Altered expression of the Fas-Fas ligand apoptotic pathway leads to lymphoproliferative and autoimmune diseases. Inlpr/lpr mice and children with autoimmune lymphoproliferative syndrome, defective apoptosis is due to Fas mutations. Large granular lymphocyte (LGL) leukemia is a clonal lymphoproliferative disorder associated with rheumatoid arthritis. Leukemic LGLs are resistant to Fas-dependent apoptosis despite expressing high levels of Fas. Such resistance can be overcome by activating leukemic LGLs in vitro, suggesting inhibition of Fas signaling in leukemic cells. We report that sera from patients with LGL leukemia contain high levels of soluble Fas. Ten of these 33 patients with LGL leukemia also had rheumatoid arthritis. Cloning and sequencing revealed expression of multiple Fas messenger RNA variants in leukemic LGL. These Fas variants, including 3 newly described here, encode soluble Fas molecules. Supernatants from cells transfected with these Fas variants blocked Fas-dependent apoptosis of leukemic LGLs. These results suggest that blockade of Fas-signaling by soluble Fas may be a mechanism leading to apoptosis resistance in leukemic LGLs.
Introduction
Apoptosis, or programmed cell death, is a process fundamental to the normal development and hemostasis of multicellular organisms.1-4 Dysregulation of apoptosis has been implicated in a number of human diseases, including cancer, leukemia, neurodegenerative disorders, and acquired immunodeficiency syndrome.3-5 One apoptotic pathway involves the Fas/Fas ligand system. Fas, also designated as Apo-1 or CD95, is a member of the tumor necrosis factor (TNF) receptor family of cell surface proteins, and Fas ligand is a member of the TNF family of proteins.6,7 Fas has a broad distribution on normal tissues, whereas Fas ligand is expressed primarily only on activated cytotoxic lymphocytes.8-11 Interaction of Fas with Fas ligand induces apoptotic cell death.7-12 Altered levels of expression of Fas and Fas ligand have been implicated in the pathogenesis of diseases associated with immune regulation.2,5 13-15
Large granular lymphocyte (LGL) leukemia is a neoplastic disorder of either CD3+ or CD3-LGLs.16 The T-cell form of this disease is associated with neutropenia and autoimmune disorders such as rheumatoid arthritis. CD3+ LGL leukemia cells are thought to represent in vivo–activated cytotoxic T lymphocytes (CTLs) of unknown antigen specificity.17 The mechanisms involved in prolonging survival of these leukemic LGLs are not entirely understood. A primary role of Fas-mediated apoptosis is deletion of peripheral antigen-activated T cells in the normal immune system.18 We have found that leukemic LGLs, like antigen-activated CTLs, express high levels of both Fas and Fas ligand. Unlike normal activated CTLs, however, leukemic LGLs are resistant to Fas-mediated apoptosis.19
Animal models of lymphoproliferative disorders associated with autoimmune features are characterized by Fas resistance.15Serologic features of the autoimmune disorder include antibodies to rheumatoid factor, high levels of circulating immune complexes, and polyclonal hypergammaglobulinemia. These serologic abnormalities are also common features of LGL leukemia. Defective apoptosis inlpr/lpr mice is due to mutations in Fas13; a similar pathogenetic mechanism has been discovered in children with autoimmune lymphoproliferative syndrome.20,21 In contrast, mutations in the death domain of Fas in leukemic LGLs have not been detected.19 A second mechanism of Fas resistance is production of soluble Fas receptors, which can act as decoys for Fas ligand.22 Soluble forms of cell surface receptors can be produced either by proteolytic cleavage of membrane-bound receptors or by alternative splicing. Normal activated T cells (peripheral blood mononuclear cells [PBMCs]) express alternative splicing messenger RNA (mRNA) variants coding for soluble forms of Fas proteins.23-25 Furthermore, expression of soluble Fas in mice leads to an autoimmune syndrome and elevated levels of soluble Fas have been found in some patients with autoimmune diseases.22 To investigate the mechanism of apoptosis resistance in LGL leukemia, we cloned and sequenced the Fas gene from leukemic LGLs. We describe several Fas mRNA variants, which functionally block Fas-mediated apoptosis. Our results suggest that blockade of Fas function by decoy Fas receptors may contribute to the pathogenesis of LGL leukemia.
Patients, materials, and methods
Patients with LGL leukemia
All patients met clinical criteria of T-cell/LGL leukemia with LGL counts ranging from 600/μL to 27 000/μL (normal, 223 ± 99/μL) and evidence of clonal T-cell receptor gene rearrangement.16
RNA extraction, complementary DNA synthesis, and reverse transcription-polymerase chain reaction
The PBMCs were purified by Ficoll-Hypaque density gradient centrifugation from 7 patients with LGL leukemia and from normal leukocyte buffy coats obtained from the Southwest Florida Blood Bank. Total RNA was prepared using a rapid RNA isolation kit (RNA STAT-60; Tell-Test, Inc, Friendswood, TX). RNA was also extracted from lymph nodes of 4 patients with B-cell non-Hodgkin lymphoma. One microgram total RNA was used in a first-strand complementary DNA (cDNA) synthesis with oligo deoxythymidine (dT) and Moloney murine leukemia virus using a standard protocol in 20 μL reactions. Two microliters of the cDNA reaction mixture was used as a template in the polymerase chain reaction (PCR). The following synthetic oligonucleotides, previously described,23 were used to amplify the Fas open reading frame starting from the initiation codon at positions 195-197 and ending at the termination codon at position 1201-1203:[U12] 5′-ATGCTGGGCATCTGGACCCT-3′ and 5′-TCTAGACCAAGCTTTGGATTTC-3′. Cycling parameters were 94°C for 30 seconds; 55°C for 1 minute 30 seconds; and 73°C for 1 minute 30 seconds for 30 cycles, with the final extension step at 72°C for 5 minutes. The PCR products were stained with 1 μg/mL ethidium bromide after migration on a 1% agarose gel, purified, and cloned in the pGEM-T Easy Vector systems (Promega, Madison WI). At least 6 clones from 2 different PCR reactions of RNA samples from each of 7 patients with LGL leukemia and from 2 healthy individuals were sequenced by the dideoxy chain termination method with an automatic DNA sequencing system (Pharmacia, Piscataway, NJ). Fas cDNA sequence was read from independent clones at least 2 times in each direction. Sequence data were analyzed using the GCG gene program (Genetics Computer Group, Madison, WI).
Soluble Fas enzyme-linked immunosorbent assay
Detection of soluble Fas in the serum of LGL leukemia patients was performed using a soluble Fas enzyme-linked immunosorbent assay (ELISA) kit according to the manufacturer's instructions (Bender Med Systems, Vienna, Austria).
Construction of expression vectors
Cloned Fas gene transcripts (the Fas full-length, FasExo6Del, and FasExo4,6Del cDNAs) were digested from pGEM-T easy vectors withEcoRI restriction enzyme and subcloned into a mammalian expression vector, pcDNA3.1Zeo(+) (Invitrogen, San Diego, CA). The orientation of each of these cDNA in pcDNA3.1Zeo(+) was verified by restriction mapping. These pcDNA3.1Zeo(+) vectors containing the Fas full-length, FasExo6Del, and FasExo4,6Del cDNAs in correct orientation were then transfected into COS cells by the superfact transfection reagent (Qiagen, Valencia, CA). After 48 hours of transfection, COS cells were selected in RPMI medium containing 400 μg/mL Zeocin for 2 weeks. Individual Zeocin-resistant colonies were then isolated and further expanded in the Zeocin-containing medium. To examine the surface expression of human Fas molecules, transfected cells were stained with phycoerythrin (PE)–labeled anti-Fas antibody (UB2; Kamiya, Tukwila, WA) or PE-labeled isotype control IgG1 (Kamiya), as described previously.19 Briefly, one million cells were incubated with 10 μL of the directly labeled antibodies for 30 minutes at 4°C in 100 μL phosphate-buffered saline (PBS) supplemented with 2% fetal calf serum and 0.02% sodium azide. After 2 washes with the same PBS washing buffer, cells were analyzed on a FACScan (Becton Dickinson, Mountain View, CA). For cytoplasmic staining of Fas and Fas variants, COS cells were fixed with 4% paraformaldehyde for 5 minutes and permeabilized with ethanol and acetic acid. The staining was performed as described as above.
Apoptosis assay
The PBMCs from healthy donors and patients with LGL leukemia were stimulated with phytohemagglutinin (PHA; 1 μg/mL, 2 days) and interleukin 2 (IL-2; 100 U/mL, 10 days). The apoptosis-inducing anti-Fas (CH11) antibody was then added in the presence of supernatant harvested from COS cells transfected with vector alone, full-length Fas cDNA, or Fas variants. Apoptotic cell detection was determined by flow cytometry using staining with 7 amino–actinomycin D (7-AAD; Calbiochem, San Diego, CA) and propidium iodide (PI; Molecular Probes, Eugene, OR). For 7-AAD staining, cells were incubated with 7-AAD at a concentration of 20 μg/mL for 30 minutes at 4°C in the dark. The cells were then resuspended in PBS and analyzed using flow cytometry.19 For PI staining, cells were washed once with PBS, then 1 mL hypotonic PI solution 2 (50 μg/mL PI in 0.1% sodium citrate solution plus 0.1% Triton X-100) was added. Cells were kept overnight at 4°C in the dark, and then analyzed on a FACScan flow cytometer (Becton Dickinson) in their staining solution, as described previously.19
Results
Identification and characterization of Fas splicing variant mRNAs in leukemic LGLs
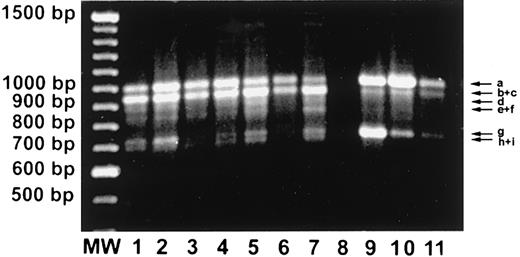
Results of reverse transcription–PCR (RT-PCR) analyses of Fas gene transcripts are shown in Figure 1. In addition to the expected fragment corresponding to the full-length cDNA, several distinct smaller products were observed (labeled a to i in Figure 1). Expression of full-length Fas gene transcript (band a) in samples from LGL leukemia patients appeared higher than expression in samples from unactivated PBMCs. These results are consistent with flow cytometric analyses showing high levels of Fas expression on leukemic LGLs.19 Expression of band b, which represents a transmembrane-deleted (exon 6 deletion) Fas mutant (see below), appeared higher in samples from patients with LGL leukemia than from samples of either unactivated or activated normal PBMCs. Bands c to i representing a variety of even smaller Fas mRNA splicing products were found primarily in samples from patients with LGL leukemia. Similar analysis of RNA samples from 4 patients with B-cell non-Hodgkin lymphoma showed the presence of only full-length Fas transcripts (not shown).
Leukemic LGLs express high levels of variant gene transcripts representing soluble Fas.
RT-PCR analyses of the Fas mRNA were performed by synthesizing cDNA with oligo dT and performing PCR amplifications with primers described in “Patients, materials, and methods.” The full-length Fas mRNA is a predicted 1009–base pair (bp) (195-1203) fragment. Lanes 1-7, unstimulated PMBCs from LGL leukemia patients; lane 8, negative control; lane 9, Jurkat cells; lane 10, normal PHA/IL-2–activated PBMCs; lane 11, unstimulated normal PBMCs. Arrow a represents full-length Fas mRNA, and arrows b through i represent alternatively spliced mRNA.
Leukemic LGLs express high levels of variant gene transcripts representing soluble Fas.
RT-PCR analyses of the Fas mRNA were performed by synthesizing cDNA with oligo dT and performing PCR amplifications with primers described in “Patients, materials, and methods.” The full-length Fas mRNA is a predicted 1009–base pair (bp) (195-1203) fragment. Lanes 1-7, unstimulated PMBCs from LGL leukemia patients; lane 8, negative control; lane 9, Jurkat cells; lane 10, normal PHA/IL-2–activated PBMCs; lane 11, unstimulated normal PBMCs. Arrow a represents full-length Fas mRNA, and arrows b through i represent alternatively spliced mRNA.
To further characterize the smaller fragments, the Fas RT-PCR products from 7 individuals with LGL leukemia and 1 healthy donor were cloned into the pGEM-T Easy Vector systems (Promega), and the nucleotide sequence was determined with T7 and SP6 primers by an automatic DNA sequencing system (Pharmacia). We found that all of these smaller transcripts were Fas cDNA variants with deletion of the transmembrane region of Fas. These Fas cDNA variants are represented schematically in Figure 2. A common descriptive nomenclature is used based on which exons are deleted in the Fas gene structure.25 Bands a (Fas), b (FasExo6Del), d (FasExo4Del), e (FasExo4,6Del), g (FasExo3,4Del), and h (FasExo3,4,6Del) have been previously found only in activated normal PBMCs.25 Band b was previously called Fas TM De122,23 and lacks exon 6, the transmembrane region of Fas. Band d encodes for a mature protein of 133 amino acid residues, 38 of which at the C-terminal end (box A in Figure 2) differ from those of the Fas protein.25 These same 38 amino acids are seen at the C-terminal end of band g, which also contains a deletion in exon 3. Band e contains 2 deletions; the second deletion results in an altered reading frame with a new termination codon at position 765. The last 21 amino acids at the C-terminal end differ from those of the Fas protein (box B). A similar structure is seen at the C-terminal end of band h, which contains an additional deletion of exon 3. Bands c (FasExo6,7Del), f (FasExo4,6,7Del), and I (FasExo3,4,6,7 Del) have not been described previously. These Fas cDNA variants are slightly different from bands b, e, and h, respectively, in that they contained an extra deletion in the 5′ end of exon 7. They thus contain an additional 20 amino acid residues at the C-terminal end (box C in Figure 2), which are different from those of the Fas protein. As a result, a new stop codon is generated at the C-terminal end of box C. The sizes of cDNA's b and c, e and f, and h and i, respectively, are very similar and they comigrate on 1% agarose gel, as shown in Figure 1.
Schematic representation of human Fas cDNAs structure.
The coding regions are represented as boxes; hatched, signal sequence (SP); dark shaded, transmembrane (TM) region; cross-hatched, signal transducing domain (ST); white boxes, sequences corresponding to the Fas protein; white boxes labeled A, B, and C correspond to sequences generated by a different reading frame. The nomenclature used to describe the variant Fas cDNAs corresponds to terminology proposed previously.25
Schematic representation of human Fas cDNAs structure.
The coding regions are represented as boxes; hatched, signal sequence (SP); dark shaded, transmembrane (TM) region; cross-hatched, signal transducing domain (ST); white boxes, sequences corresponding to the Fas protein; white boxes labeled A, B, and C correspond to sequences generated by a different reading frame. The nomenclature used to describe the variant Fas cDNAs corresponds to terminology proposed previously.25
High levels of soluble Fas in LGL leukemia sera
Because the variant Fas transcripts containing deletions in the transmembrane region of Fas were highly expressed in LGL leukemia samples, we were interested in measuring levels of soluble Fas in these patients. The amount of soluble Fas in sera from patients with LGL leukemia (n = 30) was compared to levels found in healthy donors (n = 10) using an ELISA kit (Figure 3). Each sample was assayed in duplicate and intra-assay variation was less than 5%. Sera from most LGL leukemia patients expressed high amounts of soluble Fas ranging up to 519 U/mL. In contrast, most of the control sera from healthy donors contained negligible quantities of soluble Fas with maximal value being 86 U/mL. The levels of soluble Fas in sera of LGL leukemia patients were significantly higher than control sera (P < .0001, using 2-sided Wilcoxon analyses).
Elevated levels of circulating soluble Fas in sera of patients with LGL leukemia.
The concentration of soluble Fas in sera from patients with LGL leukemia (n = 30) was measured with a soluble Fas ELISA and compared to values measured in sera from healthy donors (n = 10). The horizontal lines indicate the mean value of the samples.
Elevated levels of circulating soluble Fas in sera of patients with LGL leukemia.
The concentration of soluble Fas in sera from patients with LGL leukemia (n = 30) was measured with a soluble Fas ELISA and compared to values measured in sera from healthy donors (n = 10). The horizontal lines indicate the mean value of the samples.
Splicing variant mRNA are translated as soluble Fas
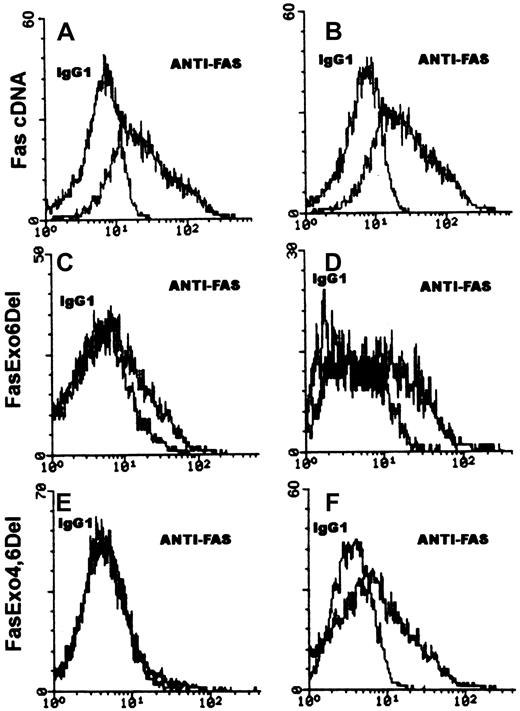
To confirm the existence of the soluble Fas translation products, Fas, FasExo6Del, and FasExo4,6Del coding regions were inserted into a mammalian expression vector, pcDNA3.1Zeo(+) and transfected into COS cells. The transfected COS cells were first grown in nonselected regular medium for 2 days and selection was begun on day 3 by the addition of 400 μg/mL Zeosin (Invitrogen) for 2 to 4 weeks. About 20 single colonies were isolated from each transfection. Evidence for production of proteins encoded by the clones was obtained by flow cytometry analyses with anti-Fas monoclonal antibody (mAb). These analyses showed the surface expression of Fas on COS cells transfected with the full-length Fas cDNA but not on COS cells transfected with the FasExo6Del or FasExo4,6Del expression plasmids. Production of the FasExo6Del and FasExo4,6Del proteins from transfected COS cells was shown by cytoplasmic Fas staining (Figure4).
Flow cytometry analysis of expression of Fas on COS cells transfected with Fas cDNAs.
COS cells were transfected with either the Fas cDNA, FasExo,6Del cDNA, or FasExo4,6Del cDNA and individual colonies were isolated. Single-cell suspensions were stained with either fluorescein isothiocyanate (FITC)–conjugated IgG1 isotype control mAb or FITC-conjugated anti-Fas mAb. Cytoplasmic staining was carried out by fixation of the cells with 0.5% paraformaldehyde and permeabilization with 0.1% Triton X-100 to allow entry of the antibody. Panels on the left represent surface staining; panels on the right represent cytoplasmic staining. The representative histograms show the specific binding (anti-Fas) and nonspecific binding (IgG1) as indicated.
Flow cytometry analysis of expression of Fas on COS cells transfected with Fas cDNAs.
COS cells were transfected with either the Fas cDNA, FasExo,6Del cDNA, or FasExo4,6Del cDNA and individual colonies were isolated. Single-cell suspensions were stained with either fluorescein isothiocyanate (FITC)–conjugated IgG1 isotype control mAb or FITC-conjugated anti-Fas mAb. Cytoplasmic staining was carried out by fixation of the cells with 0.5% paraformaldehyde and permeabilization with 0.1% Triton X-100 to allow entry of the antibody. Panels on the left represent surface staining; panels on the right represent cytoplasmic staining. The representative histograms show the specific binding (anti-Fas) and nonspecific binding (IgG1) as indicated.
Prevention of programmed cell death by soluble Fas variants
High levels of soluble Fas can block Fas-mediated apoptosis of normal activated T cells. We wanted to know whether such a mechanism mediated Fas resistance of leukemic LGLs. Sera from patients with LGL leukemia contain high levels of soluble Fas. We found that in contrast to normal sera, LGL leukemia sera abrogated Fas-induced apoptosis of normal activated T cells (Figure 5). We showed previously that Fas resistance in leukemic LGLs could be reversed by activation with IL-2.19 In this study, we found that sera from LGL leukemia patients would also protect activated leukemic LGL from apoptosis. Sera from these patients blocked apoptosis by 67% to 82%, whereas serum from a fourth patient inhibited apoptosis by 30% (Figure6).
LGL serum blocks Fas-induced apoptosis of normal activated T cells.
Panel on left shows spontaneous apoptosis. Panel in middle shows apoptosis of normal activated cells induced by anti-Fas mAb, with normal human serum added. Panel on right shows blocking of apoptosis by serum from an LGL leukemia patient. Similar results were obtained when studying sera from an additional 7 patients with LGL leukemia.
LGL serum blocks Fas-induced apoptosis of normal activated T cells.
Panel on left shows spontaneous apoptosis. Panel in middle shows apoptosis of normal activated cells induced by anti-Fas mAb, with normal human serum added. Panel on right shows blocking of apoptosis by serum from an LGL leukemia patient. Similar results were obtained when studying sera from an additional 7 patients with LGL leukemia.
LGL leukemia sera blocks Fas-induced apoptosis of leukemic LGLs.
PBMCs from patients with LGL leukemia were activated with PHA and IL-2, then incubated with CH11 antibody in the presence of 20% normal serum or patient's autologus serum.
LGL leukemia sera blocks Fas-induced apoptosis of leukemic LGLs.
PBMCs from patients with LGL leukemia were activated with PHA and IL-2, then incubated with CH11 antibody in the presence of 20% normal serum or patient's autologus serum.
We then investigated the hypothesis that the Fas variants cloned from leukemic LGLs could protect both normal PBMCs and leukemic LGLs from apoptotic death. Activated cells were cultured with apoptosis-inducing Fas mAb in the presence of serial dilutions of supernatants from transfected COS cells. We found that supernatants from COS cells transfected with Fas Exo6Del and Fas Exo4,6 Del blocked apoptosis of activated PBMCs from both healthy donors and from patients with LGL leukemia in a dose-dependent fashion (Figure7). In contrast, supernatants from COS cells transfected with full-length Fas cDNA or with control constructs did not inhibit Fas-mediated apoptosis.
Soluble Fas variants block apoptosis of leukemic LGLs.
(A) PBMCs from a patient with LGL leukemia were stimulated with PHA and IL-2, then incubated with CH11 in the presence or absence of 40% concentration of supernatant harvested from COS cells transfected with control construct, pcDNA3.1, full-length Fas cDNA, or various Fas mutants. The concentration of soluble Fas in these supernatants is indicated as measured using soluble Fas ELISA. (B) The activated PBMCs from the same patient with LGL leukemia were incubated with CH11 in the presence of serial dilutions of supernatant obtained from COS cells transfected with the Exo4,6Del Fas mutant. The concentration of soluble Fas in this dilution series is indicated. The percentages of apoptotic cells are indicated at the left upper corner. Similar data were obtained in experiments using PBMCs from 3 other patients with LGL leukemia.
Soluble Fas variants block apoptosis of leukemic LGLs.
(A) PBMCs from a patient with LGL leukemia were stimulated with PHA and IL-2, then incubated with CH11 in the presence or absence of 40% concentration of supernatant harvested from COS cells transfected with control construct, pcDNA3.1, full-length Fas cDNA, or various Fas mutants. The concentration of soluble Fas in these supernatants is indicated as measured using soluble Fas ELISA. (B) The activated PBMCs from the same patient with LGL leukemia were incubated with CH11 in the presence of serial dilutions of supernatant obtained from COS cells transfected with the Exo4,6Del Fas mutant. The concentration of soluble Fas in this dilution series is indicated. The percentages of apoptotic cells are indicated at the left upper corner. Similar data were obtained in experiments using PBMCs from 3 other patients with LGL leukemia.
Discussion
We found that sera from most patients with LGL leukemia contained high levels of soluble Fas. The amount of soluble Fas in most normal sera was less than 50 U/mL. In contrast, levels of up to 519 U/mL were observed in sera from patients with LGL leukemia. Elevated levels of soluble Fas have been reported in other hematologic malignancies as well as some autoimmune diseases.26 27 Whether elevated levels of soluble Fas contribute to the pathogenesis of these disorders is not known.
A central feature of LGL leukemia is a dysregulated Fas/Fas ligand apoptotic pathway. Despite expressing high levels of Fas, leukemic LGLs are Fas-resistant.19 Fas resistance underlies the pathogenesis of autoimmune lymphoproliferative diseases in both animal models and humans. In these circumstances, defects in Fas-mediated apoptosis are due to mutations in Fas.13,20,21 In contrast, however, we had shown a normal-sized Fas gene transcript in patients with LGL leukemia, without any function-ablating mutations.19 Previous studies indicated that Fas resistance can be reversed by activating leukemic LGLs in vitro.19 These data suggest that inhibition of Fas signaling rather than defective Fas was responsible for Fas resistance. High levels of soluble Fas can block Fas-mediated apoptosis.22 We found that LGL sera abrogated Fas-mediated apoptosis of normal PBMCs as well as leukemic LGLs activated in vitro. These results suggested that high levels of soluble Fas might be one mechanism responsible for Fas resistance in leukemic LGLs.
We identified and characterized 8 variant Fas mRNA transcripts encoding for soluble Fas from LGL leukemia samples. Of these 8 Fas variants, 5 have been described previously only in activated normal PBMCs.25 Constitutive expression of Fas variants in leukemic LGL further supports the idea that leukemic LGLs have been activated in vivo and most likely represent antigen-activated CTLs.19 Three of the identified Fas variants are novel sequences resulting from deletions in exon 7. The common feature of the splice variants is that they all contain the first 391 nucleotides (exons 1 and 2). This region appears functionally important in the ability of soluble Fas to block Fas-mediated apoptosis.25We demonstrated that 2 of the Fas variants cloned from leukemic LGL samples could be translated into protein, which functionally blocked Fas-mediated apoptosis. These results suggest that such decoy Fas receptors might contribute to the pathogenesis of LGL leukemia by blocking apoptosis of the leukemic cells. Previously, we found that constitutive signal transducer and activator of transcription 3 (STAT3) activation resulting in up-regulation of antiapoptotic geneMcl-1 might explain apoptotic resistance in leukemic LGL.28 Taken together, these results suggest that inhibition of apoptotic signaling may be a common pathway of disease causation in this disorder.
Autoimmune manifestations are a prominent and characteristic feature of LGL leukemia. Serologic abnormalities are also frequent, including autoantibodies such as rheumatoid factor and antinuclear antibody, circulating immune complexes, and polyclonal hypergammaglobulinemia. Autoimmune disease, particularly rheumatoid arthritis, occurs frequently in LGL leukemia. Indeed, 10 patients in this study had classic rheumatoid arthritis. These serologic features are also observed in animal models of Fas deficiency resulting from function-ablating mutations in Fas.13 The precise mechanisms leading to autoimmunity in these animal models of Fas deficiency are not entirely resolved. Therefore, it is not possible to conclude from our study that high levels of decoy Fas molecules contribute to the observed autoimmune features in our patients with LGL leukemia. Nevertheless, our results suggest that a similar autoimmune lymphoproliferative phenotype may also occur in diseases associated with defective Fas apoptosis secondary to blockade by soluble Fas.
The Flow Cytometry, Molecular Biology, and Biostatitics Cores at H. Lee Moffitt Cancer Center and Research Institute were used in the course of this work.
Supported by a Veterans Administration Merit Review Grant, by National Cancer Institute grants CA83947 and CA90633, and the American Cancer Society's Institutional Research Grant 93-032.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Thomas P. Loughran, Jr., MRC-4047, H. Lee Moffitt Cancer Center and Research Institute, 12902 Magnolia Dr, Tampa, FL 33612; e-mail: loughrat@moffitt.usf.edu.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal