Abstract
In this study the blood cells of 4 male patients from 2 unrelated families with chronic nonspherocytic anemia and recurrent bacterial infections were investigated. The activity of glucose-6- phosphate dehydrogenase (G6PD) in the red blood cells and in the granulocytes of these patients was below detection level. Moreover, their granulocytes displayed a decreased respiratory burst upon activation. Sequencing of genomic DNA revealed a novel 3–base pair (TCT) deletion in the G6PD gene, predicting the deletion of a leucine at position 61. The mutant G6PD protein was undetectable by Western blotting in the red blood cells and granulocytes of these patients. In phytohemagglutinin-stimulated lymphocytes the G6PD protein was present, but the amount of G6PD protein was strongly diminished in the patients' cells. Purified mutant protein from an Escherichia coli expression system showed decreased heat stability and decreased specific activity. Furthermore, we found that the messenger RNA of G6PD180-182delTCT is unstable, which may contribute to the severe G6PD deficiency observed in these patients. We propose the name “G6PD Amsterdam” for this new variant.
Introduction
In cells lacking mitochondria, such as red blood cells (RBCs), the only source of reduced nicotinamide adenine dinucleotide phosphate (NADPH) is the hexose monophosphate pathway.1 In the first step of this pathway, glucose-6-phosphate (G6P) is converted into 6-phosphogluconolactone, catalyzed by glucose-6-phosphate dehydrogenase (G6PD), and accompanied by the reduction of nicotinamide adenine dinucleotide phosphate (NADP) into NADPH. A sufficient amount of NADPH is essential for the integrity of RBCs, because NADPH reduces glutathione, which protects these cells against oxidative stress. G6PD deficiency therefore leads to hemolytic anemia, ranging from mild hemolytic anemia induced by infections or drugs to chronic nonspherocytic anemia with attacks of severe anemia induced by infections or drugs.1
Because the G6PD gene is located on the X chromosome, G6PD deficiency usually becomes manifest in hemizygous men. The severity of the G6PD deficiency depends on the effects of the mutation on protein stability and activity. So far, only missense and small in-frame deletions are known; nonsense mutations or large deletions have not been described, consistent with the idea that complete G6PD deficiency is incompatible with life. G6PD deficiency is usually restricted to RBCs, because these cells have a relatively long survival time (3 months) after release from the bone marrow but lack protein synthesis. Thus, G6PD protein instability is first manifested in RBCs.1
Although rare, severe G6PD deficiency can lead to symptoms of chronic granulomatous disease (CGD), such as recurrent bacterial and fungal infections.2-5 This disease is normally caused by a defect in one of the components of the NADPH oxidase. This enzyme catalyzes the generation of superoxide in phagocytes, which is used by the phagocytes to kill ingested microorganisms.6 In severe G6PD deficiency, superoxide cannot be formed in the phagocytes due to a lack of NADPH in these cells, an impairment that is seen when G6PD activity in the phagocytes is less than 5% of normal values.
Here, we investigated the genetic defect underlying the severe G6PD deficiency found in patients of 2 different families with mild chronic hemolytic anemia and recurrent bacterial or fungal infections. In both families a novel deletion of 3 nucleotides, the TCT triplet at position 180-182 in the G6PD gene, was found. This deletion predicts a G6PD protein that lacks a leucine at position 61. Our results indicate that the deletion of this leucine leads to the expression of an unstable and less active G6PD protein. These altered enzyme properties and the unstable messenger RNA (mRNA) of this variant probably cause the chronic nonspherocytic anemia and CGD-like symptoms in these patients. We propose to call this variant enzyme “G6PD Amsterdam.”
Patients, materials, and methods
Clinical histories
Patient I1 (born in 1981 to Caucasian parents) had an unremarkable medical history (no reports of chronic anemia) until he was admitted to an external hospital at age 15 years with recurrent episodes of fever, jaundice, gastroenteritis, and coughing. Three months after the first symptoms, he presented with cerebral convulsions. Diagnostic evaluation demonstrated invasive disseminated aspergillosis in the lungs, the nervous system (brain, right parietal side, cervical epidural abscess), and soft tissues (tumor of the right thigh, excision 1 week before admission). During the infectious episodes the patient developed icterus that appeared due to hemolysis (hemoglobin [Hb] 85 g/L [8.5 g/dL]) and for which he received an RBC transfusion. G6PD deficiency was detected. Aspergillosis was successfully treated with amphotericin B and flucytosine, followed by liposomal amphotericin B and itraconazole. Thereafter, the Hb level was normal (140 g/L [14.0 g/dL]) but reticulocytosis was still present (75‰). One of his brothers (I2) has known G6PD deficiency and presented with prolonged neonatal jaundice and episodes of acute hemolysis, but he has no known disposition to infections. In a period without clinical problems, he had a normal Hb level but 56‰ reticulocytes. His other 2 brothers are clinically healthy, although one of them (I3) was found during the course of these investigations also to be G6PD deficient. This brother also had a normal Hb value but mild reticulocytosis (32‰), as had his mother (39‰).
Patient II1 (born in 1993 to Hindustan parents) was healthy until the age of 3.5 years, when he was admitted to the hospital with high fever, coughing, tachypnea, and tachycardia. A chest x-ray showed pneumonia of the right basal lobe. Blood cultures revealed Chromobacterium violaceum, an uncommon human pathogen that can cause serious infections in patients with neutrophil dysfunction. He also had anemia (Hb 75 g/L [7.5 g/dL]), which was thought to be due to the septicemia. After initial treatment with cephalosporin (cefuroxime), the therapy was changed upon the antibiogram into meropenem, which was continued for 14 days. He responded well, although the anemia persisted. One day after antibiotic discontinuation, he relapsed with sepsis. Again Chromobacterium violaceum was cultured from the blood. Magnetic resonance imaging showed osteomyelitis of the thoracic spine at T10. Meropenem was started again in combination with ciprofloxacin, both intravenously for 28 days, followed by ciprofloxacin orally for another 2 weeks. He recovered without developing sequelae. During a follow-up of 4 years without prophylactic antibiotic treatment, he had no serious infections. His Hb levels fluctuated around 109 g/L (10.9 g/dL), with 35‰ to 67‰ reticulocytes.
Purification and culture of cells
RBCs were obtained as described from citrated blood by centrifugation and aspiration of plasma and buffy coat, followed by washing 3 times with saline.7 Leukocytes were prepared from heparinized blood as described8 by centrifugation, aspiration of plasma, and lysis of RBCs with isotonic ammonium chloride. Neutrophils and lymphocytes were purified from leukocytes by centrifugation over isotonic Percoll.7Lymphocytes were cultured in Iscoves modified Dulbecco medium supplemented with 10% fetal calf serum, penicillin/streptomycin,l-glutamine, and phytohemagglutinin (PHA).
Enzyme determinations
G6PD activity in RBCs and granulocytes was determined as described previously.7
Neutrophil function tests
Purified neutrophils were suspended in a medium that contained 138 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4, 0.6 mM CaCl2, 1.0 mM MgCl2, 5.5 mM glucose, and 0.5% (wt/vol) human albumin (pH 7.4). Oxygen consumption was measured with an oxygen electrode.8
Molecular genetic studies
Genomic DNA and complementary DNA (cDNA) sequencing were performed as described earlier5 using an ABI prism 377 automated sequencer (Applied Biosystems, Foster City, CA). Primers for polymerase chain reaction (PCR) amplification and DNA sequencing are described in Table 1.
Primers used for PCR amplification and DNA sequencing
| Primer . | Primer sequence . | Amplified fragment, bp (size) . | Annealing site . | G6PD protein part . |
|---|---|---|---|---|
| PRs | 5′ CTCTGCAGGCCCGCGGAAGCTCGTT 3′ | − 1 336 to − 908 (428) | − 1 336 to − 1 312 from ATG | Promoter region |
| PRas | 5′ CCGCTGCCGCTGCTCTGCATCCCCA 3′ | − 932 to − 908 from ATG | ||
| 1s | 5′ CGGCGATGGGGATGCGGGAGCACTA 3′ | − 987 to − 578 (409) | − 987 to − 963 from ATG | Exon I, no coding sequence |
| 1as | 5′ GCGGAGCGCGGGACAGTACGCTCCT 3′ | Intron I 56 to 32 | ||
| 2s | 5′ AGGACCTCTCAAGAAAGGGGCTAAC 3′ | − 87 to 182 (269) | Intron I − 78 to − 54 | Exon II, Met1-Ser40 |
| 2as | 5′ AAAAGCTGAGGCATGGAGCAGGCAC 3′ | Intron II 63 to 39 | ||
| 3s | 5′ AAGGGTGGAGGATGATGTATGTAGG 3′ | 9 902 to 10 272 (370) | Intron II − 73 to − 49 | Exon III-IV, Gly41-Lys88 |
| 4as | 5′ TGGGGGCTGGTAGAGAGGGCAGAAC 3′ | Intron IV 54 to 30 | ||
| 5s | 5′ CTGGGGCAGAACACACACGGACTCA 3′ | 10 691 to 11 052 (361) | Intron IV − 76 to − 52 | Exon V, Ala89-Ile162 |
| 5as | 5′ ATAGAGTGGTGGGAGCACTGCCTGG 3′ | Intron V 68 to 44 | ||
| 6s | 5′ TGGGAGGGCGTCTGAATGATGCAGC 3′ | 11 569 to 11 876 (307) | Intron V − 87 to − 63 | Exon VI, Gly163-Arg215 |
| 6as | 5′ GGCCAGGTGAGGCTCCTGAGTACCA 3′ | Intron VI 61 to 37 | ||
| 7s | 5′ GGGTGACCCCTCACATGTGGCCCCT 3′ | 11 917 to 12 166 (249) | Intron VI − 74 to − 50 | Exon VII, Phe216-Arg257 |
| 7as | 5′ GGCTCTGCCACCCTGTGCCAGCCT 3′ | Intron VII 49 to 26 | ||
| 8s | 5′ GTTTGGGGTCCCCATGCCCTTGAAC 3′ | 12 405 to 12 628 (223) | Intron VII − 77 to − 53 | Exon VIII, Asp258-Lys288 |
| 8as | 5′ CAGATGGGCCTGCGACAGGGCATGC 3′ | Intron VIII 52 to 28 | ||
| 9s | 5′ TGCACATCTGTGGCCACAGTCATCC 3′ | 12 943 to 13 268 (325) | Intron VIII − 80 to − 56 | Exon IX, Val289-Asp350 |
| 9as | 5′ TGCCCGCACACAGGGCATGCCCAGT 3′ | Intron IX 58 to 34 | ||
| 10s | 5′ GCTCCCACTGAGACACTCACGCACT 3′ | 13 273 to 13 631 (358) | Intron IX − 76 to − 52 | Exon X, Gly351-Lys429 |
| 10as | 5′ GGCCCAGGCCGCCCACCCTCCACA 3′ | Intron X 46 to 23 | ||
| 11s | 5′ CTGGGGCCCGGGGGACTCCACATGGT 3′ | 13 624 to 13 811 (187) | Intron X − 67 to − 41 | Exon XI, Asn430-Ser455 |
| 11as | 5′ ACCCCATAGCCCACAGGTATGCAG 3′ | Intron XI 45 to 22 | ||
| 12s | 5′ GGGGTGGCCTTTGCCCTCCCTCC 3′ | 13 806 to 14 037 (231) | Intron XI − 65 to − 43 | Exon XII, Asp456-Ser486 |
| 12as | 5′ GGCATGAGGTAGCTCCACCCTCAC 3′ | Intron XII 73 to 50 | ||
| 13s | 5′ AGGAAAGGGTGGGGGCTGGGGACAGA 3′ | 13 967 to 14 239 (272) | Intron XII − 94 to − 69 | Exon XIII, Arg487-Leu515 |
| 13as | 5′ GTCAATGGTCCCGGAGTCCTCCCGA 3′ | Exon XIII 177 to 153 | ||
| 3′UTRs | 5′ TTTCCAGTATGAGGGCACCTACAAG 3′ | 14 104 to 14 802 (698) | Exon XIII 43 to 66 | Exon XIII, 3′UTR |
| 3′UTRas | 5′ AAGTGGGTCCTCAGGGAAGCA 3′ | 3′UTR − 42 to − 21 | ||
| CDNAs | 5′ ATATTCATCATCATGGGTGC 3′ | 96 to 262 (166) | Exon II 96 to 115 | Exon II-III, Ser40-Ile80 |
| CDNAas | 5′ GAAGGGCTCACTCTGTTTGC 3′ | Exon III 262 to 243 |
| Primer . | Primer sequence . | Amplified fragment, bp (size) . | Annealing site . | G6PD protein part . |
|---|---|---|---|---|
| PRs | 5′ CTCTGCAGGCCCGCGGAAGCTCGTT 3′ | − 1 336 to − 908 (428) | − 1 336 to − 1 312 from ATG | Promoter region |
| PRas | 5′ CCGCTGCCGCTGCTCTGCATCCCCA 3′ | − 932 to − 908 from ATG | ||
| 1s | 5′ CGGCGATGGGGATGCGGGAGCACTA 3′ | − 987 to − 578 (409) | − 987 to − 963 from ATG | Exon I, no coding sequence |
| 1as | 5′ GCGGAGCGCGGGACAGTACGCTCCT 3′ | Intron I 56 to 32 | ||
| 2s | 5′ AGGACCTCTCAAGAAAGGGGCTAAC 3′ | − 87 to 182 (269) | Intron I − 78 to − 54 | Exon II, Met1-Ser40 |
| 2as | 5′ AAAAGCTGAGGCATGGAGCAGGCAC 3′ | Intron II 63 to 39 | ||
| 3s | 5′ AAGGGTGGAGGATGATGTATGTAGG 3′ | 9 902 to 10 272 (370) | Intron II − 73 to − 49 | Exon III-IV, Gly41-Lys88 |
| 4as | 5′ TGGGGGCTGGTAGAGAGGGCAGAAC 3′ | Intron IV 54 to 30 | ||
| 5s | 5′ CTGGGGCAGAACACACACGGACTCA 3′ | 10 691 to 11 052 (361) | Intron IV − 76 to − 52 | Exon V, Ala89-Ile162 |
| 5as | 5′ ATAGAGTGGTGGGAGCACTGCCTGG 3′ | Intron V 68 to 44 | ||
| 6s | 5′ TGGGAGGGCGTCTGAATGATGCAGC 3′ | 11 569 to 11 876 (307) | Intron V − 87 to − 63 | Exon VI, Gly163-Arg215 |
| 6as | 5′ GGCCAGGTGAGGCTCCTGAGTACCA 3′ | Intron VI 61 to 37 | ||
| 7s | 5′ GGGTGACCCCTCACATGTGGCCCCT 3′ | 11 917 to 12 166 (249) | Intron VI − 74 to − 50 | Exon VII, Phe216-Arg257 |
| 7as | 5′ GGCTCTGCCACCCTGTGCCAGCCT 3′ | Intron VII 49 to 26 | ||
| 8s | 5′ GTTTGGGGTCCCCATGCCCTTGAAC 3′ | 12 405 to 12 628 (223) | Intron VII − 77 to − 53 | Exon VIII, Asp258-Lys288 |
| 8as | 5′ CAGATGGGCCTGCGACAGGGCATGC 3′ | Intron VIII 52 to 28 | ||
| 9s | 5′ TGCACATCTGTGGCCACAGTCATCC 3′ | 12 943 to 13 268 (325) | Intron VIII − 80 to − 56 | Exon IX, Val289-Asp350 |
| 9as | 5′ TGCCCGCACACAGGGCATGCCCAGT 3′ | Intron IX 58 to 34 | ||
| 10s | 5′ GCTCCCACTGAGACACTCACGCACT 3′ | 13 273 to 13 631 (358) | Intron IX − 76 to − 52 | Exon X, Gly351-Lys429 |
| 10as | 5′ GGCCCAGGCCGCCCACCCTCCACA 3′ | Intron X 46 to 23 | ||
| 11s | 5′ CTGGGGCCCGGGGGACTCCACATGGT 3′ | 13 624 to 13 811 (187) | Intron X − 67 to − 41 | Exon XI, Asn430-Ser455 |
| 11as | 5′ ACCCCATAGCCCACAGGTATGCAG 3′ | Intron XI 45 to 22 | ||
| 12s | 5′ GGGGTGGCCTTTGCCCTCCCTCC 3′ | 13 806 to 14 037 (231) | Intron XI − 65 to − 43 | Exon XII, Asp456-Ser486 |
| 12as | 5′ GGCATGAGGTAGCTCCACCCTCAC 3′ | Intron XII 73 to 50 | ||
| 13s | 5′ AGGAAAGGGTGGGGGCTGGGGACAGA 3′ | 13 967 to 14 239 (272) | Intron XII − 94 to − 69 | Exon XIII, Arg487-Leu515 |
| 13as | 5′ GTCAATGGTCCCGGAGTCCTCCCGA 3′ | Exon XIII 177 to 153 | ||
| 3′UTRs | 5′ TTTCCAGTATGAGGGCACCTACAAG 3′ | 14 104 to 14 802 (698) | Exon XIII 43 to 66 | Exon XIII, 3′UTR |
| 3′UTRas | 5′ AAGTGGGTCCTCAGGGAAGCA 3′ | 3′UTR − 42 to − 21 | ||
| CDNAs | 5′ ATATTCATCATCATGGGTGC 3′ | 96 to 262 (166) | Exon II 96 to 115 | Exon II-III, Ser40-Ile80 |
| CDNAas | 5′ GAAGGGCTCACTCTGTTTGC 3′ | Exon III 262 to 243 |
Immunoblotting of G6PD
For immunodetection, 105 cells were boiled in sodium dodecyl sulfate sample buffer (125 mM Tris, pH 6.8; 20% [wt/vol] sodium dodecyl sulfate; and 12.5% [vol/vol] β-mercaptoethanol) and were loaded on a 12.5% polyacrylamide gel, according to Laemmli, in a gel apparatus (Mini-Protean II, BioRad, Hercules, CA). Western blotting was performed (Mini Trans-Blot cell, BioRad) according to the manufacturer's recommendations. Detection of proteins was performed as described previously with a polyclonal antibody against recombinant G6PD.
Expression of G6PD in E coli
G6PD in Escherichia coli was expressed as described by Roos et al.5 Purification of recombinant G6PD was performed by affinity chromatography as previously reported.9,10 The electrophoretic mobility of the recombinant enzymes was determined by means of electrophoresis in native polyacrylamide in Tris buffer (pH 8.8) at 4°C,11followed by G6PD activity staining with phenazine methosulfate and methylthiotetrazole. Thermostability assays were performed according to the World Health Organization protocol.12 Briefly, samples with purified recombinant enzyme were incubated for 60 minutes at 51°C; aliquots were drawn every 20 minutes and assayed for G6PD activity.
Results
G6PD activity in the RBCs and granulocytes of the patients from both families was below the detection limit of our assay (Table2). Both mothers of the patients displayed about half the enzyme activity in their RBCs and granulocytes compared with healthy controls. Because the patients suffered from recurrent infections, a defect in granulocyte function due to the impaired G6PD activity was suspected. Indeed, oxygen consumption of the granulocytes of the patients after addition of opsonized zymosan particles was drastically diminished (Table 2). Granulocytes of both mothers consumed about half the amount of oxygen compared with healthy controls. The granulocyte NADPH oxidase defect was confirmed with other tests (nitroblue tetrazolium [NBT] test, dihydrorhodamine [DHR] test, and cytochrome c reduction; not shown). In the NBT test, the mothers showed a mosaic of formazan-positive and -negative cells (not shown). Cytochrome b558spectrum, reaction with a monoclonal antibody against gp91phox, and sequencing of CYBB (the X-linked gene encoding gp91phox) revealed no abnormalities in the patients.
G6PD activity and oxidative capacity of purified blood cells
| . | Control . | Mother I . | Patient I1 . | Patient I2 . | Patient I3 . | Mother II . | Patient II1 . |
|---|---|---|---|---|---|---|---|
| G6PD activity in RBCs, U/g Hb | 4.8-7.2 | 2.9 | 0.1 | 0.1 | 0.1 | 2.8 | 0.1 |
| G6PD activity in granulocytes, mU/106cells | 9.7-20 | 8.2 | 0 | 0 | 0 | 7.9 | 0 |
| O2consumption (nM/106 granulocytes per minute) | 6.1-11.7 | 3.9 | 1.2 | 1.3 | 1.2 | 4.4 | 2.0 |
| . | Control . | Mother I . | Patient I1 . | Patient I2 . | Patient I3 . | Mother II . | Patient II1 . |
|---|---|---|---|---|---|---|---|
| G6PD activity in RBCs, U/g Hb | 4.8-7.2 | 2.9 | 0.1 | 0.1 | 0.1 | 2.8 | 0.1 |
| G6PD activity in granulocytes, mU/106cells | 9.7-20 | 8.2 | 0 | 0 | 0 | 7.9 | 0 |
| O2consumption (nM/106 granulocytes per minute) | 6.1-11.7 | 3.9 | 1.2 | 1.3 | 1.2 | 4.4 | 2.0 |
Values are given as mean of 3 determinations with patient cells. Range obtained with 10 healthy controls is also given.
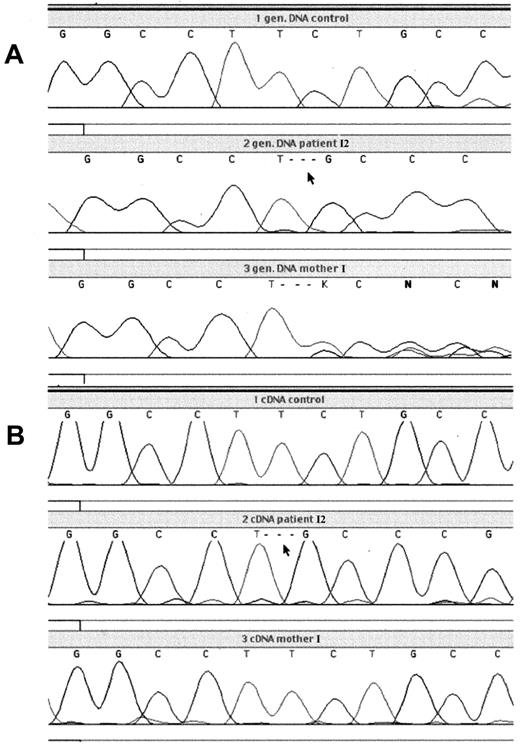
To identify the mutation responsible for the G6PD defect, DNA sequencing was performed on genomic DNA of the patients and their mothers (Figure 1). This revealed a deletion of 3 nucleotides (TCT) at position 180-182 in the coding sequence of the G6PD gene, predicting deletion of leucine at position 61 in the protein. This mutation was found in all patients, while both mothers were heterozygous for the deletion. No other mutations were found in the G6PD gene of the patients, including 236 base pairs (bp) in the promoter region, 545 bp of the 5′ untranslated region (UTR), and 638 bp of the 3′UTR.
Analysis of genomic and cDNA from patient I2 with G6PD Amsterdam and his mother.
PCR product, containing the region around the nucleotide 180-182 deletion, was generated from genomic DNA or cDNA obtained from blood leukocytes and was analyzed by dye primer cycle sequencing. (A) Genomic DNA from the leukocytes of a healthy control, patient I2, and his mother; (B) cDNA from the leukocytes of a healthy control, patient I2, and his mother.
Analysis of genomic and cDNA from patient I2 with G6PD Amsterdam and his mother.
PCR product, containing the region around the nucleotide 180-182 deletion, was generated from genomic DNA or cDNA obtained from blood leukocytes and was analyzed by dye primer cycle sequencing. (A) Genomic DNA from the leukocytes of a healthy control, patient I2, and his mother; (B) cDNA from the leukocytes of a healthy control, patient I2, and his mother.
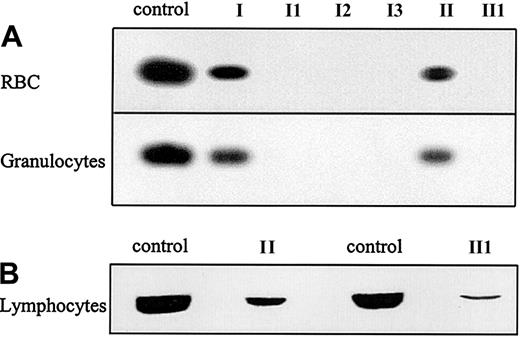
To determine whether this new deletion in the G6PD gene allows expression of protein, Western blotting was performed on the RBCs and granulocytes of the patients and their mothers. G6PD protein was not detected in the RBCs or the granulocytes of any of the patients (Figure 2A). Although diminished compared with healthy controls, G6PD was detected in the RBCs and granulocytes of the patients' mothers, mother I and mother II. Because complete G6PD deficiency is probably incompatible with life, it was investigated whether the deletion would lead to expression of an unstable protein. Therefore, rapidly dividing, PHA-stimulated lymphocytes of patient II1 and his mother were analyzed for G6PD expression. A low level of G6PD expression was found in PHA-stimulated lymphocytes of patient II1 and a decreased level of G6PD protein in those from his mother (Figure 2B).
G6PD protein expression in purified blood cells.
(A) Western blot analysis of G6PD protein expression in RBCs and granulocytes of the patients suffering from G6PD Amsterdam and their mothers. (B). Western blot analysis of G6PD protein expression in PHA-stimulated lymphocytes of patient II1 and his mother, mother II. For all Western blots, cells from a healthy donor were taken as a control.
G6PD protein expression in purified blood cells.
(A) Western blot analysis of G6PD protein expression in RBCs and granulocytes of the patients suffering from G6PD Amsterdam and their mothers. (B). Western blot analysis of G6PD protein expression in PHA-stimulated lymphocytes of patient II1 and his mother, mother II. For all Western blots, cells from a healthy donor were taken as a control.
To test the properties of the mutant protein, it was expressed in anE coli expression system and purified. Compared with the wild-type enzyme, G6PD Amsterdam showed reduced specific activity as well as reduced thermal stability (Table3 and Figure3). G6PD Amsterdam had normalKm values for both NADP and G6P. The utilization of 2-deoxy-G6P and galactose-6-phosphate was approximately doubled. The mutant enzyme used a normal percentage of deamino-NADP, and the Ki for NADPH was normal. Electrophoretic mobility of G6PD Amsterdam was increased 10% compared with the wild-type enzyme. However, the altered enzymatic properties of the recombinant G6PD Amsterdam were unlikely to account for the observed severe G6PD deficiency in the patients' cells. Therefore, the stability of the mRNA of G6PD Amsterdam was assayed. This was done by comparing the amount of mRNA encoding wild-type G6PD with the amount of mutant G6PD mRNA by cDNA sequencing (Figure 1). The mRNA was isolated from white blood cells of mother I and her 3 sons, patient I1, patient II2, and patient II3, and was subsequently sequenced. Although mother I is a carrier for the nucleotide 180-182 deletion, as confirmed by genomic sequencing, this deletion was not detected in the cDNA of her white blood cells. In contrast, the nucleotide 180-182 deletion was detected in the cDNA of her 3 sons. This proves that the mRNA encoding G6PD Amsterdam is less abundantly expressed than the mRNA coding for the wild-type protein in the mother of these 3 patients. This is strong evidence that the mutant mRNA is less stable than the wild-type G6PD mRNA, a property that is very likely to diminish the expression of G6PD Amsterdam protein.
Biochemical characterization of recombinant G6PD Amsterdam
| . | G6PD B . | G6PD Amsterdam . |
|---|---|---|
| Km G6P, μM | 64 ± 4 | 55 ± 5 |
| KmNADP, μM | 11 ± 1 | 11 ± 1 |
| Thermostability | Normal | Decreased |
| % utilization dG6P | 4 | 7.5 |
| % utilization GalP | 8 | 17 |
| % utilization 2-deoxy-NADP | 59 | 62 |
| % electrophoretic mobility | 100 | 110 |
| Specific activity, IU/mg | 210 ± 20 | 95 ± 3 |
| Ki NADPH, μM | 15 ± 2 | 14 ± 3 |
| . | G6PD B . | G6PD Amsterdam . |
|---|---|---|
| Km G6P, μM | 64 ± 4 | 55 ± 5 |
| KmNADP, μM | 11 ± 1 | 11 ± 1 |
| Thermostability | Normal | Decreased |
| % utilization dG6P | 4 | 7.5 |
| % utilization GalP | 8 | 17 |
| % utilization 2-deoxy-NADP | 59 | 62 |
| % electrophoretic mobility | 100 | 110 |
| Specific activity, IU/mg | 210 ± 20 | 95 ± 3 |
| Ki NADPH, μM | 15 ± 2 | 14 ± 3 |
Values are means ± SD of 3 independent measurements.
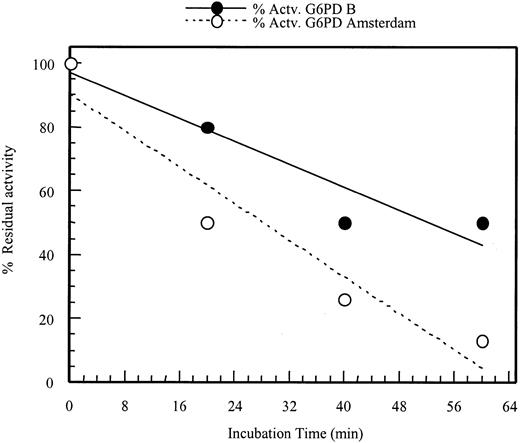
Retention of enzyme activity of G6PD Amsterdam.
The enzyme activity of G6PD Amsterdam after different incubation times at 51°C was compared with the enzyme activity of G6PD B (wild type). The activity is plotted as percentage residual activity of the enzymes at different incubation times compared with the activity of the enzymes at time zero.
Retention of enzyme activity of G6PD Amsterdam.
The enzyme activity of G6PD Amsterdam after different incubation times at 51°C was compared with the enzyme activity of G6PD B (wild type). The activity is plotted as percentage residual activity of the enzymes at different incubation times compared with the activity of the enzymes at time zero.
Discussion
In this study we describe the finding of a novel deletion of 3 nucleotides in the G6PD gene in 4 patients with mild chronic nonspherocytic anemia and CGD-like symptoms from 2 different families, predicting the deletion of leucine at position 61. This deletion leads to severe G6PD deficiency, confirmed by the absence of residual G6PD activity and of G6PD protein expression in the RBCs and granulocytes of the patients. However, a low level of G6PD protein expression was detected in rapidly dividing PHA-stimulated lymphocytes of one of the patients, consistent with the idea that the deletion of leucine on position 61 in the G6PD protein leads to the expression of an unstable protein.
This theory was partially confirmed by expression of the mutant protein in an E coli expression system. The recombinant enzyme proved to be less stable and less active than the wild-type enzyme, as shown by increased heat lability and reduced specific activity (Table 3and Figure 3). However, the enzymatic properties of G6PD Amsterdam were not severely altered but comparable to another G6PD variant, G6PD A−.13 The secondary structure of G6PD Amsterdam is predicted to differ from the wild-type enzyme in the proximity of the mutation (Table4).13 14
Prediction of secondary structure
| . | G6PD B . | G6PD Amsterdam . |
|---|---|---|
| Loop | RDGLLPEN | RDGLPEN |
| 15% α helix | 100% coil | |
| 85% coil | ||
| Loop + β sheet | RDGLLPENTFIVGYA | RDGLPENTFIVGYA |
| 40% β sheet | 28% β sheet | |
| 60% coil | 72% coil |
| . | G6PD B . | G6PD Amsterdam . |
|---|---|---|
| Loop | RDGLLPEN | RDGLPEN |
| 15% α helix | 100% coil | |
| 85% coil | ||
| Loop + β sheet | RDGLLPENTFIVGYA | RDGLPENTFIVGYA |
| 40% β sheet | 28% β sheet | |
| 60% coil | 72% coil |
The expression of G6PD Amsterdam and the activity in purified blood cells was much lower than those observed in G6PD A− and not in agreement with the enzymatic properties of the recombinant G6PD Amsterdam protein. Also, in the E coli expression system, the recovery was very low—about 2% of the protein yield found with wild-type G6PD. For comparison, the yield of G6PD A− in this system is 10% to 20% of that obtained with wild-type G6PD. The yield found with G6PD Amsterdam is in the range found with G6PD Med (2%-4%). Moreover, the clinical manifestation of the G6PD Amsterdam mutation, ie, the increased susceptibility to infections in addition to mild chronic nonspherocytic anemia, is more severe than is observed in patients with G6PD A− but comparable to another G6PD mutant previously described by us, G6PD Volendam.5 However, compared with G6PD Volendam as well as other G6PD mutants resulting in class I hemolytic anemia, the activity and stability of recombinant G6PD Amsterdam are considerably better.5 15 Therefore, we suspected that another factor influences the expression of G6PD Amsterdam in vivo. The cDNA sequencing revealed that the mRNA encoding G6PD Amsterdam was hardly detectable in a carrier of the G6PD Amsterdam mutation, proving that the mRNA of G6PD Amsterdam is less stable than the mRNA coding for the wild-type enzyme. This instability, rather than the altered enzyme properties, is likely to be the basis of the severe G6PD deficiency observed in our patients. To our knowledge, this is the first mutation in the G6PD gene that leads to mRNA instability as a significant contribution to the severity of the disease.
Remarkably, in the family with the 3 G6PD-deficient brothers, only 1 presented with recurrent microbial infections, whereas the other 2 had no known disposition to infections. This might be due to a different exposure to risk factors or to differences in disease-modifying polymorphisms in other genes,16 but that remains to be investigated.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
D. Roos, Dept of Experimental Immunohematology, Central Laboratory of the Netherlands Blood Transfusion Service, Plesmanlaan 125, 1066 CX Amsterdam, The Netherlands; e-mail:d_roos@clb.nl.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal