Abstract
The 558-565 loop region in the A2 subunit of factor (F) VIIIa forms a direct interface with FIXa. We have expressed and purified B-domainless FVIII (FVIIIWT) and B-domainless FVIII containing the hemophilia A–associated mutations Ser558Phe, Val559Ala, Asp560Ala, Gln565Arg, and the activated protein C cleavage site mutant Arg562Ala. Titration of FVIIIa in FXa generation assays showed that the mutant and wild-type proteins had similar functional affinities for FIXa (dissociation constant [Kd] values ∼5 nM-20 nM and ∼100 nM-250 nM in the presence and absence of phospholipid, respectively). The catalytic activities of the factor Xase complex composed of the hemophilia A–associated FVIII species were markedly reduced both in the presence and absence of phospholipid. FVIIIWT and FVIIIArg562Ala showed catalytic rate constant (kcat) values of approximately 60 minute−1 in the presence of phospholipid, whereas the hemophilia A–associated mutants showedkcat values ranging from 3.3 minute−1 to 7.5 minute−1. In the absence of phospholipid, all kcat values were reduced but FVIIIWT and FVIIIArg562Ala retained higher activities as compared with the hemophilic mutant FVIII forms. Fluorescence anisotropy experiments using fluorescein-modified FIXa confirmed that all FVIII forms interacted with FIXa. However, the presence of factor X yielded minimal increases in anisotropy observed with the mutant factor VIII forms, consistent with their reduced activity. These results show that residues within the 558-565 loop are critical in modulating FIXa enzymatic activity but do not contribute significantly to the affinity of FVIIIa for FIXa.
Introduction
The generation of a fibrin clot in response to vascular injury is mediated by the regulated and sequential activation of a series of serine proteases and their cofactors.1 The glycoprotein factor VIII (FVIII) in its activated form, FVIIIa, acts as a cofactor to the serine protease FIXa, in the conversion of the zymogen FX to the active enzyme (FXa). Both FVIII and FIX are essential for normal coagulation; deficiencies of either are associated with the bleeding diatheses hemophilia A and hemophilia B, respectively.
FVIII is synthesized as a 2332-residue single-chain glycoprotein, composed of 3 distinct domain types in the arrangement (NH2) A1-A2-B-A3-C1-C2 (COOH).2,3 As a result of intracellular proteolysis at the B-A3 junction plus additional sites within the B domain, FVIII circulates as a heterodimer of a variable length heavy chain (A1-A2-B domains) and light chain (A3-C1-C2).4-6FVIII is activated by thrombin cleavage at residues 372 (A1-A2 junction) and 740 (A2-B junction) in the heavy chain, and residue 1689 in the light chain.7 The resulting FVIIIa is a trimer composed of A1, A2, and A3-C1-C2 subunits.8 9
On activation FVIIIa forms a stoichiometric complex with FIXa. This complex is formed on a phospholipid surface and requires the presence of Ca++.10 The effect of phospholipid is to limit the interactions to 2 dimensions, thereby reducing theKd for the FVIIIa–FIXa interaction and theKm for substrate factor X.11 The effect of FVIIIa is to increase the catalytic rate constant (kcat) of FX conversion to FXa by several orders of magnitude.12 The mechanisms by which FVIIIa acts as a cofactor for FIXa remain unclear. In contrast to the isolated A1 and A3-C1-C2 subunits, the isolated A2 subunit can stimulate FIXa, though this ability is fractional as compared with the intact FVIIIa.13 The Ser558-Gln565 region within the A2 subunit has been shown to be critical for VIIIa–IXa interaction.14,15 This region contains the activated protein C (APC) cleavage site at Arg562.16 Interaction of FVIIIa with FIXa selectively protects this site from APC cleavage, suggesting that there is a direct interaction between FVIIIa and FIXa in this region.17 This has been confirmed by peptide studies. Synthetic peptides spanning the 558-565 residues of FVIIIa noncompetitively inhibit tenase activity.14 Fluorescence anisotropy studies of FIXa labeled with the fluorophore at the active site crevice demonstrate that the 558-565 peptide blocked the increase in anisotropy contributed by the A2 subunit of FVIIIa on interaction of the labeled FIXa, indicating the importance of this region in the A2–FIXa interaction.15
Mutations within the 558-565 region resulting in hemophilia A are described in the hemophilia A mutation database.18 The mutations Ser558Phe, Val559Ala, Asp560Ala, and Gln565Arg are described as cross reactive material–positive (CRM+), that is, having normal levels of FVIII antigen but defective FVIII activity associated with mild hemophilia A, indicating functional defects of secreted protein.
In order to determine the mechanism by which the 558-565 region contributes to the cofactor activity of FVIIIa, we have stably expressed and partially purified FVIII forms containing the mutated residues within this region. In addition, we have expressed and purified FVIII containing an Arg562Ala mutation. Using a functional assay, we demonstrate that the effect of the mutations associated with hemophilia A is not to alter the affinity of FVIIIa for FIXa but rather to directly affect the catalytic rate constant of the complex, and thus the effect of the mutations is to reduce the cofactor potential of FVIII. This conclusion is supported by fluorescence anisotropy data showing defective interaction of Fl-FFR-IXa with mutant FVIIIa in the presence of FX. In contrast, the Arg562Ala mutation has no effect on either Kd or kcat, indicating that the conservation of this residue is not critical for cofactor activity.
Materials and methods
Reagents
The reagents α-thrombin, factor IXaβ, factor X, factor Xa (Enzyme Research Laboratories, South Bend, IN); Fl-FFR-factor IXa (Molecular Innovations, Southfield, MI); and hirudin, phosphotidylethanolamine (PE), phosphotidylcholine (PC), and phospholtidylserine (PS) (Sigma, St Louis, MO) were purchased from the indicated vendors. Phospholipid vesicles composed of 40% PE, 20% PS, and 20% PC were prepared using octyl glucoside as previously described.19 FVIII-deficient plasma was prepared as previously described.20 Activated partial thromboplastin was purchased from General Diagnostics Organon Teknika (Durham, NC). The anti–FVIII antibody 10104 that recognizes the light chain of FVIII was purchased from QED Bioscience (San Diego, CA). The anti–factor VIII monoclonal antibody R8B12, which recognizes the COOH terminal region of the A2 domain, was prepared as described.9 The anti–FVIII monoclonal antibody ESH-8, which recognizes the C2 portion of the light chain, was purchased from American Diagnostica (Greenwich, CT).
The B-domainless FVIII (B-FVIII) expression construct RENeoFVIII was a gift kindly provided by Dr Pete Lollar. Baby hamster kidney (BHK) cells were obtained from American Type Culture Collection (Manassas, VA). Fetal bovine serum (FBS) was obtained from Gemini Bioproducts (Woodland, CA). All other reagents used for the BHK cell culture were obtained from Gibco BRL (Gaithesburg, MD).
Plasmid mutagenesis
B-FVIII cDNA was restricted from the RENeoFVIII expression construct using the endonucleases XhoI and NotI, and cloned into the Bluescript II K/S− vector. The Bluescript B-FVIII shuttle construct was used as template for site-directed mutagenesis. Mutations were introduced into the shuttle construct using the Stratagene QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutagenic oligonucleotides were commercially synthesized by Gibco BRL. The Ser558Phe mutant was designated FVIIISer558Phe; Val559Ala: FVIIIVal559Ala; Asp560Ala: FVIIIAsp560Ala; Arg562Ala: FVIIIArg562Ala; and Gln565Arg: FVIIIGln565Arg. B-FVIII was designated FVIIIWT. The presence of the mutations was confirmed by DNA sequencing. The mutated FVIII cDNA was then restricted and ligated back into the RENeo expression vectors. Plasmid DNA was prepared from bacterial pellets using the Qiagen Maxi-Prep kit (Valencia, CA). The presence of only the desired mutation was confirmed by DNA sequence analysis.
DNA transfection and FVIII expression
The FVIII expression constructs were stably transfected in BHK cells by liposome-mediated transfection. Plasmid DNA (1 μg) was coated with lipofectamine-plus reagent and added to the cells in serum-free Dulbecco modified Eagle medium-F12 (DMEM-F12). After incubation at 37°C, 5% CO2 for 3 hours, the cells were incubated in 10% FBS in DMEM-F12. Selection was initiated 24 hours after transfection by addition of geneticin (500 μg/mL) to the serum-supplemented DMEM-F12. Geneticin-resistant colonies were picked and expanded by plating, and FVIII expression levels determined by enzyme-linked immunosorbent assay (ELISA). The highest expressing colonies were grown and subclones prepared by limiting dilution. These were assayed for FVIII expression levels and high-expressing subclones were transferred and expanded, and aliquots frozen until required.
Approximately 27 × 106 cells were transferred to a sterile roller bottle, incubated in serum-supplemented DMEM-F12 overnight, and rotated at approximately 4 rpm. The following day the medium was replaced with 100 mL serum-free AIM-V and the bottles rotated at approximately 4 rpm at 30°C and 5%CO2(Gibco BRL). The conditioned medium was collected after 24 hours and replaced. Conditioned medium was collected daily for a total of 4 days.
Protein purification and concentration
After daily collection the conditioned medium was spun at approximately 2500g at 4°C for 10 minutes to pellet cell debris. The supernatant was then decanted, and the protein was precipitated with ammonium sulfate (40% wt/vol) overnight at 4°C. The supernatant/salt mix was centrifuged at 10 000g, 4°C for 10 minutes and the supernatant decanted. The ammonium sulfate precipitate was resuspended in 0.1 M NaCl–buffer A (20 mM MES [2-(N-morpholino) ethanesulfonic acid], pH 6.0, 5 mM CaCl2, 0.01% Tween-20) at one-tenth the original supernatant volume. The collections were pooled and dialyzed overnight in 0.2 M NaCl–buffer A (2 L) with 2 changes of buffer. FVIII was partially purified from the dialyzed protein solution after application to an SP sepharose ion–exchange column. After adsorption onto the column the protein was washed with 10 column volumes of dialysis buffer and eluted with 0.8 M NaCl–buffer A. Protein concentration of each fraction was determined by the Coomassie dye binding method of Bradford,21 and FVIII content determined by one-stage clotting assay. The partially purified FVIII was further concentrated and purified after overnight dialysis in 0.1 M NaCl–buffer B (20 mM Hepes, pH 7.5, 5 mM CaCl2, 0.01% Tween-20) and applied onto a Q-sepharose column. After application, the column was washed with 10 column volumes of 0.1 M NaCl–buffer B, and then 10 column volumes of 0.2 M NaCl–buffer B. FVIII was eluted with 0.8 M NaCl–buffer B. Protein and FVIII content were determined as described above, and the partially purified FVIII dialyzed overnight with 0.1 M NaCl–buffer B. FVIII activity and antigen were determined by one-stage clotting assay and ELISA, respectively.
FVIII activity and antigen measurement
One-stage FVIII assays were performed in chemically depleted FVIII-deficient pooled human plasma. Alternatively, FVIII activity was determined following the rate of conversion of FX to FXa using a purified system in the presence or absence of phospholipid. FVIII wild type and mutants were initially activated in 0.1 M NaCl-buffer B, 100 μg/mL bovine serum albumin, by the addition of thrombin (10 nM) in the presence of phosphatidylserine, phosphatidylcholine, and phosphatidylethanolamine (PSPCPE) vesicles (10 μg/mL), and FIXa (5 nM). Thrombin activity was inhibited after 1 minute by the addition of hirudin (2.5 U/mL). The conversion of substrate FX to FXa was initiated by the addition near Vmax (maximum kinetic velocity) levels of FX (∼300 nM). Aliquots were removed after an appropriate time to assess initial rates of FXa production and added to tubes containing ethylenediaminetetraacetic acid (EDTA) (80 mM final concentration). Rates of FXa generation were measured by the addition of the chromogenic substrate S-2765 (0.46 mM final concentration) and the reactions read at 405 nm using a Vmax microtiter plate reader (Molecular Devices, Royal Oak, MI). Factor Xa generation reactions in the absence of phospholipid were performed in 50 mM NaCl-buffer B, 100 μg/mL bovine serum albumin, as described above. Reactions were initiated by the addition of 1 μM FX.
FVIII antigen was measured by sandwich ELISA. Each well was coated overnight with 1 μg of ESH-8 monoclonal antibody (20 μg/mL, 0.15 M Na2CO3, 0.035 M NaHCO3; pH 9.6). The wells were washed with phosphate buffered saline (PBS)–0.01% Tween 20, and coated in block solution (5% nonfat dry milk dissolved in PBS-0.01% Tween 20). On removal and further washing of the plate, 50 μL of samples or standard was added and incubated at room temperature. Standards were comprised of purified FVIII of known concentration varying from 1 μg/mL to 15.6 ng/mL. After washing, 1 μg of biotinylated R8B12 (20 μg/mL) was added to each well. On further incubation and washing, horseradish peroxidase–labeled strepavidin (Calbiochem, San Diego, CA) was added to each well. After incubation and washing, the reaction was visualized by the addition of substrate containing 1 mg/mL O-phenylenediamine dihydrochloride (Sigma). The reaction was stopped by addition of 2 M H2SO4, and the absorbance measured at 490 nm using a Vmax microtiter plate reader.
Electrophoresis and western blotting
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed by the method of Laemmli,22 using a Bio-Rad minigel system (Hercules, CA). Electrophoresis was performed at 200 v for 40 minutes and protein bands were visualized using Coomassie blue staining. Proteins were transferred onto polyvinylidenefluoride (PVDF) membrane using a Bio-Rad minitransblot apparatus at 0.2 A overnight in a buffer containing 10 mM CAPS (3-(cyclohexylamino)-1-propanesulfonic acid), pH 11, and 10% (vol/vol) methanol. Western blotting was performed using the primary antibodies indicated followed by goat anti–mouse horseradish peroxidase–conjugated secondary antibody. The secondary antibody signal was produced using the enhanced chemiluminescence (ECL) system (Amersham, Arlington Heights, IL) with luminol as substrate, and the blot exposed to film for times varying between 1 minute and 10 minutes.
Fluorescence spectroscopy
Fluorescence anisotropy measurements were made using an Amico-Bowman series 2 spectrometer equipped with automatic polarizers arranged in an L-format (Spectronic Instruments, Rochester, NY). Reactions were performed at room temperature in 0.1 M NaCl-buffer B containing 100 μg/mL bovine serum albumin. Reactions contained 50 nM Fl-FFR-IXa, 200 nM FVIIIa, 50 μg/mL PSPCPE vesicles, in the absence or presence of 500 nM FX. FVIII was activated by the addition of thrombin (20 nM) for one minute, and the thrombin then inhibited by the addition of hirudin (5 U/mL). Samples were excited at 495 nm, and the emission intensity was monitored at 520 nm (4-nm band pass) for 2 seconds at each polarizer position. Each reaction was read a total of 20 times in each position, and 3 reactions performed for FVIIIWT and each mutant. Anisotropy values were calculated automatically after subtraction of blank readings and data then averaged for each measurement.
Data analysis
Data were fitted using the single-site ligand binding model where amount bound = (capacity × free)/(kd + free), using the Marquart algorithm and computed using UltraFit software (v3.04; BioSoft, Ferguson, MO). As the concentration of FVIIIa was more than the FIXa for all FVIIIa levels, the value for free factor VIIIa used the total FVIIIa concentration, therefore the Kddetermined is an apparent Kd. Probability(P) values were determined using the Studentt test.
Results
Expression and purification of recombinant factor VIII
Mutations of the 558-565 loop of FVIIIa were constructed in the expression vector RENeoFVIII and expressed in BHK cells. Between 100 μg and 300 μg of expressed protein was purified from approximately 1 L conditioned medium. The material obtained from the chromatographic steps employed was more than 30% pure as judged by gel electrophoresis and Coomassie staining (results not shown). The major contaminating protein was albumin.
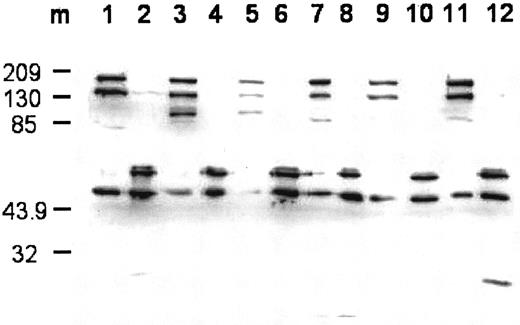
The FVIII obtained from each mutant and wild type was fully activated by thrombin. A time-course analysis of FVIIIa activity generation demonstrated peak activity at 1 minute to 2 minutes, with a 10- to 20- fold increase in activity of FVIIIa as compared with the preactivated levels (data not shown). Western blotting analysis of the intact and thrombin-cleaved FVIII forms using an anti–A2 domainal monoclonal antibody is shown in Figure 1. The intact protein demonstrated prominent high-molecular-weight bands (> 130 000) consistent with expression of B-FVIII as a single chain. A less-abundant band of approximately 90 000 represented the contiguous A1-A2 domains of the FVIII heavy chain, consistent with its absence following blotting with an anti–A3 domain-specific antibody (results not shown). Low (variable) levels of an approximately 45 000 band representing the A2 domain was observed, suggesting some degradation and/or activation of the B-FVIII. All B-FVIII forms exhibited similar peptide patterns following cleavage by thrombin, consistent with the potentiation of activity. High-molecular-weight bands (> 90 000) were converted to bands of approximately 45 000 and 50 000, representing the A2 subunit forms. The origin of the higher Mr A2 band is unclear, but may represent additional glycosylation and/or partial extension of residues at its C-terminal end.
Western blot of expressed wild-type and mutant FVIII and cleavage by thrombin.
The blot was probed using monoclonal antibody R8B12, which recognizes an epitope within the A2 domain. Lane designations are: 1 and 2, FVIIIWT and FVIIIaWT; 3 and 4, FVIIISer558Phe and FVIIIaSer558Phe; 5 and 6, FVIIIVal559Ala and FVIIIaVal559Ala; 7 and 8, FVIIIAsp560Ala and FVIIIaAsp560Ala; 9 and 10, FVIIIArg562Ala and FVIIIaArg562Ala; 11 and 12, FVIIIGln565Arg and FVIIIaGln565Arg. Lane m shows the positions for the protein markers (Mr is ×10−3).
Western blot of expressed wild-type and mutant FVIII and cleavage by thrombin.
The blot was probed using monoclonal antibody R8B12, which recognizes an epitope within the A2 domain. Lane designations are: 1 and 2, FVIIIWT and FVIIIaWT; 3 and 4, FVIIISer558Phe and FVIIIaSer558Phe; 5 and 6, FVIIIVal559Ala and FVIIIaVal559Ala; 7 and 8, FVIIIAsp560Ala and FVIIIaAsp560Ala; 9 and 10, FVIIIArg562Ala and FVIIIaArg562Ala; 11 and 12, FVIIIGln565Arg and FVIIIaGln565Arg. Lane m shows the positions for the protein markers (Mr is ×10−3).
Comparison of factor activity and antigen
The profiles of the expressed proteins and of the database entries are summarized in Table 1. Of the 5 mutants, 4 were associated with mild to severe hemophilia A. B-domainless wild-type FVIII demonstrated an FVIII activity–antigen ratio of approximately 1.57, higher than the expected value of unity. One possibility that this value is greater than unity may reflect low levels of activation in the FVIII preparation. Alternatively, this value may represent an underestimate in the concentration of FVIII protein. FVIIIArg562Ala also demonstrated a ratio greater than unity (1.86), further indicating that the mutation at residue 562 does not have a deleterious effect on FVIII function. In contrast, all the mutations associated with hemophilia A had reduced FVIII Ac/Ag ratios ranging from 2% to 10% of activity as compared with antigen, consistent with the available database entries.
FVIII activity and antigen values
| Factor VIII . | Database* . | Recombinant FVIII . | |||||
|---|---|---|---|---|---|---|---|
| Ac (U/dL) . | Ag (U/dL) . | Ratio (Ac/Ag) . | Ac (U/mL)† . | Ag (Ug/mL)‡ . | Ag (U/mL)1-153 . | Ratio (Ac/Ag) . | |
| WT | 250 | 53 | 159 | 1.57 | |||
| Ser558Phe | 21 | 175 | 0.12 | 23 | 115 | 345 | 0.07 |
| Val559Ala | 10 | 125 | 0.08 | 72.7 | 242 | 727 | 0.1 |
| Asp560Ala | 7 | – | – | 30 | 149 | 447 | 0.07 |
| Arg562Ala | 915 | 164 | 492 | 1.86 | |||
| Gln565Arg | 27/< 1 | 114/– | 0.24/– | 3.5 | 67 | 201 | 0.02 |
| Factor VIII . | Database* . | Recombinant FVIII . | |||||
|---|---|---|---|---|---|---|---|
| Ac (U/dL) . | Ag (U/dL) . | Ratio (Ac/Ag) . | Ac (U/mL)† . | Ag (Ug/mL)‡ . | Ag (U/mL)1-153 . | Ratio (Ac/Ag) . | |
| WT | 250 | 53 | 159 | 1.57 | |||
| Ser558Phe | 21 | 175 | 0.12 | 23 | 115 | 345 | 0.07 |
| Val559Ala | 10 | 125 | 0.08 | 72.7 | 242 | 727 | 0.1 |
| Asp560Ala | 7 | – | – | 30 | 149 | 447 | 0.07 |
| Arg562Ala | 915 | 164 | 492 | 1.86 | |||
| Gln565Arg | 27/< 1 | 114/– | 0.24/– | 3.5 | 67 | 201 | 0.02 |
Data were obtained from the hemophilia A mutation database (europium.csc.mrc.ac.uk/usr/WWW/WebPages/main.dir/main.htm). Ac and Ag refer to FVIII activity and antigen, respectively.
FVIII activity determined by a one-stage clotting assay as described in “Materials and methods.”
FVIII antigen determined using an ELISA as described in “Materials and methods.”
Units per milliliter were determined on the assumption that one unit of native FVIII is equivalent to 300 ng of protein.
Cofactor activity of wild-type and mutant FVIIIa forms
FVIIIa-mediated generation of FXa was performed in a purified system with varying concentrations of each of the expressed FVIII mutants and wild type, and fixed concentrations of FIXa. Experiments were performed both in the presence and absence of PSPCPE as described with near Vmax levels of FX. In all experiments FVIII was converted to FVIIIa by the addition of thrombin for 1 minute, followed by inhibition of thrombin by addition of hirudin.
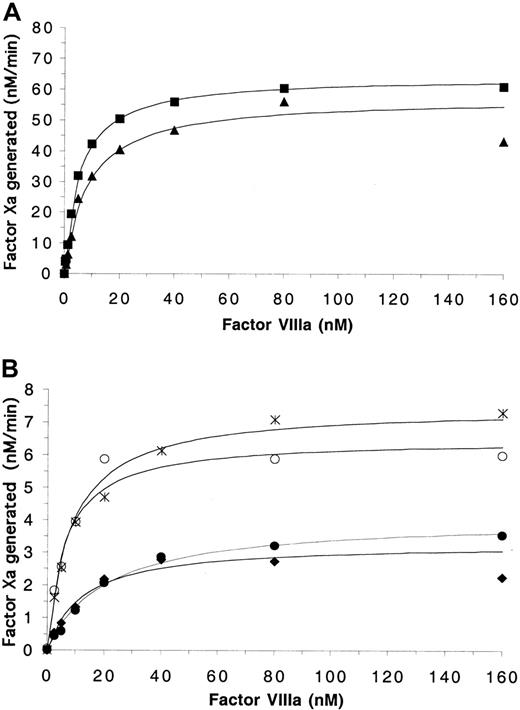
In the presence of PSPCPE, FVIIIWT and FVIIIArg562Ala demonstrated similar abilities to act as cofactors for FIXa-catalyzed generation of FXa. Similar levels of FXa were generated with increasing FVIIIa levels and saturation of FIXa was observed at similar FVIIIa concentrations (Figure2A). This was reflected in the derived functional affinity and catalytic constants. FVIIIWT and FVIIIArg562Ala yielded similar kcatvalues of 64.2 minute−1 and 57.4 minute−1, respectively (Table 2). Furthermore, the wild-type and Arg562Ala forms possessed similar affinity for FIXa (Kd of 5.4 nM and 8.5 nM, respectively). These results indicate that the mutation Arg562Ala does not affect the cofactor function of FVIIIa. The mutant proteins FVIIISer558Phe, FVIIIVal559Ala, and FVIIIAsp560Ala also demonstrated similar affinity for FIXa with saturation being reached at similar FVIIIa concentrations andKd values ranging from 6.2 nM to 12.4 nM, thus indicating that the ability to interact with FIXa is not impaired by the mutations (Figure 2B; Table 2). FVIIIGln565Argdemonstrated a reduced affinity for FIXa relative to wild type with aKd of 20.9 nM (P = .01). However, the catalytic activity of all hemophilia A–associated mutant FVIIIa–IXa complexes was diminished to between 5% to 12% that of wild type as demonstrated by the reduced FXa generated andkcat values with the mutant proteins ranging from 3.3 minute−1 to 7.5 minute−1.
Effects of mutations in the 558-565 loop on FIXa-catalyzed activation of factor X in the presence of PSPCPE.
Factor Xa generation assays contained 5 nM FIXa, 10 μM PSPCPE vesicles, variable FVIIIa concentrations, and were initiated with 300 nM FX as described in “Materials and methods.” (A) Data points obtained for FVIIIWT (▪) and FVIIIArg562Ala(▴). (B) Data generated for FVIIISer558Phe (○), FVIIIVal559Ala (♦), FVIIIAsp560Ala (∗), FVIIIGln565Arg (●). The data were plotted and fitted using a single-site ligand binding model as described in “Materials and methods.”
Effects of mutations in the 558-565 loop on FIXa-catalyzed activation of factor X in the presence of PSPCPE.
Factor Xa generation assays contained 5 nM FIXa, 10 μM PSPCPE vesicles, variable FVIIIa concentrations, and were initiated with 300 nM FX as described in “Materials and methods.” (A) Data points obtained for FVIIIWT (▪) and FVIIIArg562Ala(▴). (B) Data generated for FVIIISer558Phe (○), FVIIIVal559Ala (♦), FVIIIAsp560Ala (∗), FVIIIGln565Arg (●). The data were plotted and fitted using a single-site ligand binding model as described in “Materials and methods.”
Kinetic and binding parameters for expressed FVIII wild-type and mutant proteins
| Factor VIII . | PSPCPE present . | PSPCPE absent . | ||
|---|---|---|---|---|
| Kd* ± sd (nM) . | kcat† ± sd (min−1) . | Kd‡± sd (nM) . | kcat§ ± sd (min−1) . | |
| WT | 5.4 ± 0.8 | 64.2 ± 2.9 | 133.5 ± 63.3 | 7.96 ± 1.6 |
| Ser558Phe | 6.2 ± 3.1 | 6.5 ± 0.8 | 101.2 ± 15.8 | 0.25 ± 0.01 |
| Val559Ala | 12.4 ± 6.5 | 3.3 ± 0.6 | 90.7 ± 5.8 | 0.26 ± 0.01 |
| Asp560Ala | 8.3 ± 1.3 | 7.5 ± 0.4 | 263.3 ± 82.3 | 1.07 ± 0.14 |
| Arg562Ala | 8.5 ± 1.8 | 57.4 ± 4.1 | 134.4 ± 67.1 | 5.23 ± 0.82 |
| Gln565Arg | 20.9 ± 4.9 | 4.1 ± 0.3 | 132.0 ± 56.9 | 0.27 ± 0.02 |
| Factor VIII . | PSPCPE present . | PSPCPE absent . | ||
|---|---|---|---|---|
| Kd* ± sd (nM) . | kcat† ± sd (min−1) . | Kd‡± sd (nM) . | kcat§ ± sd (min−1) . | |
| WT | 5.4 ± 0.8 | 64.2 ± 2.9 | 133.5 ± 63.3 | 7.96 ± 1.6 |
| Ser558Phe | 6.2 ± 3.1 | 6.5 ± 0.8 | 101.2 ± 15.8 | 0.25 ± 0.01 |
| Val559Ala | 12.4 ± 6.5 | 3.3 ± 0.6 | 90.7 ± 5.8 | 0.26 ± 0.01 |
| Asp560Ala | 8.3 ± 1.3 | 7.5 ± 0.4 | 263.3 ± 82.3 | 1.07 ± 0.14 |
| Arg562Ala | 8.5 ± 1.8 | 57.4 ± 4.1 | 134.4 ± 67.1 | 5.23 ± 0.82 |
| Gln565Arg | 20.9 ± 4.9 | 4.1 ± 0.3 | 132.0 ± 56.9 | 0.27 ± 0.02 |
Experimental conditions for reactions run in the presence of PSPCPE (10 μM) and absence of PSPCPE are described in the legends to Figures 2 and 3, respectively. sd refers to standard deviation based on values from the fitted curves. No significant differences were observed for Kd values between mutants 558-562 and wild type (WT) (P = .1-.8* and P = .2-.99‡), whereas the value for Gln565Arg in the presence of PSPCPE is significant (P = .01). Differences in kcatvalues were significant when comparing the hemophilia A mutants with WT (P < .0001† and P < .001§). No significant difference was noted in comparingkcat values for Arg562Ala and WT (P = .2† and P = .15§).
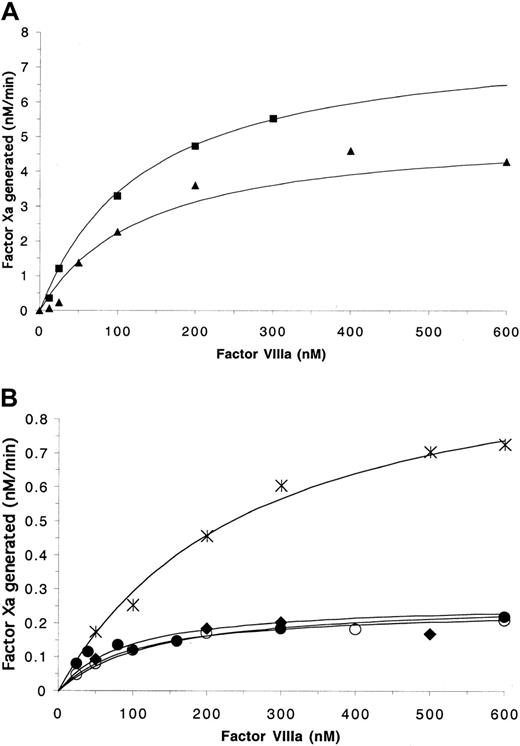
Surface-independent experiments were performed in the absence of PSPCPE in order to evaluate the direct protein–protein interaction of FVIIIa with FIXa. Experiments were performed at low salt concentration (50 mM NaCl) in order to minimize the dissociation of the FVIIIa A2 subunit from the A1/A3-C1-C2 dimer.23 The ability of the FVIIIaWT-FIXa complex to convert FX to FXa was considerably reduced in the absence of PSPCPE. The Kd andkcat values for FVIIIWT of 133.5 nM minute−1 and 7.96 nM minute−1, respectively, were similar to those previously described.23 For the mutants FVIIISer558Phe, FVIIIVal559Ala, FVIIIArg562Ala, and FVIIIGln565Arg, saturation of FIXa was reached at similar FVIIIa concentrations (Figure3) to FVIIIWT and yielded similar Kd values (Table 2; Figure 3A-B). Thus the mutations did not affect the phospholipid-independent interaction of FVIIIa with FIXa.
Effects of mutations in the 558-565 on FIXa-catalyzed activation of factor X in the absence of PSPCPE.
Factor Xa generation assays contained 5 nM FIXa, variable FVIIIa concentrations, and were initiated with 1 μM FX as described in “Materials and methods.” (A) Data points obtained for FVIIIWT (▪) and FVIIIArg562Ala (▴). (B) Data generated for FVIIISer558Phe (○), FVIIIVal559Ala (♦), FVIIIAsp560Ala (∗), FVIIIGln565Arg (●). The data were plotted and fitted using a single-site ligand binding model as described in “Materials and methods.”
Effects of mutations in the 558-565 on FIXa-catalyzed activation of factor X in the absence of PSPCPE.
Factor Xa generation assays contained 5 nM FIXa, variable FVIIIa concentrations, and were initiated with 1 μM FX as described in “Materials and methods.” (A) Data points obtained for FVIIIWT (▪) and FVIIIArg562Ala (▴). (B) Data generated for FVIIISer558Phe (○), FVIIIVal559Ala (♦), FVIIIAsp560Ala (∗), FVIIIGln565Arg (●). The data were plotted and fitted using a single-site ligand binding model as described in “Materials and methods.”
Similar to results obtained in the presence of surface, the mutation at residue 562 did not markedly affect the relative ability of FVIIIArg562Ala to act as a cofactor in the generation of FXa in the absence of phospholipid. Similar FXa levels were generated upon increasing FVIIIaArg562Ala concentration and thekcat derived approached that obtained for wild-type FVIIIa. FXa generated in the presence of saturating FVIIIaSer558Phe, FVIIIaVal559Ala, FVIIIaAsp560Ala, or FVIIIGln565Arg was fractional as compared with FVIIIWT and FVIIIArg562Ala. The determined kcatvalues for FVIIISer558Phe, FVIIIGln565Arg, and FVIIIVal559Ala were reduced to approximately 3% of that for FVIIIWT, whereas that for FVIIIAsp560Alawas approximately 13% of that of FVIIIWT. Taken together, these results demonstrate that FVIII molecules possessing mutations within the 558-565 loop region form FVIIIa-FIXa complexes with marked defects in transducing this binding event to yield enhanced enzymatic activity of FIXa. However, with the exception of FVIIIaGln565Arg, all the mutants form complexes with wild-type–like affinities for FIXa. FVIIIaGln565Argdemonstrated moderately reduced ability to form the FVIIIa-FIXa complex in the presence of phospholipid, but this reduction is marginal in comparison with its inability when in complex with FIXa to promote increased catalytic activity.
Effect of mutations in the 558-565 region of FVIII on fluorescence anisotropy of Fl-FFR-FIXa
The above results indicate that mutations in the 558-565 loop region modulate the cofactor effect of FVIIIa. In order to gain additional insights into the mechanism(s) for altered function of the mutant proteins, local topographical effects of the FVIIIa-dependent modulation of the FIXa active site crevice were assessed by fluorescence anisotropy. Reactions were performed in the presence of PSPCE, both in the absence and presence of FX. As the fluorophore is attached via a tripeptide to the active site Ser, any changes in anisotropy are a direct reflection of changes in the local conformation of the active site cleft of FIXa, as detected by altered rotational freedom of the fluorophore.
Results in Table 3 demonstrate the change in anisotropy on addition of FVIIIa in the absence or presence of further addition of FX. In the presence of FVIIIaWT, there was an increase of 0.033 in Fl-FFR-IXa anisotropy as compared with Fl-FFR-IXa alone. Variable increases ranging from 0.021 to 0.064 were detected upon addition of the mutated FVIIIa forms. The significance of these differences for the cofactor–enzyme interaction is unclear, but suggests that amino acid substitutions in this region differentially modulate the mobility of the active site–specific probe upon binding of cofactor to enzyme.
Effects of 558 loop mutations on the FVIIIa-dependent fluorescence anisotropy of Fl-FFR-IXa
| FVIII . | Fl-FFR-IXa alone3-150 . | Fl-FFR-IXa and FX3-151 . | ||
|---|---|---|---|---|
| Anisotropy ± sd . | Δr . | Anisotropy ± sd . | ΔrFX . | |
| No FVIIIa | 0.210 ± 0.009 | – | 0.215 ± 0.008 | – |
| WT | 0.243 ± 0.003 | 0.033 | 0.287 ± 0.003 | 0.072 |
| Ser558Phe | 0.257 ± 0.004 | 0.047 | 0.276 ± 0.001 | 0.061 |
| Val559Ala | 0.231 ± 0.005 | 0.021 | 0.253 ± 0.010 | 0.038 |
| Asp560Ala | 0.258 ± 0.006 | 0.048 | 0.252 ± 0.003 | 0.037 |
| Arg562Ala | 0.274 ± 0.005 | 0.064 | 0.287 ± 0.001 | 0.072 |
| Gln565Arg | 0.254 ± 0.002 | 0.044 | 0.262 ± 0.013 | 0.047 |
| FVIII . | Fl-FFR-IXa alone3-150 . | Fl-FFR-IXa and FX3-151 . | ||
|---|---|---|---|---|
| Anisotropy ± sd . | Δr . | Anisotropy ± sd . | ΔrFX . | |
| No FVIIIa | 0.210 ± 0.009 | – | 0.215 ± 0.008 | – |
| WT | 0.243 ± 0.003 | 0.033 | 0.287 ± 0.003 | 0.072 |
| Ser558Phe | 0.257 ± 0.004 | 0.047 | 0.276 ± 0.001 | 0.061 |
| Val559Ala | 0.231 ± 0.005 | 0.021 | 0.253 ± 0.010 | 0.038 |
| Asp560Ala | 0.258 ± 0.006 | 0.048 | 0.252 ± 0.003 | 0.037 |
| Arg562Ala | 0.274 ± 0.005 | 0.064 | 0.287 ± 0.001 | 0.072 |
| Gln565Arg | 0.254 ± 0.002 | 0.044 | 0.262 ± 0.013 | 0.047 |
Reactions contained 50 nM Fl-FFR-FIXa, 50 μM PSPCPE vesicles, 200 nM FVIIIa form, and were run as described in “Materials and methods.” Values represent the mean of 3 separate experiments ± standard deviation (sd).
Reactions were performed as described above and were supplemented with 500 nM FX.
Upon addition of FX to the complex of Fl-FFR-IXa and FVIIIaWT, there was an increase in anisotropy of 0.072 as compared with FX added to Fl-FFR-FIXa alone. In contrast, only the reaction containing FVIIIaArg562Ala yielded a similar increase in anisotropy observed on addition of FX. No increase to a similar level of FX-dependent anisotropy was detected in any of the hemophilia A–associated mutant FVIIIa–FIXa interactions, although this value was approached with FVIIISer558Phe (0.061). These results suggest that there is modulation of the FIXa active site region by the mutant FVIIIa forms. However, with the exception of FVIIIaArg562Ala, which possesses essentially wild-type activity, this modulation is ineffective in presenting the optimum conformation for FIXa–FX interaction in the catalytic region of FIXa.
Discussion
The 558-565 region of FVIII is a conserved region. It is identical across canine, murine, and porcine FVIII-A2 domains,24-26and shares sequence conservation across homologous A domains.27 The 558-565 loop of activated FVIII has been shown by various studies to be an FIXa interactive site. Initial studies demonstrated that FIXa interaction selectively protected FVIIIa from cleavage by APC at Arg562.17 Further studies demonstrated that interaction with FIXa could be blocked by peptides spanning the 558-565 region, as measured by FXa generation assays and by the loss of the A2-dependent increase in fluorescence anisotropy.14,15 In addition to being an FIXa interactive site and containing an APC cleavage site, the 558-565 site interacts with heparan sulfate proteoglycans as demonstrated recently by peptide analysis.28 Thus the FVIIIa Ser558-Gln565 loop represents a key interactive region of FVIIIIa. Its importance is highlighted by the association of missense mutations with hemophilia A resulting in defective FVIII molecules.29
By stable expression and subsequent partial purification of 558-565 mutant proteins we have been able to perform kinetic studies on the effects of mutations in this region on the interaction of FVIIIa with FIXa and subsequent generation of FXa. The mutant proteins FVIIISer558Phe, FVIIIVal559Ala, FVIIIAsp560Ala, and FVIIIGln565Arg, all demonstrated defective FVIII function in one-stage clotting assays to 2% to 10% of activity as compared with FVIII antigen. This reduction in activity was not due to defective cleavage of FVIII to FVIIIa by thrombin, as activity and gel analysis showed full activation of FVIII. The expressed mutant proteins reproduced the patient plasma-based assays, confirming that the mutations directly cause hemophilia A as a result of the reduced ability of secreted FVIII to act as a cofactor for FIXa. Assays performed with varying concentrations of FVIIIa in the presence of phospholipid and fixed concentrations of FIXa and FX showed reduced FXa generation. This effect was directly due to impaired cofactor activity of FVIIIa as the kcat for the FXase complexes were reduced for hemophilia A–associated mutants by 5% to 12% as compared with wild type. In contrast, the affinity of cofactor forms for FIXa was similar for mutants FVIIISer558Phe, FVIIIVal558Ala, and FVIIIAsp560Ala (Kd values from 6.2 nM-12.4 nM) as compared with FVIIIWT (5.4 nM), whereas FVIIIGln565Arg showed a modest increase inKd (20.9 nM). Previous analysis by Amano et al30 performed on transiently expressed FVIII containing the Ser558Phe mutation led to the conclusion that the likely effect of the mutation was to affect FVIIIa–FIXa interaction. This was based on an indirect determination by competition of a peptide corresponding to the 558-565 sequence with mutant FVIIIa for FIXa interaction. Therefore, while a proportionally greater reduction in mutant cofactor activity was observed as compared with wild type, no direct determination of either interprotein affinity or catalytic rate constants was performed. Our results demonstrate that the interaction of FVIIIa with FIXa and ability to form a complex were not substantially affected by any of the mutations in the presence of phospholipid, whereas the catalytic activity of the tenase complex was greatly impaired by each mutation associated with CRM+ hemophilia A.
Factor VIIIa binds to phospholipid with high affinity, with reportedKd values ranging between 10−9 M and 10−11 M.31,32 The effect of binding of FVIIIa and FIXa to platelets, microparticles, or synthetic phospholipid surfaces is to limit the interaction to a 2-dimensional surface. FVIIIa binds to phospholipid via the C2 domain33,34 and FIXa binds to the phospholipid surface via the amino-terminal 3-11 residues of the Gla domain.35,36 This association facilitates the orientation and interaction of the proteins and has the effect of increasing the affinity of the FVIIIa–FIXa interaction.37-39 The FVIIIa–FIXa interaction in the absence of phospholipid is less efficient, with the rate enhancement component due to the orientation effect of phospholipid removed.23,38 39 Thus the FVIIIa–FIXa interaction demonstrates a much higher Kd and a reducedkcat. However, by studying the interaction of wild-type and mutant FVIIIa with FIXa in the absence of phospholipid, contributions of the 558-565 region of the A2 domain to FIXa interaction can be assessed more directly.
Titration of FVIIIaWT and FVIIIaArg562Ala in FXa generation experiments in the absence of phospholipid with fixed FX and FIXa concentrations gave similar Kd andkcat values to those previously reported of 90 nM minute−1, and 5.5 nM minute−1.23 The Kdvalues determined for the hemophilia A mutant FVIII species were similar to that of FVIIIaWT. However,kcat values for these mutants were reduced by approximately 90% or more. Thus the mutations do not severely affect the affinity of FVIIIa with FIXa, but appear to have a direct effect on the ability of FVIIIa to act as a cofactor for FIXa. While the isolated A2 subunit binds FIXa, the affinity for this interaction (∼300 nM) is approximately 1% that observed with intact FVIIIa.13Thus, it is likely that key interactions in ensuring binding to FIXa are modulated at the high-affinity binding site within the light chain40 as well as possibly by other regions within the A2 domain.41
Fluorescence anisotropy measurements of interactions of Fl-FFR-IXa with wild-type and mutant FVIIIa in the absence and presence of FX lends further support to the data obtained by the functional assays. In the absence of FX, inclusion of all FVIII forms yielded increased anisotropy values compared with FIXa alone, suggesting formation of cofactor-enzyme complexes. The variability in these values suggests that changes in composition of the 558-565 loop directly influence constraints imposed on the active site label. Of particular interest are results obtained in the presence of FX. Earlier studies by Lollar et al42 showed that presence of substrate FX had little influence on the anisotropy of Fl-FFR-FIXa alone, but a marked increase was observed in the complete system consisting of surface-bound enzyme, cofactor, and substrate. Under these conditions, we observed maximal increases in anisotropy with FVIIIWT and FVIIIArg562Ala, consistent with maximal rates of FXa generation, suggesting that constraints on fluorophore rotation may represent an indicator of catalytic activity. Interestingly, under these conditions a significant increase in anisotropy was obtained for the FVIIISer558Phe mutation. We speculate that the presence of the bulky Phe residue may in itself contribute to rotational constraints in the fluorophore in the complex of enzyme, cofactor, and substrate.
In addition to FVIII residues 558-565 comprising an interactive region with FIXa, a FIXa-interactive region has been identified in the A3 domain of the light chain.43 Inhibition studies by monoclonal antibody demonstrated the location of a high-affinity FIXa interactive site within residues Gln1778-Asp1840, and subsequent peptide inhibition studies have localized the minimum sequence required for this interaction to Glu1811-Lys1818.40 TheKd for the FVIIIa light chain–FIXa interaction is similar at 14 nM to the intact FVIIIa. Thus, interaction of FIXa likely occurs with 2 regions of FVIIIa, via the high-affinity binding site with FVIIIa light chain, and the much lower affinity site within the A2 domain. The finding that mutations within the 558-565 loop region do not greatly affect FIXa binding supports the hypothesis that in the presence of phospholipid the binding of FVIIIa and FIXa is a critical first step followed by high-affinity binding involving the FVIIIa A3 domain to FIXa.44 The low-affinity binding of FVIIIa A2 to the serine protease domain, and the effect of mutations in the 558-565 loop on the kcat of the complex support the proposal that this region is key in modulating FIXa activity.
The residues 330-339 of the serine protease of FIXa represent an FVIIIa-interactive site,45,46 and more recent studies have demonstrated that this region interacts with the 558-565 loop of FVIIIa.41 The presence of hemophilia B mutations in this region also demonstrates the importance of these residues.45 As a result of studies of wild-type and mutant FIXa interaction with the isolated A2 domain of FVIIIa, an interface model of the surface of the A2 domain and FIXa protease domain has been proposed.41 In this model, FVIIIa residues Asp560, Gln561, and Arg562 are shown as interacting with Arg338 and Asp332 of FIXa, respectively. The moderately increased Kd for FVIIIAsp560Ala in the absence of phospholipid suggests that Asp560 has a role in stabilizing the interaction in this region, but that in the presence of phospholipid this interaction is not a major contribution to the binding energy of FVIIIa in its interaction with FIXa. The comparable Kd for FVIIIArg562Ala and FVIIIWT both in the presence and absence of phospholipid suggests that any conservation of FVIIIaArg562 interaction with Asp332 of FIXa is not critical in maintaining either the interaction with FIXa or modulating cofactor activity of FVIIIa. Substitution of Arg562 has previously been shown not to cause a defect in cofactor activity.47 The lack of effect of the Arg562 substitution on FVIII cofactor activity is analogous to the FVArg506Gln, referred to as factor V Leiden. The Leiden mutation is the most common inherited defect associated with venous thrombosis as a result of resistance to APC cleavage, but the substitution does not affect FVa cofactor activity.48Similarly, the substitution of Arg562 affects cleavage by APC.47
Hemophilia B–associated mutations within the helical 330 region of FIXa have been expressed and interaction with subsaturating levels of FVIIIa studied by 2 separate groups.45,46 In one study,45 8 separate residue substitutions were expressed and purified. In 7 of the 8, the result was decreased cofactor-mediated, FIXa activity, attributed primarily to the result of an increased Kd of FVIIIa–FIXa interaction of between 10- and 100-fold as compared with wild-type FIXa. In another report,46 2 residues within the 330 helix, FIX-Arg333 and FIX-Leu337, were substituted with Gln and Phe, respectively, which differed from the Leu and Ile substitutions made in the prior study. In addition to the residue substitutions, these investigators also substituted the residues FIXa 333-339 for the corresponding FX sequence. Results from these experiments indicated that the point mutations caused a decrease in FIXa-catalyzed FXa generation as compared with FIXa wild type. However, this effect resulted from a decrease in kcat for the tenase complex to between 3% and 4% of wild type, with a relatively small increase inKd detected. The differing mechanisms indicated above may reflect the different mutations studied, and their effect on this highly structured region.
In contrast to the FIXa 330-339 region that contains a helical secondary structure, the FVIII 558-565 region is likely an exposed loop region containing a number of highly exposed side chains.49 Substitutions within this region are not predicted to affect secondary structure. Our data suggest that the primary feature of this region is that it is critical in modulating cofactor activity through direct interaction near the active site of FIXa, rather than making substantial contribution to the binding energy with FIXa per se. The mutations associated with hemophilia A lead to a defective ability to act as cofactor for FIXa, and thus a dramatically reduced ability to generate FXa.
The authors thank Dr Pete Lollar and John Healey for kind provision of the RENeoB-FVIII construct and many helpful discussions. The authors acknowledge the input of Dr Noelene Quinsey and Jennifer Chandler in the early stages of this project.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2001-12-0361.
Supported by grants HL 30616 and HL 38199 from the National Institutes of Health and a Judith Graham Pool Postdoctoral Award to P.V.J. from the National Hemophilia Foundation.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Philip J. Fay, Department of Biochemistry and Biophysics, PO Box 610, University of Rochester Medical Center, 601 Elmwood Ave, Rochester, NY 14642; e-mail:philip_fay@urmc.rochester.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal