The hallmark of chronic myeloid leukemia (CML) is theBCR-ABL fusion gene, which is usually formed as a result of the t(9;22) translocation. Patients with CML show considerable heterogeneity both in their presenting clinical features and in the time taken for evolution to blast crisis. In this study, metaphase fluorescence in situ hybridization showed that a substantial minority of patients with CML had large deletions adjacent to the translocation breakpoint on the derivative 9 chromosome, on the additional partner chromosome in variant translocations, or on both. The deletions spanned up to several megabases, had variable breakpoints, and could be detected by microsatellite polymerase chain reaction in unfractionated bone marrow and purified peripheral blood granulocytes. The deletions were likely to occur early and possibly at the time of the Philadelphia (Ph) chromosome translocation: deletions were detected at diagnosis in 11 patients, were found in all Ph-positive metaphases, and were more prevalent in patients with variant Ph chromosomes. Kaplan-Meier analysis showed a median survival time of 36 months in patients with a deletion; patients without a detectable deletion survived > 90 months. The survival-time difference was significant on log-rank analysis (P = .006). Multivariate analysis demonstrated that the prognostic importance of deletion status was independent of age, sex, percentage of peripheral blood blasts, and platelet count. Our data therefore suggest that an apparently simple, balanced translocation may result not only in the generation of a dominantly acting fusion oncogene but also in the loss of one or more genes that influence disease progression.
Chronic myeloid leukemia (CML) is a clonal hematologic malignant disease arising in the stem-cell compartment.1-3The hallmark of CML is the formation of a BCR-ABL fusion gene, usually as a result of the Philadelphia (Ph) chromosome translocation. The BCR-ABL fusion gene is thought to play a central role in the pathogenesis of CML. This concept is strongly supported by retroviral transduction experiments in which p210 BCR-ABL was expressed in murine bone marrow cells and resulted in a myeloproliferative disorder resembling CML.4-9
CML is a biphasic disease with an initial chronic phase during which the disorder is readily controlled. However, chronic-phase CML is followed by a terminal blastic phase that resembles acute leukemia and is usually refractory to treatment. Transformation of chronic phase to blast crisis is accompanied by secondary cytogenetic changes in approximately 85% of cases.10 However, the genetic events responsible for the transformation of CML are poorly understood. Homozygous deletions in the p16Ink gene have been reported in lymphoid blast crisis.11,12 Mutations in the p53 and N-ras genes have been reported but are rare.13,14Allelotype analysis has identified loss of heterozygosity at multiple genomic regions in a varying proportion of patients in blast crisis.15 Some of these regions may contain tumor suppressor genes that influence the evolution to CML blast crisis.
Current options for treatment of patients with CML include hydroxyurea, α-interferon (with or without cytosine arabinoside), and allogeneic or autologous progenitor-cell transplantation. Identification of prognostic factors may be useful in guiding management. Patients with CML show considerable clinical heterogeneity in the chronic phase, and some clinical features, such as spleen size, blast cell count, and platelet count, have been incorporated into the Sokal prognostic score.16 The molecular basis for this clinical variability is not understood but may involve several mechanisms, including differences in the precise translocation breakpoints of the Ph chromosome, different genetic background, and environmental factors that modulate “competition” between normal and malignant stem cells.
The first of these possibilities has been investigated in considerable detail over the past 10 years and was reviewed by Melo.17The position of the translocation breakpoint in the BCR gene alters the amount of BCR protein in the BCR-ABL fusion protein. This in turn influences the phenotype of the associated leukemia, with p190, p210, and p230 proteins usually accompanying acute lymphoblastic leukemia, CML, and chronic neutrophilic leukemia (CNL), respectively. It was also reported that the position of the breakpoint within the M-bcr region may influence prognosis,18 although larger studies failed to confirm this idea.19-22 The position of the breakpoint relative to the ABL coding sequence is less variable. The few cases with fusions to the a3 exon seem indistinguishable from those with the more usual a2 fusion.17 The ABL-BCR fusion gene is expressed in only about two thirds of patients with CML,23 and the presence of the ABL-BCR transcript does not influence cytogenetic response to interferon.24
We here present the first evidence of a previously unrecognized form of genetic heterogeneity among patients with CML. We found that a substantial minority of patients with CML had large acquired genomic deletions resulting in loss of chromosome 9 and 22 sequences flanking the translocation breakpoint on the derivative 9, on additional partner chromosomes, or on both. Furthermore, such deletions were associated with a worse prognosis. Our results suggest that the loss of genes adjacent to the translocation breakpoint may influence the progression of CML.
Methods
Patients' clinical and laboratory data
Fixed cytogenetic preparations from cultured bone marrow samples were obtained from 56 patients with CML seen between January 1990 and March 1997 in the Department of Hematology at Addenbrooke's Hospital, Cambridge, United Kingdom (UK), and in the Department of Oncological Cytogenetics, Christie's Hospital, Manchester, UK. Patients who had either standard (n = 34) or variant (n = 22) Ph translocations were included in the study. The patients were unselected for stage of disease or type of therapy and were treated at various centers in the Cambridge and Manchester regions. Risk categories were determined as described previously.16
Fluorescence in situ (FISH) probes and detection systems
Triple-probe/3-color system.
This system is based on the following 3 probes (Figure1): ASS probe, a 350-kilobase (kb) cosmid contig that contains the ASS gene, the gene 8604 Met (which maps to a region about 20 kb proximal to ABL exon 1b), and the most centromeric breakpoints reported in patients with CML25; ABL probe (Vysis Inc, Downer's Grove, IL), a 300-kb cosmid contig that contains the 3′ region of theABL gene (ABL exon 3 to the last exon of ABL); and BCR probe (Vysis Inc), an approximately 300-kb contig that begins between BCR exon 13 and 14 and extends well beyond the M-bcr region.
Structure of ABL and BCR genes and the probes used in the triple-probe/3-color and D-fluorescence in situ hybridization (FISH) systems.
Structure of ABL and BCR genes and the probes used in the triple-probe/3-color and D-fluorescence in situ hybridization (FISH) systems.
As described previously,26 the 3 probes, labeled with Spectrum Green, Spectrum Orange, and Aqua, were used in combination for FISH experiments to create the triple-probe/3-colorBCR-ABL detection system. Images were captured and pseudocolored to obtain the signal pattern of the original triple-probe system.
D-FISH BCR-ABL detection system.
This system is available commercially (Appligene Oncor, UK) and consists of the following 2 probes (Figure 1): ABL probe, a 600-kb contig spanning the breakpoint region on ABL labeled with a green fluorochrome (fluorescein isothiocyanate, conjugated [FITC]); and BCR probe, a 500-kb contig containing the major and minor breakpoint regions of BCR labeled with a red fluorochrome (Texas red). Images were captured and analyzed using a Smart Capture View Point multicolor imaging station (Digital Scientific, Cambridge, UK), as with the triple-probe/3-color system.
Chromosome 22 and 9 locus-specific probes.
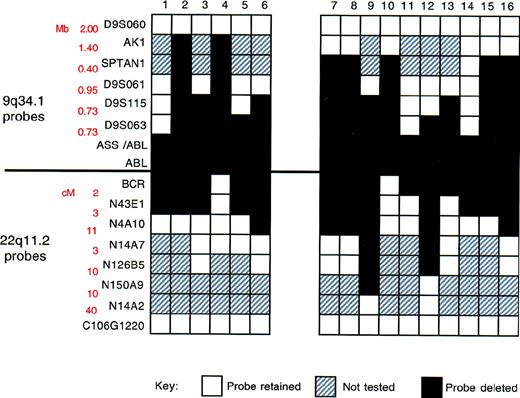
The cosmid C106G1220, isolated from the telomeric region of chromosome 22, was provided by D. Ledbetter (University of Chicago). All other locus-specific probes (cosmids) were obtained from the Sanger Centre, Wellcome Trust Genome Campus, Hinxton, Cambridgeshire, UK (Figure 5). Approximate distances between the locus-specific probes, the BCR gene, and the 22q telomere are given in Figure 5.27 Distances between the locus-specific probes from chromosome 9 used in this study (Figure 5) were based on a physical map of the 9q34 region.28 C106G1220 and D9S60 were labeled with Spectrum Orange by using a Vysis nick-translation kit (catalogue 32-801 300). All other locus-specific probes were labeled with biotin by using a Gibco-BRL (Life Technologies, Inc, Gaithersburg, MD) BioNick labeling system (catalogue 18 247-015). The 2 probes labeled with Spectrum Orange were used to mark the position of 9q and 22q in metaphase cells. Combinations of either C106G1220 or D9S60 and each of the biotinylated probes were applied to cytogenetic preparations from patients known to have deletions of the region between ABL andASS. For these experiments, the biotinylated probe was detected with an avidin/FITC detection system. After capture, Spectrum Orange signals were pseudocolored in red and FITC signals in green.
Hybridization and detection procedures
The triple-probe system was dissolved in hybridization buffer to a concentration of 10 ng/μL for each probe with 200 × Cot 1 DNA, denatured, and preannealed as previously described.29The hybridization and washing conditions used were those recommended for the Vysis dual-color BCR-ABL detection system. The Oncor D-FISH and M-bcr/m-bcr–specific probes were applied and detected according to the manufacturer's instructions. Combinations of biotinylated and Spectrum Orange–labeled single-locus probes were denatured with Cot 1 DNA, preannealed, hybridized, washed, and detected as previously described.30 All FISH images were captured and analyzed with a Smart Capture View Point multicolor imaging station (Digital Scientific). For each marker, consistent data were obtained from a minimum of 10 Ph-positive metaphase cells in which the normal chromosomes 9 and 22 displayed normal signal patterns.
Microsatellite polymerase chain reaction (PCR)
Granulocytes and T cells were prepared from peripheral blood as previously described.31 Microsatellite PCR amplification was performed as previously described31 by using primers corresponding to a sequenced tagged site (STS) within intron 14 of the ASS gene32 or to D9S159 (described at http://www.gdb.org).
Statistical analysis
All calculations were performed with the SPSS statistical package (SPSS Inc, Chicago, IL). Means and SDs were calculated for age, sex, and clinical and laboratory findings at diagnosis for patients with and without deletions and tested for significance with the Mann-WhitneyU test (continuous variables) or χ2 analysis and Fisher's exact tests (categorical variables). Survival time was calculated from month of presentation to month of death, with a median follow-up of 23 months (range, 4-90 months). Two patients who died from transplantation-related complications and one who died in chronic phase for reasons unrelated to CML were censored at date of death. Survival time in patients with deletions was compared with that in patients without deletions. Survival data were available for a total of 55 patients. Values were calculated with the Kaplan-Meier estimator; significance was tested with the log-rank test. The analysis was performed both by time censoring at the time of transplantation and also by excluding patients who underwent transplantation. Survival data from the 2 groups of patients were also used to construct a Cox proportional hazard model from which significance by χ2analysis, hazard ratio, and 95% confidence intervals (CI) was derived. Possible confounding effects of age, sex, percentage of peripheral blood blasts, and platelet count on the prognostic importance of deletion status were assessed with logistic regression analysis.
Results
Deletion of chromosome 9 sequences on the derivative 9 chromosome
We previously described a triple-probe/3-color FISH system for detection of BCR-ABL–positive cells.26 The introduction of a probe for the ASS locus, which lies upstream of ABL, permits identification of the derivative 9 chromosome in Ph-positive cells (Figure 1) and thus allows highly sensitive detection of residual disease.
The normal pattern observed in a Ph-positive metaphase is shown in Figure 2A. Colocalization of red and blue signals (ABL and ASS) was seen on the normal chromosome 9 and a single green signal (BCR) was seen on the normal chromosome 22. In contrast, colocalization of red and green signals identified the Ph chromosome, and an isolated blue signal was observed on the derivative chromosome 9. While extending our original observations to a larger number of patients, we noticed loss of theASS signal on the derivative 9 chromosome in 16 of 56 patients with CML (Figure 2B). Deletions of ASS were found in 6 of 34 patients with a standard Ph chromosome and in 10 of 22 patients with a variant Ph chromosome. No particular pattern of additional chromosome involvement was noticed in the patients with variant Ph chromosomes. The deletions were observed in diagnostic samples in 11 of 16 patients, indicating that the deletions usually occur early in the chronic phase of CML.
FISH analysis of Ph-positive metaphase cells.
(A) Triple-probe/3-color signal pattern observed in a Ph-positive metaphase cell with no detectable deletion of chromosome 9 sequences.ABL is in red, BCR in green, and ASS in blue. A colocalized red/green signal marks the Ph chromosome, the der(9) chromosome is identified by a single blue signal, the normal 22 by a green signal, and the normal 9 by a blue/red doublet. (B) Triple-probe/3-color signal pattern in a Ph-positive metaphase cell with a deletion of chromosome 9 sequences. The der(9) chromosome does not show a blue ASS signal (arrow). (C) D-FISH signal pattern in a Ph-positive metaphase cell with no detectable loss of chromosome 9 or 22 sequences. ABL is in green and BCR in red. A colocalized red/green signal marks both the Ph and der(9) chromosomes, a green signal identifies the normal 9, and a red signal marks the normal 22. (D) D-FISH signal pattern in Ph-positive metaphase with loss of sequences from chromosome 9 and 22. ABL is in green andBCR in red. There is a colocalized red/green signal on the Ph chromosome but not on the der(9) (arrow).
FISH analysis of Ph-positive metaphase cells.
(A) Triple-probe/3-color signal pattern observed in a Ph-positive metaphase cell with no detectable deletion of chromosome 9 sequences.ABL is in red, BCR in green, and ASS in blue. A colocalized red/green signal marks the Ph chromosome, the der(9) chromosome is identified by a single blue signal, the normal 22 by a green signal, and the normal 9 by a blue/red doublet. (B) Triple-probe/3-color signal pattern in a Ph-positive metaphase cell with a deletion of chromosome 9 sequences. The der(9) chromosome does not show a blue ASS signal (arrow). (C) D-FISH signal pattern in a Ph-positive metaphase cell with no detectable loss of chromosome 9 or 22 sequences. ABL is in green and BCR in red. A colocalized red/green signal marks both the Ph and der(9) chromosomes, a green signal identifies the normal 9, and a red signal marks the normal 22. (D) D-FISH signal pattern in Ph-positive metaphase with loss of sequences from chromosome 9 and 22. ABL is in green andBCR in red. There is a colocalized red/green signal on the Ph chromosome but not on the der(9) (arrow).
Deletion of chromosome 22 sequences from the derivative 9 chromosome
Our study found a loss of chromosome 9 sequences from the derivative 9 chromosome in a proportion of patients with CML. To assess whether chromosome 22 sequences were also lost, we used the D-FISH system (Appligene Oncor). The D-FISH system entails the use of 2 large probes spanning the translocation breakpoints in the BCR andABL loci (Figure 1). The normal pattern observed in a Ph-positive metaphase is shown in Figure 2C. Colocalized red and green signals appeared on both the Ph chromosome and the derivative 9 chromosome. Single red and green signals were seen on normal chromosome 22 and normal chromosome 9, respectively. In contrast, loss of the colocalized signal on the derivative 9 chromosome was observed in 14 of 16 patients in whom deletion of ASS was observed (Figure 2D and Figure 5). These data showed that deletions involving both chromosome 9 and chromosome 22 sequences were present in a substantial minority of patients with CML.
Detection of deletions by microsatellite PCR analysis of peripheral blood granulocytes
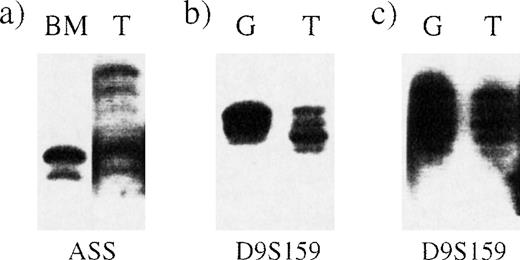
The Ph chromosome is present in peripheral blood granulocytes in most patients with chronic-phase CML although, in some, the percentage of BCR-ABL–positive granulocytes may be significantly lower than the percentage of BCR-ABL–positive bone marrow cells.26 We confirmed the existence of the deletion by using a polymorphic STS that lies within the ASS gene. Loss of heterozygosity could clearly be seen in bone marrow from a patient with a deletion (Figure 3A). To investigate whether the deletion was detectable in peripheral blood cells, we purified granulocytes and T cells from 2 other patients, one with and one without a deletion. Microsatellite PCR using primers for the D9S159 locus, which lies between D9S115 and D9S63, clearly showed loss of heterozygosity in granulocytes from the patient in whom our FISH studies detected a deletion (Figure 3B and Figure 3C).
Deletions detectable by microsatellite PCR in bone marrow and peripheral blood granulocytes.
Microsatellite PCR was performed on samples from 3 different Ph-positive patients by using primers for the loci indicated below each panel. BM indicates bone marrow; G, granulocytes; and T, T cells. (A) Loss of heterozygosity at the ASS locus in bone marrow from patient 5. (B) Loss of heterozygosity at the D9S159 locus in peripheral blood granulocytes from patient 3. (C) No loss of heterozygosity at the D9S159 locus in granulocytes from a patient with no detectable deletion.
Deletions detectable by microsatellite PCR in bone marrow and peripheral blood granulocytes.
Microsatellite PCR was performed on samples from 3 different Ph-positive patients by using primers for the loci indicated below each panel. BM indicates bone marrow; G, granulocytes; and T, T cells. (A) Loss of heterozygosity at the ASS locus in bone marrow from patient 5. (B) Loss of heterozygosity at the D9S159 locus in peripheral blood granulocytes from patient 3. (C) No loss of heterozygosity at the D9S159 locus in granulocytes from a patient with no detectable deletion.
Size of and variations in deletion breakpoints
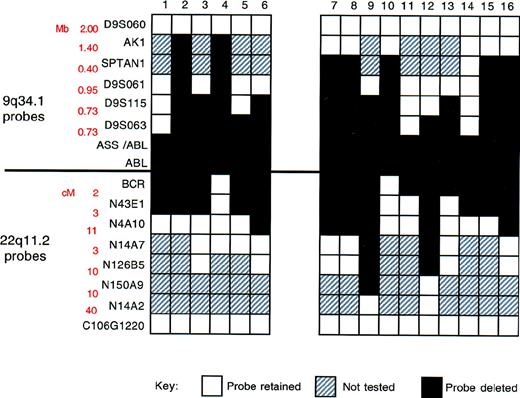
To assess the extent of the deletion of chromosome 9 and 22 sequences, we used a panel of locus-specific FISH probes to map the deletions in 16 patients found to lack the ASS signal (Figure4 and Figure5). For each probe, a minimum of 10 Ph-positive metaphases were analyzed. For each patient, all metaphases showed the same hybridization pattern; in no instance did we find clonal variation (ie, some metaphases carrying a deletion and others lacking a deletion) within the population of Ph-positive metaphases.
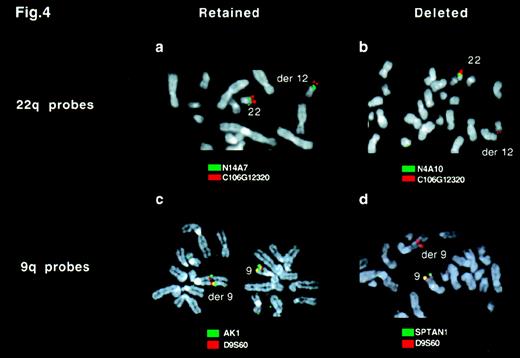
FISH analysis of deletions by using locus-specific probes.
Ph-positive metaphases from a patient with a variant Ph translocation t(9;12;22)(q34;q11;q11) were hybridized with probes from chromosome 9 (A and B) or 22 (C and D). (A) C106G1220 (red) and N14A7 (green) signals were observed on both the normal 22 and derivative 12 chromosome. (B) C106G1220 and N4A10 signals were seen on the normal 22, but only the C106G1220 signal was detectable on the derivative 12 chromosome. (C) D9S60 (red) and AK1 (green) signals were observed on both the normal 9 and the derivative 9 chromosome. (d) D9S60 and SPTAN1 signals were seen on the normal 9 chromosome, but only the D9S60 signal was detectable on the derivative 9 chromosome.
FISH analysis of deletions by using locus-specific probes.
Ph-positive metaphases from a patient with a variant Ph translocation t(9;12;22)(q34;q11;q11) were hybridized with probes from chromosome 9 (A and B) or 22 (C and D). (A) C106G1220 (red) and N14A7 (green) signals were observed on both the normal 22 and derivative 12 chromosome. (B) C106G1220 and N4A10 signals were seen on the normal 22, but only the C106G1220 signal was detectable on the derivative 12 chromosome. (C) D9S60 (red) and AK1 (green) signals were observed on both the normal 9 and the derivative 9 chromosome. (d) D9S60 and SPTAN1 signals were seen on the normal 9 chromosome, but only the D9S60 signal was detectable on the derivative 9 chromosome.
Summary of FISH mapping of deletions.
Locus-specific probes from 9q34.1 and 22q11.2, together with corresponding genomic27 and physical map28data, are shown on the left. White boxes indicate region retained (2 FISH signals/metaphase cell); black boxes, region deleted (1 FISH signal/metaphase cell); and gray boxes, not performed.
Summary of FISH mapping of deletions.
Locus-specific probes from 9q34.1 and 22q11.2, together with corresponding genomic27 and physical map28data, are shown on the left. White boxes indicate region retained (2 FISH signals/metaphase cell); black boxes, region deleted (1 FISH signal/metaphase cell); and gray boxes, not performed.
Our results are summarized in Figure 5. Two features are particularly important. First, deletion breakpoints varied among patients. Second, the deletions were very large; in some instances, they spanned several megabases. Contiguous deletion of multiple markers adjacent to the translocation breakpoint was identified in all patients.
Deletion of the derivative 9 chromosome and a poor prognosis
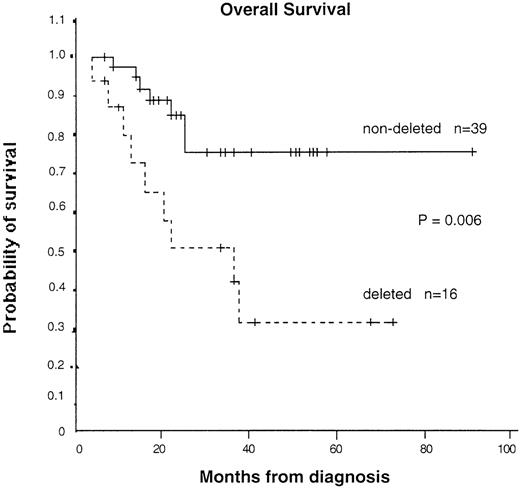
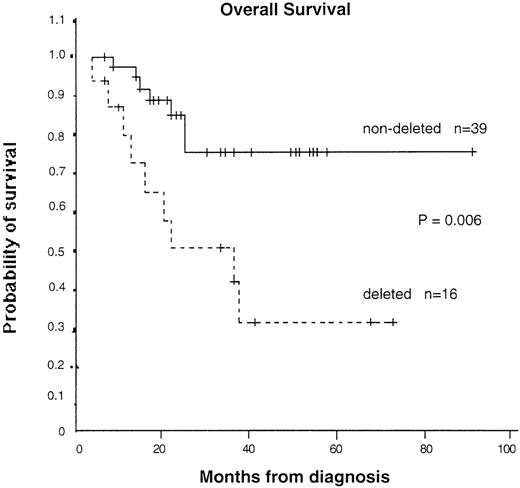
To assess the possible biologic importance of the chromosomal deletions, we collected data on clinical features, treatment history, and survival of the patients. Survival data were available for 55 patients (16 with and 39 without detectable deletions). As shown in Table 1, the clinical and laboratory features at presentation in patients with and without deletions were not significantly different. However, the median survival time was 36 months (95% CI, 13-60 months) in patients with a deletion and could not be calculated in patients without a deletion because too many are still alive. The median follow-up time was similar in patients with and without a deletion (23.5 months compared with 23 months), as was the range (4-90 months compared with 7-90 months). The difference in survival time was significant on log-rank analysis (P = .006) (Figure 6). If patients who received a bone marrow transplantation were excluded, the significance level increased (P = .004). Significance was also obtained with Cox proportional hazard analysis, with a hazard ratio of 3.81 (95% CI, 1.36-10.73) calculated for patients with a deletion compared with those without a deletion. Multivariate analysis showed that the prognostic importance of deletion status was independent of age, sex, percentage of peripheral blood blasts, or platelet count. The only possible confounding variable was treatment with interferon only: 5 of 16 patients with deletions but 28 of 40 patients without deletions received this agent. It seems unlikely that this could have accounted for the marked survival difference observed here, particularly because randomized trials have shown only a moderate benefit, at best, with interferon.33-36 Furthermore, if our Kaplan-Meier analysis is restricted to patients treated with interferon, although the numbers are small, patients with deletions have a significantly worse survival time (P = .04). Taken together, our results suggest that large genomic deletions at the translocation breakpoint on the derivative 9 chromosome are associated with a worse prognosis.
Kaplan-Meier survival curves for patients with and without detectable deletions.
Patients who underwent transplantation were censored at the time of the transplantation. P = .006 by log-rank test.
Kaplan-Meier survival curves for patients with and without detectable deletions.
Patients who underwent transplantation were censored at the time of the transplantation. P = .006 by log-rank test.
Discussion
We found that a substantial minority of patients with CML had large deletions adjacent to the translocation breakpoint of the derivative 9 chromosome. These data show the existence of previously unsuspected genetic heterogeneity among patients with CML. We also found evidence suggesting that the large deletions are associated with a poor prognosis. Our results raise the possibility that an apparently simple, balanced translocation may result not only in generation of a dominantly acting fusion oncogene but also in loss of one or more tumor suppressor genes adjacent to the translocation breakpoint.
Deletions associated with leukemia translocations have been described previously. The t(8;21) in one patient was sequenced and deletions of 5 base pairs (bp) and 18 bp were observed to flank the translocation breakpoint.37 Analysis of the inv(16) chromosome has identified larger deletions (150-350 kb) centromeric to the short-arm breakpoint,38-41 and partial deletions of MLLsequences may accompany 11q23 translocation.42
Small deletions of chromosome 22 sequences adjacent to the Ph chromosome translocation breakpoint have also been identified (by Southern blot analysis).43,44 However, these deletions were thought to be small (about 8-10 kb)43 and without pathophysiologic importance.44 Indeed, loss of small intronic regions would not usually be expected to interfere with the splicing events required for fusion RNA formation.
In contrast, the deletions we observed were large and, in many cases, spanned several megabases. We detected deletions in 16 of 56 patients, but this figure may be an underestimate for 2 reasons. First, theASS and D-FISH probes are large and may therefore miss deletions of even 100 to 200 kb. Second, our 16 patients with deletions were identified by an initial screening assessment that looked for the loss of ASS. Our data do not exclude the possibility that the patients “without deletions” may nonetheless have loss of chromosome 22 sequences.
Our data also shed light on the mechanism of the deletions. The breakpoints were variable, and therefore the deletions did not reflect single recombination hot spots within the chromosome 9 and chromosome 22 sequences on either side of the translocation breakpoint. The timing of the deletions is not clear. Six of 10 cell lines derived from patients with blast crisis carried ASS deletions (unpublished data). This prevalence is higher than what we have observed in chronic-phase samples and may indicate that deletions occur as a late event in association with the development of blast crisis. However, several other observations are consistent with the idea that deletions arise as an early event: (1) the deletions were seen in diagnostic samples from 11 patients; (2) in each patient, the deletion was present in all Ph-positive metaphases analyzed—we deliberately avoided interphase analysis because of the difficulties inherent in interpreting the loss of hybridization signals in interphase nuclei; and (3) the prevalence of deletions was higher in patients with variant Ph chromosomes, and this would be consistent with a model in which the complex recombination events that occur during the formation of variant Ph translocations are associated with an increased probability of deletion. We therefore favor the idea that the large deletions we identified occur at the time of the Ph translocation. Analysis of paired samples from patients at diagnosis and with blast crisis will be needed to confirm or refute this concept.
Our results suggest a significant association between the presence of derivative 9 deletions and a poor prognosis. This association remained significant when the analysis was restricted to patients who received interferon, although the numbers were small. These results must be confirmed in a larger study, but could possibly be the basis for a simple test to identify high-risk patients who may benefit from more intensive therapy. The use of microsatellite PCR assessment of DNA from peripheral blood granulocytes would even obviate the need for bone marrow aspirations.
How might such genomic deletions influence the progression of CML? Several mechanisms can be envisaged.45 In a 2-hit model, the deletion would result in loss of one copy of a target gene, with inactivation of the second copy by a subsequent event. The second event could involve mutation, deletion, or transcriptional silencing by methylation or imprinting.46,47 Alternatively, loss of only a single copy of the gene may itself have a pathogenetic role, as has been shown for p27Kip1.48 A variety of developmental defects in mice and human beings result from loss of one copy of a particular gene49-52 or monosomy of particular chromosome regions.53 If haploinsufficiency for individual genes can perturb fetal development, it seems likely that the same mechanism could disrupt differentiation within adult stem-cell systems such as hemopoiesis. The large size of the genomic deletions identified in our study also raises the possibility that the deletions result in loss of 2 or more target genes. In this case, inactivation of one or both copies of each critical gene may be necessary to perturb progenitor-cell behavior.
Acknowledgments
We are grateful to Toby Prevot, Brian Tom, and Sue Richards for statistical advice and to John Proffit and Kim Wieber from Vysis, Inc, Downer's Grove, IL, for preparing the ASS contig.
Reprints:A.R. Green, Department of Hematology, MRC Centre, Hills Road, Cambridge CB2 2QH, UK; e-mail: arg1000@cam.ac.uk.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 U.S.C. section 1734.