding sites is preferred to the other by the docking calculation because amino acid side-chains are held rigid in these calculations; this suggests that the orientations of side chains are more favorable in one of the monomer subunits.
Docking of heparin pentasaccharide ligands was performed using several protein structures. Docking was performed with the published dimeric NMR structure pdb code 1B53 (Figure 7B) and a monomer (A chain) derived from it (not shown), then it was performed with the IL-8 (not shown) and PF4-based dimers (Figure 7C). For all 3 protein models used in the docking calculations, the heparin-binding site was predicted to be in the same area of the protein surface (Figure 7C). This region includes basic residues Arg 17, Arg 45, and Arg 47 (in agreement with Koopmann and Krangel10) and occasionally Lys 44. In the published dimer 1B53 and the compact dimer model based on PF4, this area is exposed to solvent, and in both models the heparin-binding sites are at opposite ends of the dimer. No differences in predicted binding site resulted from the 2 iduronate conformations in the pentasaccharide ligands. An additional docking calculation was performed using an endecasaccharide (Figure 7D) rather than a pentasaccharide heparin model; this identified the same binding site.
Discussion
Studies by Hoogewerf et al13 imply that glycosaminoglycans such as heparin mediate the oligomerization of chemokines, including MIP1α, which may be important to raise local chemokine concentrations at sites of activity and possibly to aid presentation to their 7 transmembrane receptors. Using oligomerization-deficient MIP1α created by Graham et al,15 we demonstrated that there is a significantly stronger ionic interaction between the physiological ligand HS and dimeric MIP1α than monomeric or tetrameric MIP1α (Figure 1). In contrast, progressive neutralization of the C-terminal acidic acid residues, which bestows a decreased oligomerization state to these MIP1α variants, actually increased the strength of binding to the more highly sulfated glycosaminoglycan heparin,27 possibly because of the annulment of repulsion between these residues and the negatively charged sulfates of heparin. The presence of the less charged N-acetylated regions in HS may alleviate repulsion with the C-terminal of MIP1α, as has been postulated for other chemokines,28 and it is thought to confer increased flexibility on the HS chain,29 which could enable it to adopt a configuration more compatible with the MIP1α dimer. These ideas are supported by preliminary experiments using CD34+cord blood cells in a serum-free clonogenic assay,30 which showed a 12% to 37% enhancement of the predominantly dimeric BB100-10 MIP1α activity with the addition of 1 μg/mL HS and little or no effect for the same concentration of heparin (personal communication Erica de Wynter, Leeds University, United Kingdom). The apparent reduced capacity of heparin to interact with dimeric MIP1α compared to HS may also be responsible for its lack of ability to support MIP1α-mediated LTC-IC maintenance.8
MIP1α is postulated to have a role in the activation of selected leukocyte populations from the blood during the inflammatory response and a role in hemopoietic stem cell proliferation inhibitor. Thus its localization on the endothelial cell surface and in the bone marrow stroma, possibly through binding to HS proteoglycans,8,13is of paramount importance to its biologic activity. Our results demonstrate that MIP1α BB100-10 binds preferentially to bone marrow stromal and 3T3 fibroblast HS when compared with (bovine aortic) endothelial HS. Cell type–specific sulfation pattern, length, frequency, and spacing of the highly sulfated S-domains of HS polysaccharide chains are thought to be critical for HS ligand binding and HS proteoglycans biologic activity.12 Structural analysis of 3T3 fibroblast and bovine aortic endothelial HS demonstrated that there was a lower frequency of the larger dodecasaccharide and tetradecasaccharide S-domains and an overall decreased level of 6-O-sulfation in the endothelial HS,31though the composition of this may change during the inflammatory response. Subsequent experiments confirmed the importance of these larger S-domains, more prevalent in fibroblast HS for MIP1α binding (see below). In addition, 6-O-sulfation of HS appears to be important for MIP1α biologic activity, specifically for its role in LTC-IC maintenance.8
As might be expected the S-domains of HS, which are enriched in O-sulfation, appeared to be important for MIP1α binding because heparinase I digestion resulted in a significant decrease in the ionic interaction (Figure 3B). We found that MIP1α bound more strongly to decasaccharide, dodecasaccharide, and tetradecasaccharide S-domains than to the smaller S-domains (Figure 3D), possibly because of the increased occurrence of 6-O-sulfate groups in longer S-domains32 33 and a specific size dependence of binding. However, S-domains alone, isolated by heparinase III digestion, were unable to reproduce the binding properties of the parent molecule. These findings are in contrast to those for basic fibroblast growth factor and for the extracellular matrix molecule fibronectin, in which the optimum binding sites appear to be contained within one extended sulfated domain.
Another striking difference with a number of previously characterized binding sites on HS is the large molecular mass (8.3 kDa, equivalent to approximately 17 disaccharides) of the MIP1α-protected fragment isolated from murine fibroblast HS. This size is similar to that of the binding sites isolated in recent years for other multimeric cytokines, such as PF4 (9.3 kDa), IL-8 (approximately 6 kDa), and interferon-γ (IFN-γ) (10 kDa). The size of the MIP1α-binding site was confirmed by the ability of platelet heparinase-resistant 10 kDa, but not 5 kDa, HS fragments to bind with as high affinity to MIP1α as the parent HS (Figure 5). The use of the enzyme platelet heparinase described in this paper may provide an important tool for the isolation of HS-binding sites of this nature. It enables larger quantities of the appropriately sized range of HS fragments for the binding sites of multimeric ligands to be isolated than is possible by the protection assay, which could facilitate detailed sequence analysis of the sites.
The depolymerization profiles of MPD, after specific enzyme and chemical scission, were used to determine the key structural features shown in the diagram in Figure 7. The heparinase III depolymerization profile (Figure 6C) indicates that murine 3T3 fibroblast HS, in common with human fibroblast HS,34 largely consists of blocks of 3 to 7 N-sulfated disaccharides (S-domains) spaced apart by extended N-acetylated sequences. The 34-saccharide MPD has a 5-fold enrichment of the rare dodecasaccharide and tetradecasaccharide S-domains (Figure6D) compared to the parent HS. MPD has approximately twice the level of contiguous N-sulfated disaccharides (compare Figure 6B with 6A), which affirms the presence of 2 S-domains and is consistent with the ratio of approximately 2 to 1 N-sulfated to N-acetylated disaccharides in the proposed average structure (Figure 7A). Internal IdoA residues flanked by GlcNS in fibroblast heparinase III–resistant S-domains are consistently modified by 2-O-sulfation of C2,32 and 1 or 2 6-O-sulfate groups are seen more frequently toward the center of longer fibroblast S-domains, such as those in MPD. All heparinase III–derived S-domains that have been sequenced end in GlcNAc.32 The most prevalent contiguous N-acetylated region in MPD appears to be an octasaccharide (Figure 6B), which separates the 2 S-domains. The molecular weight of the predicted structure (Figure 7A) containing these features is almost exactly 8.3 kDa, the average size of MPD.
The motif of 2 S-domains separated by an N-acetylated region is emerging as a common structure for multimeric cytokines binding to HS and is thought to enable simultaneous interaction with 2 or more binding sites on the proteins. The PF418IFN-γ35 and IL-836 binding sites had this type of structure. However, in contrast to the dodecasaccharide or tetradecasaccharide S-domains of MPD, hexasaccharide and octasaccharide S-domains were present in the IFN-γ and IL-8 HS-binding sites. PF4-protected domains such as MPD had predominantly N-sulfated tetradecasaccharide regions at either end, but in PF4 each of these regions appeared to be formed from adjacent hexasaccharides and octasaccharides separated by a single heparinase III cleavage site. The difference between the MIP1α and PF4 sites may be indicative of the MIP1α requirement for 6-O-sulfation, which tends to be more prevalent within the longer S-domains, whereas 6-O-sulfation did not appear to be critical for PF4 binding. The MIP1α binding site appears to contain a particularly short octasaccharide N-acetylated stretch compared to these regions of approximately 15 saccharides for PF4 and IL-8 and a stretch of approximately 30 predominantly N-acetylated saccharides for IFN-γ. The length of this region may be critical for correct spacing and positioning of the S-domains to bind optimally to basic regions on the cytokines.
An MPD model was constructed using the coordinates of the solution structure of heparin (1 HPN) to form the S-domains (Figure 7A) because it has been shown that variations in sulfate substitution have relatively minor effects on the overall conformation of the polysaccharide chain.37 The unsulfated middle region of alternating GlcA and GlcNAc residues was modeled as an extended chain in which glycosidic linkages adopt sterically allowed conformations. The iduronate-containing S-domains such as heparin can be expected to adopt a relatively well-defined conformation, but there is no reason to suppose that the unsulfated GlcA-containing regions are similarly constrained.38 The GlcA-GlcNAc (cellobioselike) and GlcNAc-GlcA (maltoselike) linkages may adopt more than one low-energy conformation,39 40 leading to flexibility in this region. Binding sites face outward in opposite directions, so some flexibility of the HS fragment is necessary if each terminal S-domain is to bind to one of the 2 sites. The unsulfated central region of the MPD fragment measures approximately 40 Å in length when fully extended, similar to the end-to-end distance between the 2 heparin-binding sites of the dimer of 30 to 40 Å (for both the NMR structure 1B53 and the compact dimer modeled here). The overall length of the extended MPD fragment is more than 140 Å, which would be ample to wrap around 3 faces of the MIP1α dimer in a horseshoe shape.
Heparin probes were used to model where the S-domains of MPD were most likely to interact with the MIP1α dimer. Docking of pentasaccharide and endecamer heparin probes to MIP1α dimer structures identified the basic residues Arg 17, Arg 45, and Arg 47 and occasionally Lys 44 (in agreement with results from site-directed mutagenesis10) as the binding site. Although the S-domain–like heparin endecamer model was long enough to bridge both heparin-binding sites in the PF4-based dimer and there is no steric hindrance to its interacting with both sites across the β-sheet face of the dimer, it is noteworthy that the systematic docking procedure did not identify this as an energetically favored binding mode (Figure 7D). Therefore, the large size of the S-domains prevalent in MPD may be critical for the presence and correct positioning of 6-O-sulfate group(s) thought to be important for binding (see above discussion), as opposed to any additional electrostatic interactions occurring at the peripheries of the S-domains. Site-directed mutagenesis demonstrated that the same arginine residues—Arg17, 45, and 47—in a conserved 3-dimensional cleft were essential for heparin binding to MIP1α, with Lys44 playing a lesser role in heparin binding and the single basic residue in the α-helix, Lys60, playing no detectable role.10 This is in contrast to the α-chemokines IL-8 and PF4, in which lysine residues in the α-helix were thought to be involved in heparin binding41,42 though in the latter case mutation studies suggested arginine residues outside the helix may be more critical.43
Mutation of the key basic heparin-binding residues in MIP1α did not appear to affect receptor binding or signaling by MIP1α, nor was HS required for binding to CCR1 in HS-deficient CHO cells.9,10 Therefore, HS does not appear to be critical for ligand presentation to the receptor, as has been proposed for the fibroblast growth factors. MIP1α interaction with HS proteoglycans may be more important for appropriate localization and concentration of this chemokine, such as in bone marrow stem cell niches. Knowledge of the structure of the HS binding site for MIP1α may enable us to design agents to enhance the myeloprotective or peripheral blood stem cell mobilization properties of MIP1α that could be used to improve cancer chemotherapy treatments.6
We thank Erika de Wynter (Leeds University, United Kingdom) and members of the Haematology Group (Paterson) for their continued support with this project.
Supported by Cancer Research UK.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Sally E. Stringer, Drug Development Group, Paterson Institute for Cancer Research, Christie Hospital NHS Trust, Wilmslow Rd, Withington, Manchester, M20 4BX, United Kingdom; e-mail:sallyelizabethstringer@yahoo.co.uk.




This feature is available to Subscribers Only
Sign In or Create an Account Close Modal