Abstract
The activated form of prothrombin plays pivotal roles in the regulation of crucial coagulation, fibrinolytic, and cellular processes. Among several congenital genetic defects affecting the prothrombin gene, a G→A mutation at position 20210—the accepted polyadenylation site—has been linked to hyperprothrombinemia and a corresponding increase in venous and arterial thrombotic risk. The current study substantiates the hypothesis that the 20210A mutation effects posttranscriptional dysregulation of the prothrombin messenger RNA (mRNA). Moreover, data from experiments carried out in fresh liver tissue indicate that the 20210A mutation does not affect prothrombin mRNA stability but, rather, effects a change in the location of the 3′-cleavage/polyadenylation reaction. Based upon this evidence, we propose an alternate model for the dysregulated expression of the prothrombin 20210A gene that does not require a change in the stability of its mRNA.
Introduction
The regulation of prothrombin expression is crucial to maintaining normal hemostatic function. Mutations that negatively affect the level or activity of prothrombin are uncommon but typically manifest as a correspondingly severe hemorrhagic phenotype. In contrast, a mutation in the prothrombin gene that has been linked to its overexpression1 exhibits a frequency exceeding 4% in some populations.2 This mutation is a G→A transversion at position 20210, the accepted prothrombin messenger RNA (mRNA) polyadenylation site.3 Heterozygotes for the 20210A gene variant express prothrombin at levels that are about 25% higher than average.1,4,5 The physiological importance of the 20210A mutation is evidenced by the significant risk of venous1,6and arterial thromboembolism5,7,8 displayed by 20210G/A heterozygotes, which can be particularly severe in specific subpopulations and target organs.9-11
The molecular basis for the hyperprothrombinemia of the 20210A mutation is poorly understood although potentially important from at least 2 perspectives. First, naturally occurring mutations that up-regulate gene expression are relatively uncommon. Second, its position in the 3′-untranslated region (3′UTR) suggests that the 20210A mutation affects prothrombin gene expression through a process that is unrelated to its transcription. A number of studies have tested the effect of the 20210A mutation on the stability of prothrombin mRNA. With a single exception,12 studies utilizing cultured cells have failed to demonstrate any clear difference in stability resulting from a G→A exchange at position 20210.13,14 Although carefully done, the experiments from the single positive study were performed using heterologous HeLa cells, despite previous demonstration that mechanisms affecting mRNA stability can display cell-type specificity.15 Moreover, important high-order structures that form in the 3′UTR of the native, full-length prothrombin mRNA may be inaccessible in commonly used reporter mRNAs comprising the prothrombin 3′UTR fused to a heterologous transcribed region (typically from human β-globin).
The current work evaluates the effect of the 20210A mutation on prothrombin gene expression by establishing the relative levels of prothrombin 20210G and 20210A mRNAs in fresh liver tissue obtained from a 20210G/A heterozygote. We observe that the 2 prothrombin mRNAs are present at equal levels, indicating that the 20210A mutation does not materially affect the stability of full-length prothrombin mRNA in intact hepatocytes. In addition, our studies demonstrate that the 20210A mutation affects the position of the 3′-cleavage/polyadenylation reaction, an event that may lead to its abnormal mRNA function.
Materials and methods
PCR-based screening of genomic DNA
Archival paraffin-embedded blocks containing recipient liver tissue from individuals undergoing orthotopic transplantation were obtained from the Cooperative Human Tissue Network (Eastern Division) under a University of Pennsylvania institutional review board–approved protocol. Total genomic DNA, prepared as described,16 was polymerase chain reaction (PCR) amplified using a variation of a previously described method.17 18 In this method, the forward DNA primer complements the prothrombin gene at positions 20063-20083, while the reverse oligomer (positions 20233-20212) contains a G→A substitution at position 20214, creating aHindIII restriction site in the context of an A, but not a G, at prothrombin position 20210. The prothrombin genotype is subsequently established by HindIII digestion of the PCR product.
Preparation of full-length prothrombin RNA from primary human hepatocytes
Total RNA was prepared from fragments of OCT-embedded fresh liver tissue (Sakura, Torrance, CA) snap-frozen in liquid N2. Tissue samples were digested overnight at 52°C in buffer (17 mM Tris [pH 7.5], 17 mM ethylenediaminetetraacetic acid, 170 mM NaCl, 0.85% sodium dodecyl sulfate, 2 mg/mL proteinase K) and RNA subsequently prepared from the clarified supernatant using TRIzol reagent in a scaled-down variation of the manufacturer's recommended method (LTI, Gaithersburg, MD).
Reverse transcriptase–PCR analysis
Approximately 1 μg purified RNA was annealed with 200 pmol oligomer (PT-T20: 5′-ATCGATGAATTCT20-3′) and first-strand complementary DNAs generated using avian myeloblastosis virus (AMV) reverse transcriptase under conditions recommended by the manufacturer (Boehringer Mannheim, Germany). Reaction products were diluted 1:25 in a 50 μL reaction (200 μM each deoxyribonuclease triphosphate, 3 mM MgCl2, 5 units Taq polymerase) in 1 × buffer supplied by the manufacturer (Applied Biosystems, Foster City, CA), using 100 pmol PT-T20 and 100 pmol of a forward primer (PT-26347: 5′-AAACGAGGGGATGCCTGTG-3′), and then amplified for 30 cycles under standard conditions. Approximately 2 μL of this reaction was then reamplified using primers PT-T20 and PT-26378 (5′-TCTAGAGGATCCGGGGACCCTTTGTCATGAAG-3′).BamHI/EcoRI-digested PCR products were ligated into the cognate site of plasmid pSP72 and library efficiency DH5α competent cells transformed under conditions recommended by the manufacturer (LTI). Plasmid DNAs prepared with a Qiaprep Spin Kit (Qiagen, Valencia, CA) were provided to the Nucleic Acid/Protein Core Facility at The Children's Hospital of Philadelphia for dideoxysequencing.
Results and discussion
A PCR cloning assay establishes the relative levels of prothrombin 20210G and 20210A mRNAs in primary liver tissue
The approximate 25% increase in prothrombin expression that is observed in 20210G/A heterozygotes1,4,5 is hypothesized to result from an increase in the stability of the prothrombin 20210A mRNA. This degree of hyperprothrombinemia would require a 1.5-fold increase in the steady-state levels of the prothrombin 20210A mRNA, a prediction we directly tested by assessing the relative levels of prothrombin mRNAs in fresh liver tissue obtained from a prothrombin 20210G/A heterozygote. A genotypically suitable liver was identified from more than 60 archival paraffin-embedded liver samples using a PCR-based screening method,17 18 and the corresponding OCT-embedded sample was obtained for subsequent analysis.
The low-level expression of both the prothrombin 20210G and 20210A mRNAs in intact liver precluded the use of standard Northern or primer extension analyses for their accurate quantitation. Consequently, the relative levels of the 2 prothrombin mRNAs were determined using an adaptation of a highly sensitive reverse transcriptase–PCR method (Figure 1).15 First-strand complementary DNAs, generated from total cellular RNA using an oligo(dT) primer, were PCR amplified using nested prothrombin-specific oligomers. The resulting DNA products were directionally inserted into a suitable vector that was used to transform competentEscherichia coli bacteria. The cloned PCR fragments from independent colonies were then directly sequenced to establish the ratio of prothrombin 20210G and 20210A transformants, reflecting the ratio of the cognate mRNAs in the original liver tissue.
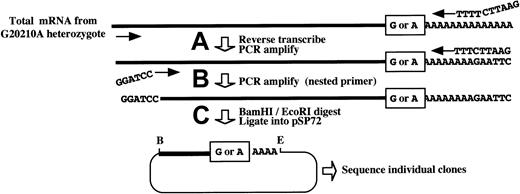
Features of a novel PCR cloning method for assessing the relative levels of 20210G and 20210A mRNAs in liver from a prothrombin 20210G/A heterozygote.
Total mRNA is prepared from the liver of a 20210G/A heterozygote using a method that does not discriminate among mRNAs on the basis of poly(A) tail length. (A) Prothrombin mRNAs, containing either a G or an A at position 20210 (boxed), are reverse transcribed and PCR amplified using a forward primer within the penultimate exon (forward arrow) and an oligo(dT) primer containing a 5′-terminal EcoRI restriction site. (B) An aliquot of the initial PCR reaction is reamplified using the oligo(dT) primer and a nested, forward primer containing a 5′-terminal BamHI restriction site (indicated). (C) The resulting complementary DNAs are digested with BamHI andEcoRI (B and E, respectively) and directionally inserted into the cognate sites of pSP72. Competent DH5α Escherichia coli are transformed, and plasmid DNAs prepared from individual ampicillin-resistant colonies are sequenced to determine the G:A ratio at position 20210, reflecting the original ratio of the cellular 20210G and 20210A mRNAs.
Features of a novel PCR cloning method for assessing the relative levels of 20210G and 20210A mRNAs in liver from a prothrombin 20210G/A heterozygote.
Total mRNA is prepared from the liver of a 20210G/A heterozygote using a method that does not discriminate among mRNAs on the basis of poly(A) tail length. (A) Prothrombin mRNAs, containing either a G or an A at position 20210 (boxed), are reverse transcribed and PCR amplified using a forward primer within the penultimate exon (forward arrow) and an oligo(dT) primer containing a 5′-terminal EcoRI restriction site. (B) An aliquot of the initial PCR reaction is reamplified using the oligo(dT) primer and a nested, forward primer containing a 5′-terminal BamHI restriction site (indicated). (C) The resulting complementary DNAs are digested with BamHI andEcoRI (B and E, respectively) and directionally inserted into the cognate sites of pSP72. Competent DH5α Escherichia coli are transformed, and plasmid DNAs prepared from individual ampicillin-resistant colonies are sequenced to determine the G:A ratio at position 20210, reflecting the original ratio of the cellular 20210G and 20210A mRNAs.
The prothrombin 20210G and 20210A mRNAs are present at equal levels in liver tissue from a 20210G/A heterozygote
Because of the known linkage between the stability of an mRNA and the length of its poly(A) tail,19 we prepared RNA using a nondiscriminatory method20 rather than poly(A)-select methods favored by others that can introduce significant ascertainment bias into the analysis. Total hepatic RNA prepared from a 20210G/A heterozygote was subsequently analyzed using the PCR cloning assay described above. Seventy prothrombin subclones were sequenced: 38 (54%) contained a G at position 20210, while 32 (46%) contained an A at the same position (Figure2A). The likelihood of obtaining this distribution from a mixture of prothrombin mRNAs containing a 1.5-fold excess of 20210A mRNAs is remote (.015 > P > .010; binomial probability), indicating that the prothrombin 20210G and 20210A mRNAs are present at equal levels in the original liver tissue. These data are not consistent with an increase in the stability of the prothrombin 20210A mRNA and indicate that the hyperprothrombinemia in 20210G/A heterozygotes must arise through a different mechanism.
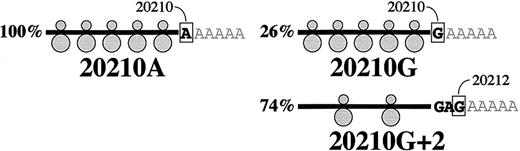
Relative levels and structural characteristics of prothrombin mRNAs prepared from the liver of a prothrombin 20210G/A heterozygote.
(A) Levels of prothrombin 20210G and 20210A mRNAs in primary hepatocytes. The expected number of colonies derives from the assumption that the average 125% prothrombin levels in 20210G/A heterozygotes arise from an approximate 1.5-fold excess of 20210A mRNA. The observed number of colonies, representing the ratio of 20210G and 20210A mRNAs in the liver of a 20210G/A heterozygote, were determined using the method described in Figure 1. (B) The 20210A mutation alters the polyadenylation pattern of prothrombin mRNA. The genomic DNA sequence bracketing position 20210 of the prothrombin gene is illustrated. The wild-type prothrombin mRNA (20210G) is polyadenylated at 2 positions: the published position (20210, 26%) and a position 2 nucleotides downstream (20212, 74%). The mutant prothrombin mRNA (20210A) is polyadenylated exclusively at position 20210.
Relative levels and structural characteristics of prothrombin mRNAs prepared from the liver of a prothrombin 20210G/A heterozygote.
(A) Levels of prothrombin 20210G and 20210A mRNAs in primary hepatocytes. The expected number of colonies derives from the assumption that the average 125% prothrombin levels in 20210G/A heterozygotes arise from an approximate 1.5-fold excess of 20210A mRNA. The observed number of colonies, representing the ratio of 20210G and 20210A mRNAs in the liver of a 20210G/A heterozygote, were determined using the method described in Figure 1. (B) The 20210A mutation alters the polyadenylation pattern of prothrombin mRNA. The genomic DNA sequence bracketing position 20210 of the prothrombin gene is illustrated. The wild-type prothrombin mRNA (20210G) is polyadenylated at 2 positions: the published position (20210, 26%) and a position 2 nucleotides downstream (20212, 74%). The mutant prothrombin mRNA (20210A) is polyadenylated exclusively at position 20210.
The 20210A mutation effects a change in prothrombin mRNA polyadenylation
The PCR cloning analysis also indicated an unanticipated but clear difference in the 3′-polyadenylation pattern of the prothrombin 20210G and 20210A mRNAs expressed in intact liver (Figure 2B). Wild-type (20210G) mRNAs were polyadenylated at 2 sites: position 20210 (26% of RNAs) and position 20212 (74%). In contrast, all 32 of the mutant (20210A) mRNAs were polyadenylated at position 20210. The variability in prothrombin 20210G mRNA processing was subsequently verified using mRNA purified from the liver of a 20210G/G homozygote (not shown), indicating that alternate mRNA 3′-terminal cleavage is a normal feature of prothrombin gene expression. Hence, a G→A mutation at prothrombin position 20210 results in defined secondary changes in the structure of approximately three quarters of the encoded mRNA transcripts. Although prothrombin mRNA stability does not appear to be affected by allele-specific polyadenylation patterns, these results raise the possibility that the site specificity of 3′-end processing may affect the translational efficiency of prothrombin mRNA. In this context it is worth noting the growing body of evidence linking mRNA translational efficiency to specific structures within the 3′UTR.21-24
A model for the hyperprothrombinemia observed in prothrombin 20210G/A heterozygotes
In contrast to previous analyses,12 our PCR cloning method indicates a high likelihood that the 20210G and 20210A mRNAs are equally stable in primary hepatic cells (Figure 2A) and exhibit allele-specific patterns of 3′-end processing and polyadenylation (Figure 2B). These findings can be accommodated by a straightforward model for the posttranscriptional regulation of prothrombin gene expression in which the prothrombin 20210G and 20210A mRNAs are present at equal levels but translate with efficiencies that are dictated by differences in their structures (Figure 3). Prothrombin mRNAs that are polyadenylated at position 20210 translate with higher efficiency than mRNAs polyadenylated at position 20212; consequently, 20210A mRNAs (which are all polyadenylated at the translationally active 20210 site) generate more prothrombin than an equal quantity of 20210G mRNAs (most of which are polyadenylated at the translationally quiescent 20212 site). The mechanism underlying mRNA-specific translational efficiencies would undoubtedly reflect differences in their high-order structures, including potential 3′-processing site-dependent effects on the lengths of their poly(A) tails12 or alterations in the binding efficiencies of functionally important trans-acting factors.13 This model would account for the equal stabilities of the prothrombin 20210G and 20210A mRNAs, for their allele-specific polyadenylation patterns and, ultimately, for the hyperprothrombinemia displayed by prothrombin 20210G/A heterozygotes.
A model for the hyperprothrombinemia in prothrombin 20210G/A heterozygotes.
Prothrombin 20210G/A heterozygotes transcribe equal quantities of RNA from the wild-type (right) and mutant genes (left). Nascent prothrombin 20210G mRNA is polyadenylated at positions 20210 and 20212 in the indicated ratio; by comparison, the 20210A mRNA is polyadenylated exclusively at position 20210. Prothrombin mRNAs that are polyadenylated at positions 20210 and 20212 translate with high and low efficiencies, respectively (illustrated by dense and sparse ribosomal loading).
A model for the hyperprothrombinemia in prothrombin 20210G/A heterozygotes.
Prothrombin 20210G/A heterozygotes transcribe equal quantities of RNA from the wild-type (right) and mutant genes (left). Nascent prothrombin 20210G mRNA is polyadenylated at positions 20210 and 20212 in the indicated ratio; by comparison, the 20210A mRNA is polyadenylated exclusively at position 20210. Prothrombin mRNAs that are polyadenylated at positions 20210 and 20212 translate with high and low efficiencies, respectively (illustrated by dense and sparse ribosomal loading).
Despite the recognition that posttranscriptional control of mRNAs may be highly cell specific,15 existing models for the control of prothrombin gene expression are based on studies carried out in heterologous cultured cells,12 under the assumption that crucial functional and structural characteristics of prothrombin mRNA are unaffected. In contrast, we elected to characterize prothrombin mRNAs at steady state in intact human liver, with the expectation that these studies might more accurately assess relevant molecular processes. In addition to suggesting structural and mechanistic bases for the pathophysiology of the prothrombin 20210A mutation, the results may provide important insights into mechanisms that may be crucial to the regulated expression of evolutionarily related coagulation factors, including factors VII, IX, and X, and protein C.25 These factors are encoded by genes derived from a common prothrombinlike progenitor gene and possess related functional domains, raising the intriguing possibility that their expression might be regulated through conserved molecular mechanisms. Further delineation of the molecular pathophysiology of the prothrombin 20210A mutation may provide valuable insight into the role of the 3′UTR in regulating the expression of one or more of these factors.
The authors thank Emma E. Furth for generously providing OCT-fixed liver and acknowledge the technical assistance of Rama Kudaravalli and Huyen Tran.
Prepublished online as Blood First Edition Paper, April 17, 2002; DOI 10.1182/blood-2002-02-0412.
Supported in part by NIH K08 HL03661 (E.S.P.) and NIH P30 DK50306 (J.E.R.). E.S.P. is a Doris Duke Clinical Scientist Awardee (T98062B).
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
J. Eric Russell, Abramson Research Building, Room 316F, The Children's Hospital of Philadelphia, 34th St and Civic Center Blvd, Philadelphia, PA 19104; e-mail:jeruss@mail.med.upenn.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal