Most cases of hereditary hemochromatosis are due to a single nucleotide mutation in the hemochromatosis gene(HFE) that results in a Cys to Tyr conversion at amino acid 282 (Cys282Tyr) in the protein.1 Sequencing revealed a second mutation (His63Asp) in the HFE protein, but the penetrance of this mutation is much lower compared to Cys282Tyr. Although a candidate early in the search for the hemochromatosis gene, the transferrin receptor (TFR) was not found to be mutated in hemochromatosis. Several years ago, we discovered a second human transferrin receptor termed transferrin receptor 2 (TFR2).2 Recently, a hemochromatosis pedigree was discovered in Sicily, where a non–sense mutation at position 250 in the TFR2 gene was found.3 Affected individuals lacked the Cys282Tyr HFE mutation. The carrier frequency of this mutation is 0.9% among a cohort of southern and central Europeans.4 Roetto and colleagues recently reported 2 new mutations (at exon 2, 84-88insC, resulting in Glu60Xaa; and at exon 4, Met172Lys) in iron overload patients having what has been termed hemochromatosis type 3.5 A fourth inactivating mutation ofTFR2 (a 4–amino-acid loss Ala-Val-Ala-Gln at 594-597) has recently been reported.6
Here, we investigated the genomic DNA from individuals having atypical hemochromatosis with the aim to look for a correlation between mutations of the TFR2 gene and an altered iron phenotype. We also asked whether differences in penetrance of the Cys282Tyr mutation were associated with mutations in TFR2. The study included several selected cohorts: (1) Sibling pairs homozygous for HFECys282Tyr with a discordant phenotype. The most common discordance between homozygote siblings was serum ferritin concentration. Many of these patients, however, also exhibited significant differences in liver fibrosis and aminotransferase levels (11 patients); (2) Non-Cys282Tyr HFE homozygotes with evidence of iron excess (7 patients), which included patients having evidence of iron overload but lacking the Cys282Tyr mutation (3 patients), a patient normal at the 282 position but homozygous for His63Asp (1 pt), and patients with iron overload but heterozygous for Cys282Tyr (3 patients); (3) Homozygous (Cys282Tyr) relatives of probands identified in a blood bank screen (because of elevated transferrin saturation) who have evidence of morbidity (3 patients); and (4) Cys282Tyr homozygotes under 30 years of age with iron overload (9 patients). In addition, we investigated samples from 10 healthy individuals as controls. Values for serum iron, transferrin saturation, and serum ferritin were available from all individuals.
Genomic DNA was analyzed by polymerase chain reaction–single strand conformation polymorphism (PCR-SSCP) as previously described.7 Nineteen primer pairs were designed using sequence information from the Genbank for TFR2 (GI 3135305) from the complete sequence for chromosome 7q22. Each PCR reaction contained 20 ng DNA, 10 pmol of each of the primers, 2 nmol of each of the deoxynucleoside triphosphates (dNTPs), 0.5 unitsTaq DNA polymerase, and 3 μCi (11.1 × 104Bq) α-[32P] deoxycytidine triphosphate (dCTP) in 20 μL of the specified buffer with 1.5 mM MgCl2. The PCR cycles were 30 seconds for denaturing at 94°C, 40 seconds for annealing at 60°C, and 60 seconds for extension at 72°C (35 cycles). The samples were separated on a 6% nondenaturating polyacrylamide mutation detection enhancement (MDE) gel. DNA purified from mutant candidate bands showing altered migration through SSCP analysis was directly sequenced by the ABI PRISM dye terminator cycle sequencing reaction (Perkin Elmer, Foster, CA).
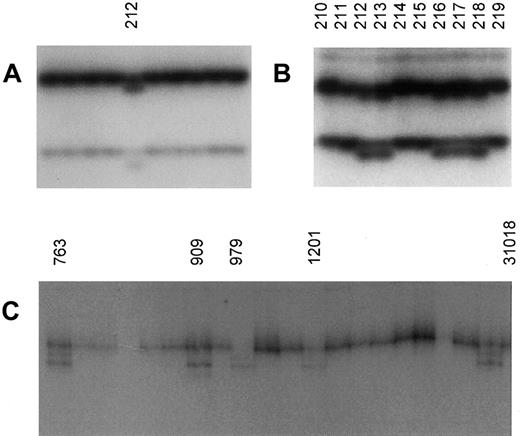
Analysis of exon 10 of TFR2 revealed an aberrantly shifted band (Figure 1A) in a sample from an individual in group A, pedigrees in which a discordance existed in phenotype between Cys282Tyr homozygous sibling pairs. Direct nucleotide sequencing found a G>A transversion at nt 1391, resulting in a substitution at codon 455 of Gln (mutant) for an Arg (normal sequence). The change was verified by sequencing from both the sense and antisense directions. The individual initially identified with the mutation was the mother of the proband. The husband and 8 children were then analyzed. Of the children, 4 were found to have the polymorphism (Figure 1B). One was normal at the HFE locus for the Cys282Tyr mutation, 2 were heterozygous, and 1 (the brother of the proband) was also homozygous for Cys282Tyr. The proband did not have the Arg455Gln mutation. Interestingly, this brother was identified in cohort A as having evidence of liver fibrosis, where his brother (the proband) did not.
Polymerase chain reaction–single strand conformation polymorphism analysis of exons 10 and 18 in atypical hemochromatosis.
(A) Sample no. 212 (mother of proband) had an aberrantly shifted band in exon 10. Direct nucleotide sequencing found a G>A transversion resulting in a substitution at codon 455 of Gln (aberrant) for an Arg (normal sequence). (B) Four of the genomic DNA samples (nos. 213, 216-218) from the children of proband no. 212 showed, by PCR-SSCP, the same DNA-migration pattern as sample no. 212. Direct DNA sequencing of these samples revealed Arg455Gln. (C) Evidence for polymorphism in the 3′ untranslated region of exon 18 of TRF2. Samples 763, 909, 979, 1201, and 31018 showed the same polymorphic pattern by SSCP. Direct sequencing of these samples identified a change of G>C at nucleotide 154513 (GI3135305).
Polymerase chain reaction–single strand conformation polymorphism analysis of exons 10 and 18 in atypical hemochromatosis.
(A) Sample no. 212 (mother of proband) had an aberrantly shifted band in exon 10. Direct nucleotide sequencing found a G>A transversion resulting in a substitution at codon 455 of Gln (aberrant) for an Arg (normal sequence). (B) Four of the genomic DNA samples (nos. 213, 216-218) from the children of proband no. 212 showed, by PCR-SSCP, the same DNA-migration pattern as sample no. 212. Direct DNA sequencing of these samples revealed Arg455Gln. (C) Evidence for polymorphism in the 3′ untranslated region of exon 18 of TRF2. Samples 763, 909, 979, 1201, and 31018 showed the same polymorphic pattern by SSCP. Direct sequencing of these samples identified a change of G>C at nucleotide 154513 (GI3135305).
In addition, analysis of exon 18 of TFR2 showed an identically shifted band (Figure 1C) in 5 samples (1 from group A, 1 from group B, 2 from group D, and 1 in the normal controls). Direct sequencing of these samples showed a change of G>C in the 3′ untranslated region (3′ UTR) of exon 18. Since this change also was present in the normal control sample, we believe that it represents a previously unreported polymorphism. No correlation was found between this alteration and any of the clinical subtypes.
In summary, a group of individuals selected for unusual iron phenotypes was analyzed for evidence of mutation in the TFR2 gene. None had mutations corresponding to those described in the Italian iron overload pedigrees. We describe a new mutation, Arg455Gln, in exon 10 of TFR2 in a pedigree containing an individual with evidence of liver fibrosis in contrast to his HFE identical brother. This mutation could represent a modifier for penetrance of the hemochromatosis phenotype when present with homozygosity for Cys282Tyr. Unlike TFR1 expression, TFR2 expression is not down-regulated in the liver of iron-loaded mice.8 Our screening for mutations in all 18 exons of TFR2 in genomic DNA from all of the individuals indicates that mutations of theTFR2 gene are rare. The polymorphism in the 3′ UTR that we detected was found in individuals with abnormal iron metabolism as well as in a healthy control. Further cohort studies are needed to determine if this polymorphism is associated with a subtype of hereditary hemochromatosis.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal