Abstract
Treatment of different human leukemia cell variants with the anthracycline adriamycin was associated with a rapid activation of the proteasome. Thus, proliferating U937, TUR, and retrodifferentiated U937 cells exhibited a 4.3-fold, 5.8-fold, and 4.3-fold proteasome activation within 15 minutes after adriamycin treatment, respectively. In contrast, little if any proteasome activation was detectable in a growth-arrested differentiated U937 population following adriamycin treatment. Further analysis of this mechanism revealed a significant reduction of adriamycin-induced proteasome activity after inhibition of poly(ADP-ribose) polymerase (PARP) by 3-aminobenzamide (3-ABA) in the proliferating leukemic cell types. These findings suggested that PARP is involved in the regulation of drug-induced proteasome activation. Indeed, anti-PARP immunoprecipitation experiments of adriamycin-treated cells revealed increasing levels of coprecipitated, enzymatically active proteasome particularly in the proliferating cell variants in contrast to the differentiated U937 cells, with a maximum after 15 minutes, and sensitivity to PARP inhibition by 3-ABA. The specific role of the PARP was investigated in U937 and TUR cell clones stably transfected with a constitutively active antisense PARP (asPARP) vector. Thus, asPARP-TUR cells developed a 25-fold increased sensitivity to adriamycin treatment. Furthermore, we investigated leukemic blasts isolated from acute myelogenous leukemia patients and obtained a similarly enhanced proteasome activity after adriamycin treatment, which was dependent on the PARP and thus could be coprecipitated with anti-PARP antibodies. Transient transfection of leukemic blasts with the asPARP vector significantly reduced the adriamycin-induced proteasome activation. These data suggest that the PARP-associated nuclear proteasome activation represents a potential target within chemotherapeutic defense mechanisms developed by leukemia cells.
Introduction
Chemotherapeutic treatment of acute myelogenous leukemia (AML) achieves complete remission in approximately 60% to 70% of adult AML. However, many patients obtaining a complete remission will still suffer from leukemic recurrence.1-3To improve the efficiency of antileukemic drugs, closer insights into the cellular defense mechanisms of leukemic cells are required.
Common chemotherapies for treatment of AML are based on DNA intercalating antitumor drugs such as the nucleoside analog cytosine-arabinoside (AraC),4 etoposide (VP-16),5 and compounds directly liberating free oxygen radicals, such as anthracyclines.6 The cytotoxic action of anthracyclines by the generation of free radicals has been demonstrated both in vitro and in vivo.7-9 Radical production can be associated with intratumor drug metabolism or redox cycling reactions,10,11 particularly in close proximity to the chromatin.7,8,11 Oxidative stress in the nucleus contributes to the formation of oxidatively damaged histones, which can modify DNA by cross-linking12 13 and therefore can alter chromatin dynamic processes such as DNA transcription, replication, and repair. Thus, the selective and efficient degradation of oxidatively damaged nuclear proteins appears to be a requirement for maintaining genomic integrity. However, such an antioxidative defense system might also be used by certain leukemia cells to cope with nuclear damages induced by radical-producing compounds. Consequently, these resistant leukemia cells survive chemotherapy and potentially develop a tumor relapse.
Usually, oxidatively damaged proteins undergo selective proteolysis via an adenosine triphosphate– and ubiquitin-independent pathway.14 The 20S proteasome, a large 700-kd multicatalytic and multisubunit proteinase located in the cytosol and in the nucleus,15 is largely responsible for the selective degradation of such damaged proteins,16,17 including oxidatively damaged DNA-bound histones.18 Previous studies have demonstrated an elevated proteasome activity in leukemia cells19 and a possible relationship to the proliferative activity.20
Other work has described an activation of the proteasome in vitro by interaction with poly(ADP-ribosyl)ated poly(ADP-ribose) polymerase (PARP).21 This enzyme was demonstrated to be highly activated by both oxidative stress and chemotherapy treatment.19,22 PARP is a nuclear enzyme that catalyzes the synthesis and transfer of poly(ADP-ribose) from NAD+molecules to various nuclear acceptor proteins as well as to itself23 and may therefore play also a role in DNA transcription and repair.24 In this context, previous studies have suggested that PARP inhibition modulates chemotherapy toxicity in the same direction.6,25,26 Proposed mechanisms for antitumor activity of PARP inhibition during chemotherapy involve disturbance of DNA repair,6,27 induction of specific resistance-related molecules,28 or a loss of amplified oncogenes.29 Interestingly, PARP is also discussed as a useful target for enhancing the cytotoxicity of several chemotherapeutic agents,4-6,30 particularly adriamycin.31 However, little is known about the possible mechanisms of adriamycin-induced chemotherapy potentiation by PARP and its relationship to a proteasome activation in vivo.
The extent of nuclear damage and cytotoxicity after application of antileukemic drugs depends on the type and possibly on the state of development of the targeted tumor cells. Therefore, we used in this study a well-established differentiation-retrodifferentiation system of human U937 myelomonocytic leukemia cells,32-34 which allows the investigation of differentiation-dependent alterations in closely related populations. Treatment of human U937 leukemia cells with 12-O-tetradecanoylphorbol-13-acetate (TPA) is accompanied by growth arrest and induction of a monocytic differentiation program.35 During this differentiation process, the cells acquire a variety of morphologic and nonmalignant functional changes associated with monocytes or macrophages.32 These TPA-differentiated U937 cells remain and survive in a transient G0 growth-arrest cycle for approximately 2 to 3 weeks.33Thereafter, the differentiated population loses all acquired differentiation markers and re-enters the proliferative cell cycle by a retrodifferentiation process, developing again the undifferentiated U937 phenotype.33 Other work has characterized a U937 subclone, termed TUR, which is unable to differentiate along the monocytic lineage and continues to proliferate in the presence of TPA.36 Moreover, human TUR leukemia cells demonstrate aggressively developing tumors in scid mice in contrast to the parental U937 cells.37 These populations of U937 cell variants, which resemble certain states of differentiation and proliferation, were used in the present study to investigate the adriamycin-induced activation of the nuclear protesome and a possible involvement of the proteasome in developing adriamycin resistance.
Materials and methods
Cell culture of human leukemic cell lines and isolated leukemic patient blasts
Human U937 myeloid leukemia cells and TUR cells were grown in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum (FCS), 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine (Gibco, Grand Island, NY). The cells were treated with 5 nM TPA (Sigma, St Louis, MO) at a density of 2 × 105/mL for up to 72 hours. The long-term culture of adherent TPA-differentiated U937 cells was performed in the absence of TPA by changing the culture medium every 3 days as described previously.32 Cell viability was assayed by trypan blue exclusion. Human leukemic blasts were isolated from peripheral blood samples by Ficoll-Hypaque density gradient centrifugation. Cells were cultivated in RPMI 1640 medium supplemented with 10% heat-inactivated FCS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM glutamine for up to 48 hours without any significant change in the cell viability as assayed by trypan blue exclusion.
Treatment with adriamycin, AraC, and VP-16
Cells were washed twice with phosphate-buffered saline (PBS) and incubated with different concentrations of chemotherapeutic compounds to investigate the toxicity after a 24-hour treatment. For these experiments sublethal concentrations (more than 90% viable cells) of 10−7 M adriamycin, 10−7 M AraC, or 10−6 M VP-16 were determined and used in cell culture medium at a density of 106/mL with or without a 30-minute preincubation using 1 mM of the specific PARP inhibitor 3-aminobenzamide (3-ABA) or using 2 μM of the selective and reversible proteasome inhibitor MG-132 (Affiniti Research Products, Exeter, United Kingdom), respectively. The applied inhibitor concentrations have been optimized for endogenous PARP and proteasome activity, respectively, and for the viability of the treated cells (data not shown). After incubation for the appropriate time points, the reaction was stopped by addition of a 10-fold volume of ice-cold PBS. Thereafter, the cells were centrifuged and washed with PBS immediately.
Isolation of nuclei and measurement of the nuclear proteasomal proteolytic activity
Highly purified nuclei without detectable contamination of other cellular compartments were obtained from the human leukemic cell lines by the procedure of Emig et al.38 The isolated nuclei were lysed by 5 freeze-thawing cycles in 10 mM HEPES (pH 7.5), 24 mM KCl, 10 mM MgCl2, and 0.5 mM dithiothreitol. The proteasome activity in nuclear lysates was measured by the peptidase activity toward the hydrophobic fluorogenic peptide succinyl-leucine-leucine-valine-tyrosine-methylcoumaryl-amide (suc-LLVY-MCA) by incubation with 50 mM Tris-HCl (pH 7.8), 20 mM KCl, 5 mM MgOAc, 0.5 mM dithiothreitol, and 200 μM suc-LLVY-MCA for 1 hour at 37°C in the presence and absence of 10 μM lactacystin. After incubation for 2 hours at 37°C, the reaction was stopped by the addition of an equal volume of ice-cold ethanol and a 10-fold volume of 125 mM sodium borate (pH 9.0). Thereafter, fluorescence intensity was determined at 380 nm excitation and 440 nm emission wavelength.
Immunoblot analysis
Sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) was performed in a 15% (for detection of proteasome subunits) and in a 10% (for detection of PARP) separation gel. Electrophoresis was standardized using prestained low molecular weight markers (Bio-Rad, Munich, Germany). Proteasome subunits were detected with a polyclonal rabbit antiproteasome antibody (Affiniti Research Products), and PARP was detected with a polyclonal rabbit anti-PARP antibody (a kind gift from Dr Schweiger, Berlin, Germany). For control blots a monoclonal mouse anti–green fluorescent protein (GFP) antibody (Clontech, Palo Alto, CA) and a polyclonal rabbit antiactin antibody (Sigma) were used, respectively. After incubation with an appropriate peroxidase-conjugated secondary antibody, the blots were developed using an enhanced chemiluminescence (ECL) detection kit (Amersham, Uppsala, Sweden).
Immunoprecipitation experiments
Immunoprecipitation experiments were performed in lysates of nuclei isolated from adriamycin-treated human leukemic cell lines and adriamycin-treated human leukemic patient blasts. For this technique a rabbit anti-PARP antibody was used with a modified procedure by Zwilling et al.39 The primary antibody reaction was performed in 50 mM Tris (pH 7.8) containing 150 mM NaCl (Tris-buffed saline), 0.1% bovine serum albumin, 100 μg nuclear lysates, and 1μL rabbit anti-PARP immunoglobulin G (IgG) (5 μg/μL) for 1 hour at 4°C. For secondary antibody reactions, 1 μL donkey antirabbit IgG (5 μg/μL) was added and incubated for 1 hour at 4°C. Binding of the secondary antibody was detected by using an ECL kit. Immunoprecipitations were initiated by adding 20 μL protein A–Sepharose (equilibrated with an equal volume of 50 mM Tris, 150 mM NaCl, and 0.1% bovine serum albumin, pH 8.0) for a 1-hour incubation at 4°C. Control immunoprecipitations were performed with the secondary antibody alone and the protein A–Sepharose. The Sepharose beads were centrifuged (14 000g for 30 minutes) and washed 5 times with Tris-buffered saline. Sepharose-bound proteins were separated by 15% SDS-PAGE, transblotted to nitrocellulose membranes, and incubated either with a polyclonal goat anti-PARP IgG or a polyclonal rabbit anticore-proteasome IgG.
Measurement of oxidatively modified nuclear proteins
Oxidatively modified nuclear proteins were determined by derivatization of protein-bound carbonyl moieties with dinitrophenylhydrazine (DNPH) according to Reznick and Packer.40 For this assay, 1.5 mg nuclear protein was derivatized with DNPH, precipitated with trichloroacetic acid, extensively washed with ethanol/ethylacetate (1:1 vol/vol), and redissolved in guanidinhydrochloride solution. Dinitrophenylhydrazones were determined spectrophotometrically at 370 nm.
Detection of covalent DNA-protein cross-links
Endogenous proteins were labeled with [3H]lysine for 5 population doublings by addition of 37 kBq (1 μCi) [3H]lysine (Amersham) to 5 × 105 U937, antisense PARP (asPARP)–U937, TUR, or asPARP-TUR cells in culture medium, respectively. The cells were washed 3 times in culture medium to remove nonincorporated radioactivity and cultured further on in the presence of 10−7 M adriamycin with or without a 30-minute preincubation of 1 mM 3-ABA for up to 24 hours. At the time points indicated, the nuclei of the labeled and treated cells were isolated and lysed as described above. Covalent DNA-protein cross-links were separated by nonreducing SDS–urea electrophoresis in a 3% stacking gel and a 2-phase separation gel system (5%/20%) as described previously.41 Whereas noncovalently bound proteins as well as other proteins were detectable in the lower 20% gel phase, the DNA and proteins covalently associated with DNA could be detected by silver stain and were quantified by scintillation counting of the ethidium bromide–stained DNA bands.
Alkaline single-cell electrophoresis
DNA damage was investigated by alkaline single-cell gel electrophoresis (comet assay), which has been proven as a sensitive method for the detection of low levels of DNA damages.42,43Alkaline single-cell gel electrophoresis was performed as described by Singh et al.44 Briefly, the cells were centrifuged, resuspended in PBS/0.5% low melting point agarose at 37°C, and 75 μL of the mixture was added onto a frosted glass microscope slide precoated with 100 μL of 1% melting point agarose. The slides were incubated 4°C for 10 minutes to allow polymerization of the agarose. The slides were immersed in cell lysis buffer (2.5 M NaCl, 100 mM ethylenediaminetetraacetic acid, 10mM Tris, 300 mM NaOH, 10% DMSO, and 1% Triton X-100, pH 10) at 4°C for 1 hour in the dark and placed in a horizontal alkaline electrophoresis unit (300 mM NaOH,1 mM ethylenediaminetetraacetic acid, pH 13) for 60 minutes to allow DNA unwinding. Thereafter, electrophoresis was performed at 20 V (1 V/cm, 300 mA) for 30 minutes, and the slides were gently neutralized (0.4 M Tris-HCl, pH 7.5) for 3 × 15 minutes. The extent of DNA migration was visualized after ethidium bromide staining using an Olympus BX-50 fluorescence microscope.
Cloning of an asPARP vector
Total RNA was isolated from BV2 cells (RNeasy Maxi Kit, Qiagen, Valencia, CA) followed by a complementary DNA synthesis (cDNA Synthesis Kit, Clontech) with a random primer hexamer and an amplification of the DNA of interest by polymerase chain reaction (PCR) (MJ Research, Waltham, MA). The PCR amplification product (a 650–base pair asPARP-containing fragment) was ligated into the GFP-coding pTracer CMV2 vector (Invitrogen, Carlsbad, CA) and amplified in competentEscherichia coli cells (Genotype TOP10F′). The successful cloning of the asPARP fragment was confirmed by sequencing analysis.
Stable transfection of human leukemic cell lines with the asPARP vector
U937 and TUR cells (107/mL) were incubated with 20 μg of either the pTracer CMV2 vector alone or the asPARP-containing vector in 500 μL RPMI 1640 medium. The cells were electroporated with a 250 V/960 μF pulse and then cultured in complete culture medium for 48 hours to allow expression of the zeocin gene. Thereafter, stably transfected U937 and TUR cells were selected with culture medium containing 100 μg/mL zeocin for the next 4 weeks, in each case. Long-term culture of stably transfected cells was maintained in culture medium with 50 μg/mL zeocin.
Transient transfection of human leukemic patient blasts with the asPARP vector
Freshly isolated human blasts from AML patients were transiently transfected by a lipofectin method. Blasts were transfected in serum-free RMPI 1640 medium with 12 μg vector DNA and 60 μL Rotifect (Carl-Roth, Karlsruhe, Germany) in a total volume of 20 mL for 5 hours. After removal of the transfection solution, blasts were cultivated in RMPI 1640 medium supplemented with 10% FCS. The successful transfection was determined by detection of the GFP in a Western blot and in a fluorescence microscope.
Results
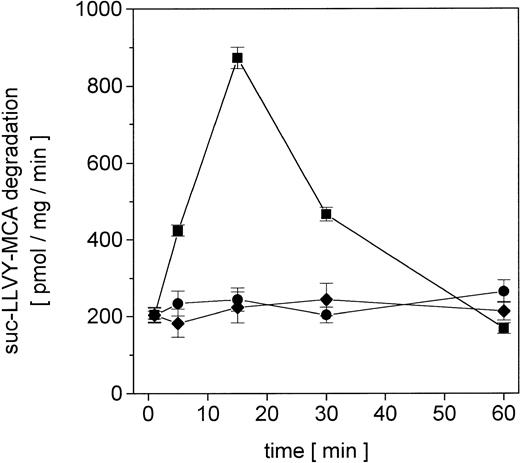
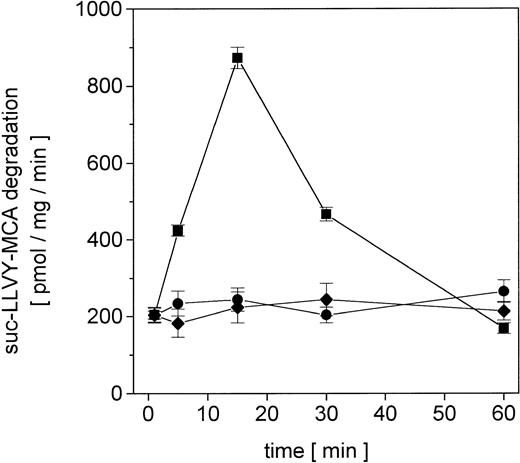
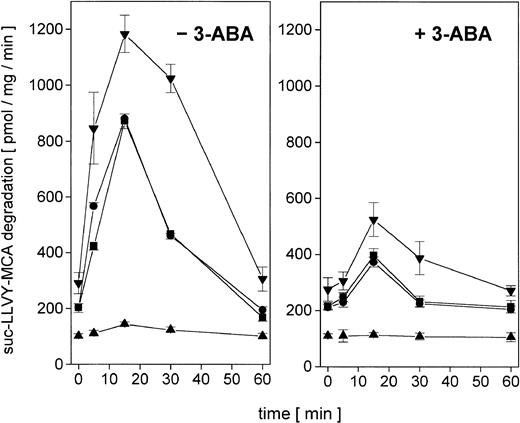
Proliferating U937 human leukemia cells were treated with sublethal concentrations of AML therapy compounds, including DNA intercalating and oxygen radical–generating drugs, to investigate metabolization effects and repair mechanisms of damaged proteins by the nuclear proteasome. The proteasome activity in the nuclei was measured as lactacystin-sensitive cleavage of the proteasome substrate suc-LLVY-MCA (Figure 1). Following adriamycin treatment of U937 cells, the proteasome activity increased about 4.3-fold after 15 minutes and subsequently returned to control level after 60 minutes (Figure 1). In contrast, treatment with DNA structure–modifying agents such as VP-16 and AraC demonstrated no significant changes in the activity of the nuclear proteasome within 60 minutes, respectively (Figure 1).
Nuclear proteasomal activity measured by suc-LLVY-MCA degradation in U937 human myelomonocytic leukemia cells after treatment with different chemotherapeutics.
Nuclei from U937 cells treated with 10−7 M adriamycin (▪), 10−7 M AraC (●), or 10−6 M VP-16 (♦), were isolated and lysed and the degradation of lactacystin-senstive suc-LLVY-MCA was quantified as proteasomal activity. Data are given as mean ± SD (n = 4).
Nuclear proteasomal activity measured by suc-LLVY-MCA degradation in U937 human myelomonocytic leukemia cells after treatment with different chemotherapeutics.
Nuclei from U937 cells treated with 10−7 M adriamycin (▪), 10−7 M AraC (●), or 10−6 M VP-16 (♦), were isolated and lysed and the degradation of lactacystin-senstive suc-LLVY-MCA was quantified as proteasomal activity. Data are given as mean ± SD (n = 4).
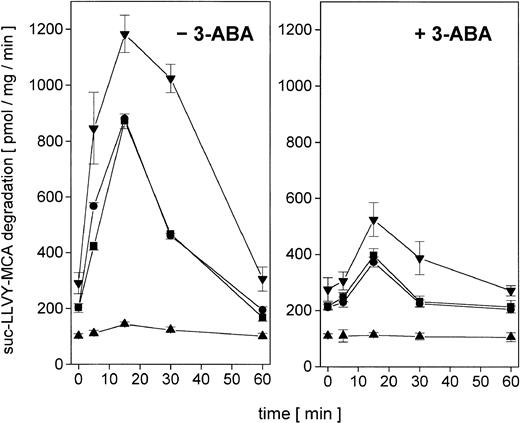
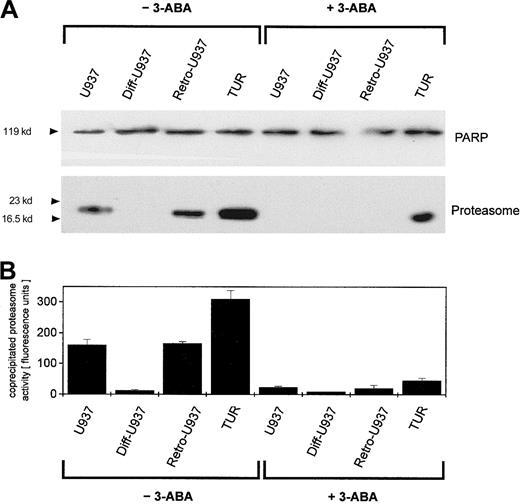
Whereas tumor cells in distinct states of development may respond differently to the same antitumor chemotherapy, we investigated the enzymatic activity and regulation of the nuclear proteasome after adriamycin treatment in certain cell variants of U937 cells. Thus, proliferating U937 cells, nonproliferating TPA-differentiated cells (Diff-U937), retrodifferentiated cells (Retro-U937), as well as the aggressively tumor-developing U937 subclone TUR were treated with sublethal concentrations of adriamycin in a time-dependent manner for up to 60 minutes. The constitutive nuclear proteasomal activity in U937 control cells increased about 4.3-fold after 15 minutes of adriamycin treatment and returned to control level after 60 minutes (Figure2, left panel). In growth-arrested Diff-U937 cells, the constitutive nuclear proteasomal activity was significantly reduced and demonstrated little if any difference after adriamycin treatment. However, when the proliferative capacity resumed in Retro-U937 cells, the proteasome activity was comparable to the level observed in U937 control cells both constitutively and after adriamycin-induced oxidative stress (Figure 2, left panel). Moreover, the highly tumorigenic U937 subclone TUR demonstrated a 1.5-fold elevated constitutive proteasomal activity as compared with U937 cells, which increased to about 5.8-fold after adriamycin treatment (Figure 2, left panel).
Nuclear proteasomal activity after addition of 10−7 M adriamycin in different U937 leukemic cell variants in the absence or presence of the PARP inhibitor 3-ABA.
(Left) −3-ABA. Nuclei from U937, Diff-U937, Retro-U937, and TUR cells were isolated and lysed after treatment with 10−7 M adriamycin for the time points indicated, respectively. Proteasome activity is expressed as lactacystin-sensitive suc-LLVY-MCA degradation. Data are given as mean ± SD (n = 4). (Right) +3-ABA. Time course of proteasome activity in lysates of nuclei from adriamycin-treated cells in the presence of 1 mM 3-ABA. Data are given as mean ± SD (n = 4). ▪, U937; ▴, Diff-U937; ●, Retro-U937; ▾, TUR.
Nuclear proteasomal activity after addition of 10−7 M adriamycin in different U937 leukemic cell variants in the absence or presence of the PARP inhibitor 3-ABA.
(Left) −3-ABA. Nuclei from U937, Diff-U937, Retro-U937, and TUR cells were isolated and lysed after treatment with 10−7 M adriamycin for the time points indicated, respectively. Proteasome activity is expressed as lactacystin-sensitive suc-LLVY-MCA degradation. Data are given as mean ± SD (n = 4). (Right) +3-ABA. Time course of proteasome activity in lysates of nuclei from adriamycin-treated cells in the presence of 1 mM 3-ABA. Data are given as mean ± SD (n = 4). ▪, U937; ▴, Diff-U937; ●, Retro-U937; ▾, TUR.
Our recent studies have suggested a possible regulatory involvement of PARP during H2O2-induced activation of the nuclear proteasome,21 and similar results were obtained in adriamycin-treated human leukemia cells (Figure 2, right panel). Thus, inhibition of PARP by addition of 3-ABA significantly reduced the adriamycin-induced proteasome activity in proliferating U937, Retro-U937, and TUR cells by about 50%, respectively (Figure 2, right panel). In contrast, the low constitutive proteasome activity in growth-arrested Diff-U937 cells remained unchanged following PARP inhibition by 3-ABA (Figure 2, right panel). Taken together, these data suggested an important role of PARP in the regulation of adriamycin-induced proteasome activity that is associated with the proliferative capacity and the state of development in leukemic cells.
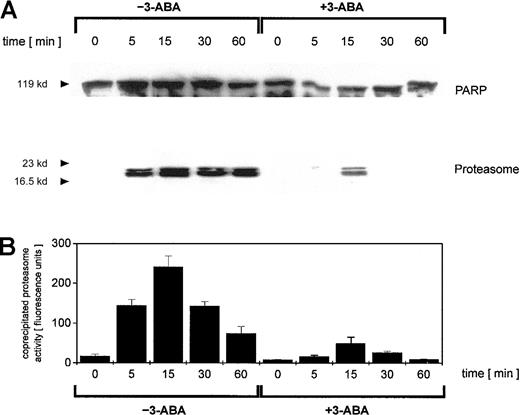
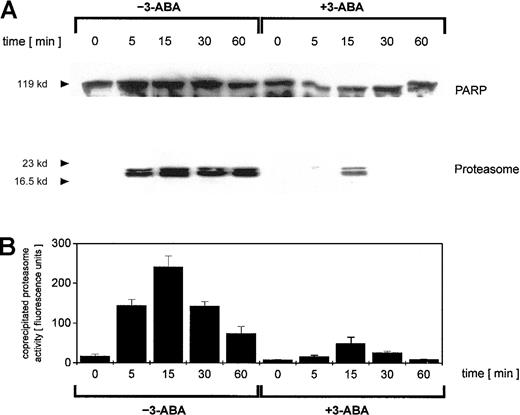
To address this point, we investigated possible interactions between PARP and the proteasome by coimmunoprecipitation experiments. Adriamycin treatment of U937 cells for up to 60 minutes was followed by an anti-PARP immunoprecipitation of nuclear proteins and subsequent anti-PARP and antiproteasome Western blotting (Figure3A). Anti-PARP immunoprecipitates demonstrated little if any differences in the level of PARP protein (Figure 3A). In contrast, proteasome-specific subunits were detected exclusively in adriamycin-treated cells, and inhibition of PARP by 3-ABA strongly reduced the coprecipitation of proteasome subunits (Figure 3A). To test if these coprecipitates contained only certain proteasome subunits or the functional active proteasome, we also measured the proteasomal enzymatic activity in the anti-PARP immunoprecipitates (Figure 3B). Thus, the proteasomal enzymatic activity demonstrated a strong correlation with the amount of coprecipitated proteasome protein (Figure 3A-B). These data suggested that after oxidative stress, increasing levels of functionally active proteasome complex become associated with PARP. Moreover, inhibition of PARP by 3-ABA significantly reduced the association with the proteasome complex and consequently diminished proteasome activity.
Immunoprecipitation of PARP in nuclei from adriamycin-treated U937 cells and proteasomal activity of PARP coimmunoprecipitates.
U937 cells were treated with 10−7 M adriamycin in the absence or presence of 1 mM 3-ABA for the time points indicated. Thereafter, the nuclei were isolated, lysed, and immunoprecipitated with a polyclonal rabbit anti-PARP antibody. (A) Immunoprecipitated proteins were separated by SDS-PAGE, transblotted, and visualized with a polyclonal anti-PARP and a polyclonal antiproteasome antibody, respectively. One experiment of 3 similar ones is presented. (B) Proteasome enzyme activity in the anti-PARP immunoprecipitated lysates was measured as suc-LLVY-MCA degrading activity. Data are given as mean ± SD from 3 parallel proteolysis assays corresponding to the Western blot in panel A.
Immunoprecipitation of PARP in nuclei from adriamycin-treated U937 cells and proteasomal activity of PARP coimmunoprecipitates.
U937 cells were treated with 10−7 M adriamycin in the absence or presence of 1 mM 3-ABA for the time points indicated. Thereafter, the nuclei were isolated, lysed, and immunoprecipitated with a polyclonal rabbit anti-PARP antibody. (A) Immunoprecipitated proteins were separated by SDS-PAGE, transblotted, and visualized with a polyclonal anti-PARP and a polyclonal antiproteasome antibody, respectively. One experiment of 3 similar ones is presented. (B) Proteasome enzyme activity in the anti-PARP immunoprecipitated lysates was measured as suc-LLVY-MCA degrading activity. Data are given as mean ± SD from 3 parallel proteolysis assays corresponding to the Western blot in panel A.
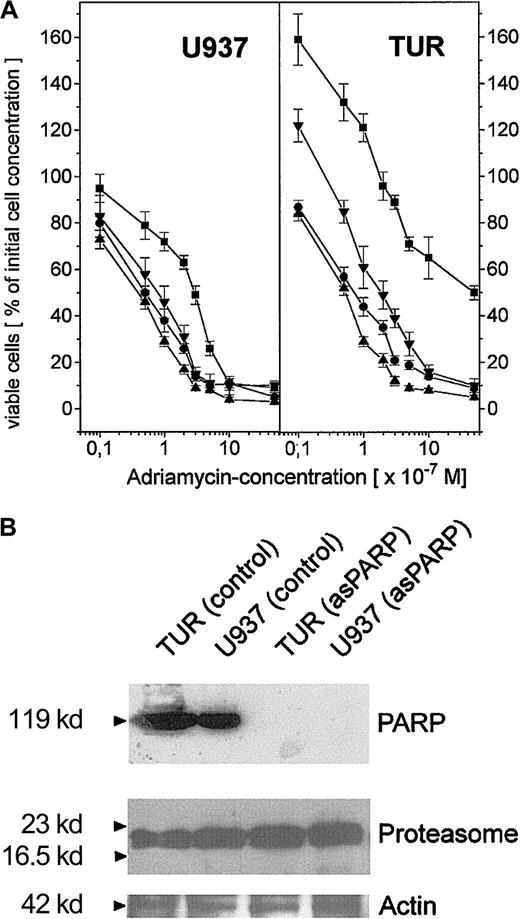
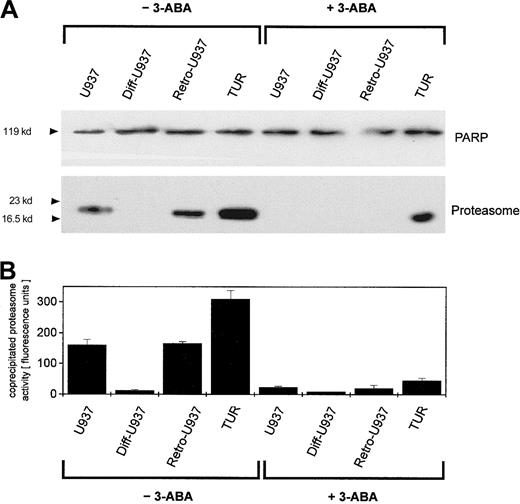
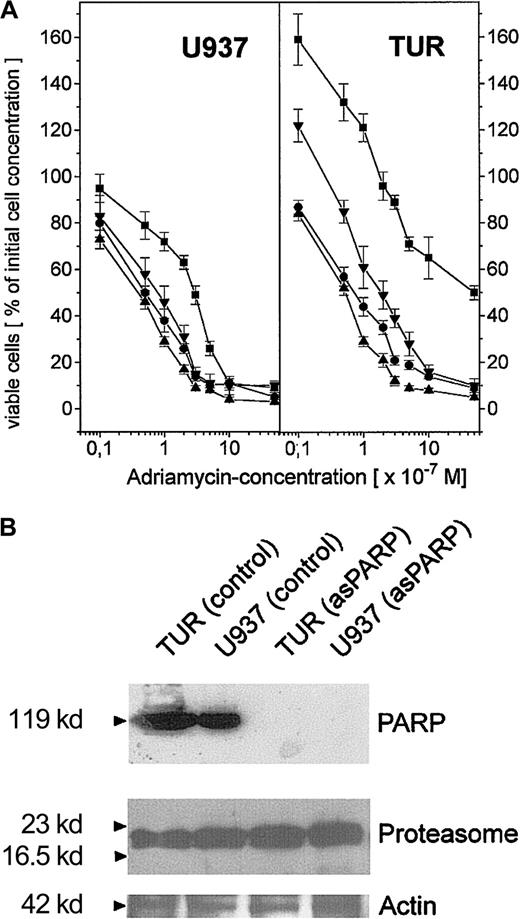
To compare the extent of this PARP-proteasome interaction in the different leukemic cell variants, U937, Diff-U937, Retro-U937, and TUR cells were treated with adriamycin for maximal nuclear proteasome activity (15 minutes) followed by extraction of nuclear proteins and anti-PARP immunoprecipitation (Figure 4). Subsequent anti-PARP Western blotting revealed little if any differences in the level of precipitated PARP protein throughout the 4 investigated cell types (Figure 4A). In contrast, most of the coprecipitated proteasome complex was demonstrated by TUR cells when compared with U937 and Retro-U937 cells, whereas the proteasome was undetectable in Diff-U937 (Figure 4A). Similar results were obtained when the proteasomal activity was measured in the anti-PARP immunoprecipitates of these 4 cell types in the absence and presence of the PARP inhibitor 3-ABA (Figure 4B). Taken together, these studies confirm the association of the functionally active proteasome complex with PARP in oxidatively stressed cells; however, the different levels of proteasome association in the 4 different cell types suggested altered antioxidative defense mechanisms depending on the state of development. We therefore investigated the adriamycin toxicity in U937 cells and in the highly chemotherapy-resistant TUR cells (Figure5). In this experiment U937 and TUR cells were preincubated with the selective proteasome inhibitor MG-132 or with the PARP-inhibitor 3-ABA and then treated with various adriamycin concentratrions. The amount of remaining viable cells after 24 hours is demonstrated in Figure 5A. In all applied concentrations, adriamycin exhibited an antiproliferative effect on U937 cells and a median lethal dose (LD50) of about 3 × 10−7 M, whereas TUR cells remained in the proliferative cycle at adriamycin concentrations below 10−7 M. Moreover, TUR cells exhibited an LD50 of about 5 × 10−6 M and thus developed an approximately 16-fold higher adriamycin resistance as compared with U937 cells (Figure 5A). However, inhibition of PARP by 3-ABA was associated with a significantly enhanced adriamycin sensitivity and a similar LD50 of about 7 × 10−8 M adriamycin in both U937 and TUR cells (Figure 5A). Similar effects were observed after preincubation with the selective proteasome inhibitor MG-132, which also abolished the initially high resistance of TUR cells to the level of U937 cells (Figure 5A). Taken together, these findings demonstrated that PARP inhibition significantly reduced the proteasome activation in both cell types and, consequently, significantly reduced the adriamycin resistance by about 16-fold in U937 cells and by about 73-fold in TUR cells (Figure 5A).
Detection of PARP-dependent proteasome activations in different U937 leukemic cell variants.
U937, Diff-U937, Retro-U937, and TUR cells were treated with 10−7 M adriamycin for 15 minutes (maximal protesome activation) in the absence and presence of 1 mM of the PARP inhibitor 3-ABA, respectively, and subsequent anti-PARP immunoprecipitation was performed. (A) SDS-PAGE and immunoblot with a polyclonal rabbit anti-PARP antibody and a polyclonal rabbit antiproteasome antibody. (B) Proteasome enzyme activity in the anti-PARP immunoprecipitated lysates was measured as suc-LLVY-MCA degrading activity. Data are given as mean ± SD from 3 parallel proteolysis assays corresponding to the Western blot in panel A.
Detection of PARP-dependent proteasome activations in different U937 leukemic cell variants.
U937, Diff-U937, Retro-U937, and TUR cells were treated with 10−7 M adriamycin for 15 minutes (maximal protesome activation) in the absence and presence of 1 mM of the PARP inhibitor 3-ABA, respectively, and subsequent anti-PARP immunoprecipitation was performed. (A) SDS-PAGE and immunoblot with a polyclonal rabbit anti-PARP antibody and a polyclonal rabbit antiproteasome antibody. (B) Proteasome enzyme activity in the anti-PARP immunoprecipitated lysates was measured as suc-LLVY-MCA degrading activity. Data are given as mean ± SD from 3 parallel proteolysis assays corresponding to the Western blot in panel A.
Adriamycin toxicity in U937 and TUR cells with inhibited PARP by 3-ABA or depleted PARP by asPARP transfectants.
(A) Adriamycin toxicity as the percentage of remaining viable U937 and TUR cells after a 24-hour incubation with 10−8 M to 10−5 M adriamycin in the absence (▪) or presence (●) of 1 mM 3-ABA and 2 μM MG-132 (▴), respectively. Moreover, stable transfectants of U937 and TUR cells containing a functional asPARP expression vector (asPARP cells, ▾) were also examined with respect to the adriamycin-induced cytotoxicity, respectively. (B) Western blot control of PARP protein expression in the constitutively functional asPARP vector of stably transfected U937 cells (U937 asPARP) and TUR cells (TUR asPARP), respectively, without impairment of proteasome protein levels. The actin immunoblot demonstrated equal loading.
Adriamycin toxicity in U937 and TUR cells with inhibited PARP by 3-ABA or depleted PARP by asPARP transfectants.
(A) Adriamycin toxicity as the percentage of remaining viable U937 and TUR cells after a 24-hour incubation with 10−8 M to 10−5 M adriamycin in the absence (▪) or presence (●) of 1 mM 3-ABA and 2 μM MG-132 (▴), respectively. Moreover, stable transfectants of U937 and TUR cells containing a functional asPARP expression vector (asPARP cells, ▾) were also examined with respect to the adriamycin-induced cytotoxicity, respectively. (B) Western blot control of PARP protein expression in the constitutively functional asPARP vector of stably transfected U937 cells (U937 asPARP) and TUR cells (TUR asPARP), respectively, without impairment of proteasome protein levels. The actin immunoblot demonstrated equal loading.
To obtain more detailed insights into these effects, we used U937 and TUR cell clones stably transfected with an asPARP expression vector, because functional knockouts of the proteasome contribute to severe cell damage and subsequent apoptosis.15 The successful and highly specific function of the asPARP transfectants was investigated by anti-PARP Western blots (Figure 5B). TUR and U937 control cells express PARP proteins in contrast to the U937 and TUR asPARP transfectants, respectively (Figure 5B). These effects occur without impairment of proteasome protein expression as demonstrated by similar protein levels in each lane of the antiproteasome Western blots (Figure 5B). Whereas U937 and TUR control vector transfectants exhibited similar effects when compared with U937 and TUR cells, respectively (data not shown), adriamycin treatment of asPARP-TUR cells demonstrated an LD50 of about 2 × 10−7 M adriamycin, which is 25-fold lower than the control TUR cells but still approximately 4-fold higher than the LD50 of 3-ABA. In adriamycin-treated asPARP-U937 cells, the LD50 was reduced to 8 × 10−8 M adriamycin and thus remained 1.6-fold higher than in the presence of 3-ABA (Figure 5A). Whereas the PARP protein was undetectable in the asPARP-transfected cells (Figure 5B), the difference compared with the chemically induced PARP inhibition by 3-ABA might be explained by a recently discovered novel PARP protein.45 This PARP-2 protein contributes to about 10% of the total nuclear PARP activity45 and may neither be recognized by our PARP antibody nor affected by the sequence-specific antisense construct.
These data suggested that functionally active PARP may contribute to protect leukemic cells from cytotoxic effects after about 24 hours of drug treatment by a transient process of proteasome activation within 15 minutes to 30 minutes. To address this question, we investigated the adriamycin-induced nuclear protein and DNA damage in U937 and TUR cells within 24 hours. DNA-protein cross-links could be detected after 15 minutes of adriamycin treatment (Figure6A,B). Whereas in U937 cells the cross-links disappeared after 15 minutes, there was a significantly progressive accumulation following inhibition of PARP by 3-ABA and in asPARP-U937 cells within 24 hours, respectively (Figure 6A-B). Moreover, the steady-state level of intranuclear oxidatively modified proteins remained unchanged in U937 cells following a transient increase after 15 minutes of adriamycin treatment (Figure 6C). In contrast, oxidatively modified proteins significantly accumulated in U937 cells with inhibited PARP and in asPARP-U937 cells within 24 hours (Figure 6C). Previous work has demonstrated that protein cross-links can inhibit the proteasome activity.41 46 Thus, nuclear proteasome activity following adriamycin treatment revealed a significantly decreasing activity in U937 cells with inhibited PARP and in asPARP-U937 after 24 hours of adriamycin treatment (Figure 6D). In parallel, alkaline single-cell electrophoresis revealed an increased DNA damage after 15 minutes of adriamycin treatment in all cell types; however, U937 cells with inhibited PARP or asPARP-U937 cells demonstrated a strong accumulation of damaged DNA between 1 hour and 24 hours (Figure 6E). Of interest, the formation and accumulation of DNA-protein cross-links preceded the accumulation of DNA damage, suggesting that these cross-links might interfere with DNA repair during continuous adriamycin-induced damage. Similar results were obtained for TUR cells and asPARP-TUR cells (data not shown).
Adriamycin-induced protein and DNA damage in U937 cells.
Proteins of U937 and asPARP-U937 cells were labeled with [3H]lysine and cultured in the presence of 10−7 M adriamycin and for U937 cells with or without a 30-minute preincubation of 1 mM 3-ABA for up to 24 hours. At the time points indicated, the nuclei were isolated, lysed, and separated by nonreducing SDS–urea gel electrophoresis. DNA cross-linked protein was visualized by silver staining (A). Following ethidium bromide staining, the DNA bands were excised and the radioactivity of the DNA–cross-linked [3H]lysine-labeled proteins was quantified. Data are given as mean ± SD (n = 4) (B). Accumulation of protein-bound carbonyls (C) and appropriate proteasome activity (D) was quantified in the nuclear proteins. Data are given as mean ± SD (n = 4), respectively. DNA damage was evaluated by the comet assay (E). Data are based on 100 cells. Total fluorescence intensity, tail intensity, and tail length were calculated by the MetaMorph-software (Version 3.6a, Universal Imaging, West Chester, PA).
Adriamycin-induced protein and DNA damage in U937 cells.
Proteins of U937 and asPARP-U937 cells were labeled with [3H]lysine and cultured in the presence of 10−7 M adriamycin and for U937 cells with or without a 30-minute preincubation of 1 mM 3-ABA for up to 24 hours. At the time points indicated, the nuclei were isolated, lysed, and separated by nonreducing SDS–urea gel electrophoresis. DNA cross-linked protein was visualized by silver staining (A). Following ethidium bromide staining, the DNA bands were excised and the radioactivity of the DNA–cross-linked [3H]lysine-labeled proteins was quantified. Data are given as mean ± SD (n = 4) (B). Accumulation of protein-bound carbonyls (C) and appropriate proteasome activity (D) was quantified in the nuclear proteins. Data are given as mean ± SD (n = 4), respectively. DNA damage was evaluated by the comet assay (E). Data are based on 100 cells. Total fluorescence intensity, tail intensity, and tail length were calculated by the MetaMorph-software (Version 3.6a, Universal Imaging, West Chester, PA).
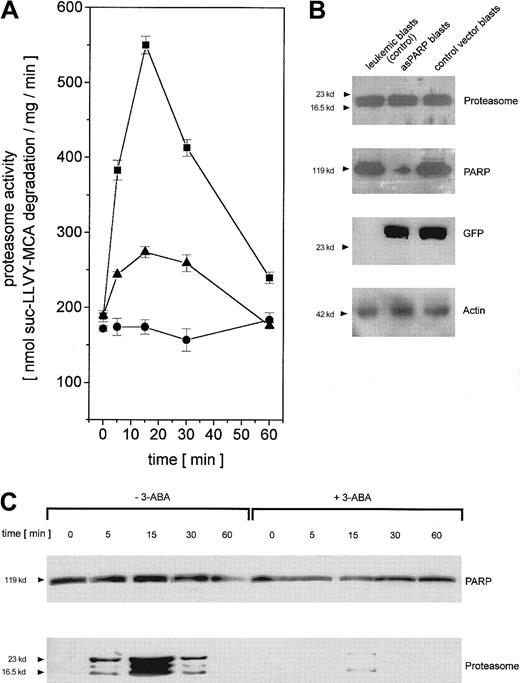
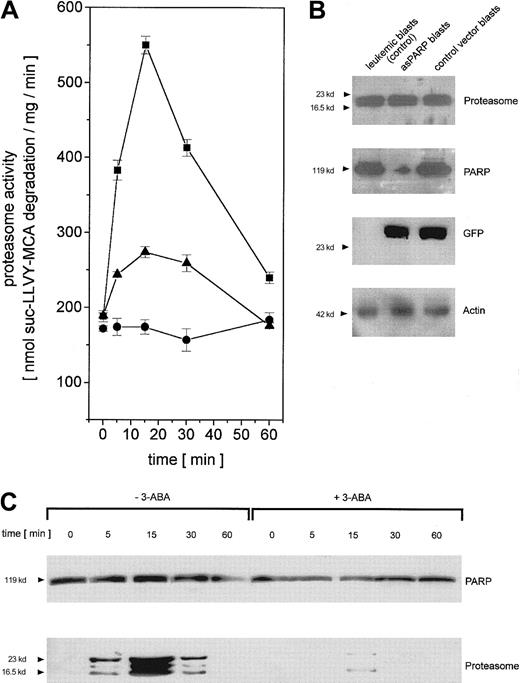
To test the functional involvement of PARP in the regulation of adriamycin-activated nuclear proteasome beyond the cell culture model, we isolated leukemic blasts from previously untreated patients suffering from AML. The leukemic blasts were transiently transfected with the control vector and the asPARP vector, respectively, and cultured for up to 60 minutes in the presence of adriamycin. In close similarity to the time course of the cell culture model, the proteasome activity in human leukemic blasts increased after in vitro adriamycin treatment and was markedly reduced after PARP inhibiton by 3-ABA or after PARP depletion by asPARP transfection (Figure 7A). The successful function of the transient asPARP transfection was investigated by Western blot analysis. Whereas proteasome protein expression was equally distributed throughout the different samples, anti-PARP Western blots demonstrated significant levels of PARP proteins in the leukemic blasts and the control vector blasts in contrast to the low levels observed in the asPARP blasts (Figure 7B). Anti-GFP Western blots documented the transfection controls, and antiactin demonstrated the equal loading controls, respectively (Figure 7B).
Regulation of proteasome activity in adriamycin-treated leukemic blasts from patients with AML.
(A) Nuclei from human leukemic blasts in the absence (▪) or presence (●) of 1 mM 3-ABA and nuclei from human leukemic blasts transiently transfected with the asPARP vector (▴) were isolated and lysed after treatment with 10−7 M adriamycin for the time points indicated, respectively. Proteasome activity is expressed as lactacystin-sensitive suc-LLVY-MCA degradation. Data are given as mean ± SD (n = 3) from one representative patient typical for all AML patients (n = 4) investigated. (B) Western blot control of PARP protein expression in leukemic blasts of an AML patient and human leukemic blasts of the same patient transiently transfected with the asPARP vector (asPARP blasts) and the control vector (control vector blasts), respectively, without impairment of proteasome protein levels. Transient transfection of the blasts is documented by the GFP blot, and the actin immunoblot demonstrated equal loading. (C) Human leukemic blasts from AML patients were treated with 10−7 M adriamycin in the absence or presence of 1 mM 3-ABA for the time points indicated. Thereafter, the nuclei were isolated, lysed, and immunoprecipitated with a polyclonal rabbit anti-PARP antibody. Immunoprecipitated proteins were separated by SDS-PAGE, transblotted, and visualized with a polyclonal anti-PARP and a polyclonal antiproteasome antibody, respectively. A representative set of data is presented from one AML patient out of all AML patients (n = 4) investigated.
Regulation of proteasome activity in adriamycin-treated leukemic blasts from patients with AML.
(A) Nuclei from human leukemic blasts in the absence (▪) or presence (●) of 1 mM 3-ABA and nuclei from human leukemic blasts transiently transfected with the asPARP vector (▴) were isolated and lysed after treatment with 10−7 M adriamycin for the time points indicated, respectively. Proteasome activity is expressed as lactacystin-sensitive suc-LLVY-MCA degradation. Data are given as mean ± SD (n = 3) from one representative patient typical for all AML patients (n = 4) investigated. (B) Western blot control of PARP protein expression in leukemic blasts of an AML patient and human leukemic blasts of the same patient transiently transfected with the asPARP vector (asPARP blasts) and the control vector (control vector blasts), respectively, without impairment of proteasome protein levels. Transient transfection of the blasts is documented by the GFP blot, and the actin immunoblot demonstrated equal loading. (C) Human leukemic blasts from AML patients were treated with 10−7 M adriamycin in the absence or presence of 1 mM 3-ABA for the time points indicated. Thereafter, the nuclei were isolated, lysed, and immunoprecipitated with a polyclonal rabbit anti-PARP antibody. Immunoprecipitated proteins were separated by SDS-PAGE, transblotted, and visualized with a polyclonal anti-PARP and a polyclonal antiproteasome antibody, respectively. A representative set of data is presented from one AML patient out of all AML patients (n = 4) investigated.
Anti-PARP immunoprecipitation of human leukemic blast lysates after adriamycin treatment revealed similar PARP protein levels throughout the 60-minute time course (Figure 7C). In contrast, the coprecipitation of distinct proteasome complexes was only detectable between 5 minutes and 30 minutes after adriamycin treatment, with a maximum level after about 15 minutes (Figure 7C). Moreover, inhibition of PARP by 3-ABA was associated with a significantly reduced coprecipitation of proteasome (Figure 7C). Taken together, these results in leukemic blasts are in agreement with our findings in the leukemic cell culture model and thus support an involvement of PARP in the proteasome activation during an adriamycin-based chemotherapy.
Discussion
Anthracyclines are part of most chemotherapeutic regimes in hematologic oncology, and resistance to these drugs causes major complications during the therapy. Anthracyclines progressively accumulate in the nuclei of hematopoietic cells and reach peak concentrations approximately 8 minutes after administration.47 Certain anthracyclines, including adriamycin, develop oxygen radicals to derail nuclear protein functions, ie, by oxidation of histones and damage of DNA. As a major component of the antioxidative defense system in the nucleus, adriamycin treatment contributed to an activation of the 20S proteasome. Previous work has demonstrated that up-regulation of several antioxidant enzyme activities like superoxide dismutase, catalase, and glutathione peroxidase is associated with adriamycin treatment.48 Moreover, enhanced proteasome activity19,20 and activation of PARP has been demonstrated in human tumor cells following chemotherapy.4-6,31 We were therefore interested if the proteasome activity is associated with PARP activation after adriamycin treatment. Indeed, inhibition of PARP by 3-ABA abolished most of the proteasome enzyme activity following adriamycin administration of proliferating leukemia cells. Interestingly, aggressive tumor-developing TUR cells revealed an enhanced constitutive and adriamycin-inducible proteasome activity when compared with the parental U937 control and Retro-U937 cells, whereas the proteasome activity in nonproliferating Diff-U937 cells was constitutively reduced and little if any change was observed after adriamycin treatment. These findings indicate that activation of an antioxidative defense system might be used particularly by proliferating tumor cells to cope with nuclear damages induced by chemotherapeutic agents because the selective and efficient degradation of oxidatively damaged nuclear proteins appears to be a requirement for maintaining genomic integrity. Thus, previous work has suggested that up-regulation of an antioxidative defense system is involved in adaptive response and resistance development against antitumor chemotherapy.49 Consequently, tumor cells can acquire a certain insensitivity or resistance against chemotherapeutic agents in the course of treatment.50,51 In this context, aggressively proliferating and tumor-developing human TUR leukemic cells exhibit such a resistance against drug-induced differentiation and apoptosis.52 Previous studies have demonstrated, however, that these resistance features developed by TUR cells are independent of a p-glycoprotein up-regulation53 and are therefore based on a novel mechanism.
To get closer insights into the cellular defense mechanisms of these leukemic cells, coimmunoprecipitation experiments revealed interactions between PARP and the nuclear proteasome activation after adriamycin administration. Thus, the coprecipitated proteasome subunit levels and subsequent enzymatic activities demonstrated a strong correlation after incubation with adriamycin. Following PARP inhibition with 3-ABA in adriamycin-treated cells, however, the coprecipitated proteasome subunits and, accordingly, the proteasomal proteolytic activity was significantly reduced in the presence of unaltered PARP protein levels. This indicates an interaction of the proteasome with PARP via poly(ADP-ribose) chains, because previous work has demonstrated in vitro that poly(ADP-ribose) chains can attach to the functionally active PARP during the automodificaton reaction and thus contribute to an enhanced proteolytic activity.23 Consequently, an increased PARP automodification in proliferating leukemic cells such as U937 and TUR provides a potential to develop chemotherapeutic resistance by elevating the proteasome metabolic activity.
To verify PARP as a possible primary target within chemotherapeutic defense mechanisms developed by certain tumor cells, we selected U937 and TUR cell clones stably transfected with a constitutively active asPARP vector. Adriamycin treatment revealed a 4-fold increased sensitivity in asPARP-U937 cells and a 25-fold increased sensitivity in asPARP-TUR cells as compared with the appropriate wild-type cells, respectively. Thus, we could show that the initially high adriamycin resistance of TUR cells can be significantly reduced by specific depletion of the PARP protein. In this context, the observed reduction and subsequent decline of proteasome activity in 3-ABA–treated or asPARP cells within 24 hours suggests a causal relationship to the protein cross-link formation following adriamycin treatment. Indeed, previous studies have documented that carbon-centered protein radicals and protein carbonyls can react with nucleobases and thus form DNA-protein cross-links.54,55 Moreover, anthracyclins can initiate the formation of DNA-protein cross-links and DNA interstrand cross-links.56,57 Furthermore, recent work has also demonstrated that DNA-protein cross-links are involved in the inhibition of the proteasome.41 Consequently, the declining proteasome activity in PARP-inhibited cell populations enhances further formation and accumulation of oxidatively damaged protein products and protein-DNA cross-links, which contribute to a progressive DNA damage and subsequent cell death. Activation of the nuclear proteasome system could therefore be considered as an intranuclear antioxidative defense system that prevents the accumulation of oxidatively damaged proteins beyond a critical threshold.
The clinical relevance of these findings was tested in isolated monocytic leukemia cells from patients with acute myelocytic leukemia (AML subtype M5) prior to initial chemotherapeutic treatment. PARP inhibition by 3-ABA and specific PARP depletion by transient transfection of these human leukemic blasts with the asPARP vector confirmed a significant reduction of proteasome activity after adriamycin treatment, respectively. Moreover, although the proteasome subunit patterns in leukemic blasts appear different to those observed in the 4 leukemic cell line variants, the time course of maximal interaction with PARP and the sensitivity to PARP inhibition are similar in the coimmunoprecipitation experiments.
Taken together, these findings suggest that the regulation of the proteasome activity in adriamycin-treated leukemic cells is associated with functionally active PARP and paralleled by a certain level of chemotherapeutic resistance. Whereas oxidatively damaged proteins induce PARP for poly-ADP-ribosylation of both the damaged proteins and PARP itself, the automodified form of PARP may contribute to activate the proteasome subunits.23 PARP could therefore be considered as a potential target for strategies to potentiate the chemotherapeutic sensitivity and thus to reduce primary or secondary resistance mechanisms.
Ö.C. and O.U. contributed equally to this work.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.
References
Author notes
Ralf Hass, Biochemistry and Tumor Biology, Clinic of Obstetrics and Gynecology (OE 6410), Medical School/Oststadtkrankenhaus, Podbielskistr 380, D-30659 Hannover, Germany; e-mail: hass.ralf@mh-hannover.de.

![Fig. 6. Adriamycin-induced protein and DNA damage in U937 cells. / Proteins of U937 and asPARP-U937 cells were labeled with [3H]lysine and cultured in the presence of 10−7 M adriamycin and for U937 cells with or without a 30-minute preincubation of 1 mM 3-ABA for up to 24 hours. At the time points indicated, the nuclei were isolated, lysed, and separated by nonreducing SDS–urea gel electrophoresis. DNA cross-linked protein was visualized by silver staining (A). Following ethidium bromide staining, the DNA bands were excised and the radioactivity of the DNA–cross-linked [3H]lysine-labeled proteins was quantified. Data are given as mean ± SD (n = 4) (B). Accumulation of protein-bound carbonyls (C) and appropriate proteasome activity (D) was quantified in the nuclear proteins. Data are given as mean ± SD (n = 4), respectively. DNA damage was evaluated by the comet assay (E). Data are based on 100 cells. Total fluorescence intensity, tail intensity, and tail length were calculated by the MetaMorph-software (Version 3.6a, Universal Imaging, West Chester, PA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2830/6/m_h80910982006.jpeg?Expires=1767909238&Signature=CvbVHkNS2Nt7ZKnTzX-H4XgWjyCWD5i-r7-RZMnCiq93FC3603oPF8S8Q~CVEDpPcDMiYkYTWnmylTHy-W3N9h0mxg~50QfyjUyI7a5l9sbVRhVbTXZNimTTaeMaHztdyJxlEniNFf1X0zLPJRTEJrBiv3dDL6DThhBAtU8T0UbFHKO29iStQg7Ol2mwNJQc5Y0XgMSucqXXgPvk9P8y0hctd8Z7H62p2HXYjLKrcuGw3xMzb8Q0LecaaevXbmidY1tTCUuQhpXlW-W5ivcOybjdHyeCZclXJ2-QzoOdy0rQ1-bS8uSz~efsFhKWrLCJrYbXjFVBl1DyzdfRyN7Yng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)

![Fig. 6. Adriamycin-induced protein and DNA damage in U937 cells. / Proteins of U937 and asPARP-U937 cells were labeled with [3H]lysine and cultured in the presence of 10−7 M adriamycin and for U937 cells with or without a 30-minute preincubation of 1 mM 3-ABA for up to 24 hours. At the time points indicated, the nuclei were isolated, lysed, and separated by nonreducing SDS–urea gel electrophoresis. DNA cross-linked protein was visualized by silver staining (A). Following ethidium bromide staining, the DNA bands were excised and the radioactivity of the DNA–cross-linked [3H]lysine-labeled proteins was quantified. Data are given as mean ± SD (n = 4) (B). Accumulation of protein-bound carbonyls (C) and appropriate proteasome activity (D) was quantified in the nuclear proteins. Data are given as mean ± SD (n = 4), respectively. DNA damage was evaluated by the comet assay (E). Data are based on 100 cells. Total fluorescence intensity, tail intensity, and tail length were calculated by the MetaMorph-software (Version 3.6a, Universal Imaging, West Chester, PA).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/97/9/10.1182_blood.v97.9.2830/6/m_h80910982006.jpeg?Expires=1767981602&Signature=WsN21V4rtBXjFZIysNslPbY2O4~Mm86GCr2hbdbkKicyIcL2ZLAGHc5u9w7BRhAVrBdIQTnQ8PxM8EwNzhAg0lysQnUN7hcE12Mo7MnbwzT4gRZWczM4NTWalrNEuHBXkUh0cJ4fAELAOqJGwMESfq7E9FdljrtyDvfkLrcSyG1xB-cf~bNfvPiJ3uEb8LfN5x6o7Mp3JX-YEjN0N8XW08PpLtNeP3So4gUdcrjggVtz~NsSKUUaqo9hFAso~DgPH70mfJebBGU9uL6gH0EBYyx0D3iqqPdqBnV-hzunZTZZlDBEqgcFc509rXjvxfFRz6oHqfhYSgBwB-H7k-cbaw__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)