To the Editor:
In a recent article in BLOOD, Kruyt and Youssoufian1 examined the cellular localization and possible interaction of the Fanconi anemia (FA) proteins, FANCA and FANCC. There are several inconsistencies between their data and our published work.2 We would like to clarify our results and offer an explanation for the new discordant data.
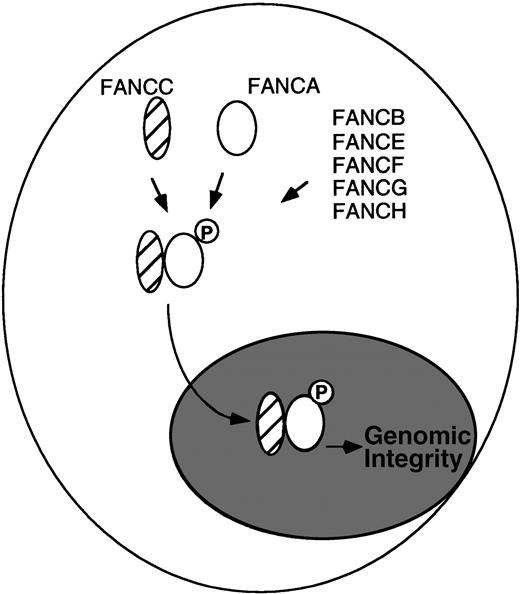
Several studies support the existence of a physical interaction between FANCA and FANCC in the nucleus. First, for lymphoblasts, primary fibroblasts, and primary bone marrow cells expressing normal (endogenous) levels of the FANCA and FANCC proteins, we detected a physical complex of FANCA and FANCC.2 The FANCA/FANCC protein complex was detected by reciprocal immunoprecipitation/Western blotting protocols, with either anti-FANCA or anti-FANCC antisera. Second, the FANCA/FANCC complex was detected in protein fractions from the cytoplasm and the nucleus of primary cells. The coimmunoprecipitation was more efficient from nuclear extracts.3 Other studies have also used confocal microscopy to localize FANCC to the nucleus.4 Third, the interaction of FANCA and FANCC in a complex is critical to the function of the proteins. For lymphoblast lines derived from FA patients, mutant FANCC proteins fail to bind to FANCA,2 and mutant FANCA proteins fail to bind to FANCC.5 Functional complementation of these cells rescued FANCA/FANCC binding. Fourth, new experimental evidence, independent of the use of anti-FANCA antisera, demonstrated a FANCA/FANCC complex (I. Garcia-Higuera, unpublished observation). For these studies, we generated an amino terminal Flag-tagged FANCA protein and expressed this protein in an FA-A cell line, GM6914. The Flag-tagged FANCA protein corrected the MMC sensitivity of the transfected cells and cofractionated with FANCC from an anti-Flag column. Fifth, we have shown that the FANCA protein is a phosphoprotein and that its phosphorylation correlates with FANCC binding.5 FANCA is not phosphorylated and the FANCA/FANCC complex is not detected in FA cells derived from other FA complementation groups (groups B, E, F, G, and H), suggesting that products of other FA genes regulate the assembly of the nuclear complex.5 According to this model (Fig1), other FA proteins may act as the kinase or adaptor proteins of the complex.
Model for the regulated binding of the FA proteins. (Data from Yamashita et al.5).
Model for the regulated binding of the FA proteins. (Data from Yamashita et al.5).
Despite this overwhelming evidence supporting an FANCA/FANCC complex, Kruyt and Youssoufian1 have concluded that no such complex exists. Their failure to detect the complex can be readily explained by their use of different anti-FANCA and anti-FANCC antisera, different immunoprecipitation protocols, and different cell lines for their analysis. Whereas the anti-FANCA antibody used in their study detected endogenous FANCA protein on Western blot of lymphoblasts, it did not appear to immunoprecipitate endogenous FANCA protein from cell lines. The antibody is therefore not suitable for immunoprecipitating endogenous cellular complexes of FANCA and FANCC. Also, the investigators performed their immunoprecipitation in RIPA buffer, which in our hands disrupts FANCA/FANCC complexes. Finally, the investigators used 293 cells overexpressing FANCA and FANCC. In our experience, transfected cells overexpressing FANCA and FANCC paradoxically yield less coimmunoprecipitation of FANCA and FANCC, because overexpressed free monomeric FANCA and FANCC compete with the endogenous FANCA/FANCC complex for antibody binding. Also, when FANCA and FANCC were overexpressed, a corresponding increase in the amount of FANCA/FANCC complex was not observed, because complex formation appears to be limited by the expression of other FA gene products.5 In short, the FANCA/FANCC complex is most readily detected in nuclear extracts of primary cells, expressing normal levels of the FANCA and FANCC protein, and with the use of high-affinity antisera capable of immunoprecipitating endogenous (low) levels of FANCA or FANCC protein. We maintain our conclusion that the FANCA and FANCC proteins physically and functionally interact, in accordance with our published work.2,3,5 It will be interesting to determine whether other FA proteins, such as the recently cloned FANCG protein,6 are also components of the nuclear FANCA/FANCC protein complex.
Response
To the Editor:
Garcia-Higuera and D’Andrea present a model in which FANCA and FANCC form a complex in the cytoplasm that is mediated by unidentified adaptor molecules.1-1-1-3 We have been unable to confirm the central element of this model, namely, an interaction between FANCA and FANCC.1-4 A number of their allusions to our work are also inaccurate.
(1) Differences in methodology. We tried to recapitulate the experimental conditions of D’Andrea et al by using a variety of conditions (ionic and nonionic detergents at different concentrations, physical methods of cell disruption, and different salt concentrations) to detect potentially low-affinity interactions. We used the same pair of mutant and cDNA-complemented FA-A lymphoblasts used by D’Andrea et al. Likewise, to perform comparative immunoprecipitations with our own antibody, we obtained their carboxy-terminal antibody that was used successfully in their laboratory. None of these strategies yielded a positive interaction. Garcia-Higuera and D’Andrea point out that this interaction can only be demonstrated by immunoprecipitation of endogenous FANCA and FANCC complexes. This argument seems to run counter to their own data, because they were clearly able to demonstrate interactions between retrovirally overexpressed FANCA and FANCC.1-2 Although it is prudent to be cautious about methodological differences, we do not believe that there is a fatal flaw in our experimental strategy.
(2) Location and functional compartments of FA proteins.Although D’Andrea has recently changed his view on this matter (compare Kupfer et al1-1 and Yamashita et al1-5), our early studies1-6,1-7 as well as theirs1-5 showed that FANCC is primarily cytoplasmic. We are aware of one other published study that addresses this issue.1-8 Using 293 cells overexpressing the FANCC cDNA, only a minor pool (∼10%) of the transfected cells showed FANCC protein in the nucleus, whereas the majority showed cytoplasmic staining. Even if there is a minor pool of FANCC in the nucleus, a more important consideration relates to the cellular site of action of FANCC. Previously, we had shown that cytoplasmic localization was essential for the complementation function of FANCC, whereas targeting to the nucleus abolished this function.1-7 More recently, we looked at the expression of FANCA and FANCC within individual cells and observed their movement.1-4 Whereas FANCC remained in the cytoplasm, FANCA was found in the nucleus, cytoplasm, or both. To the extent that FANCA is both nuclear and cytoplasmic, our data agree with that of D’Andrea. However, there is complete dyssynchrony in their spatial association. Moreover, when we enriched FANCA either in the nucleus or the cytoplasm, nuclear localization of FANCA was necessary for its complementation function.1-4 The conclusion that FANCA and FANCC work in different cellular compartments seems inescapable.
(3) Technical and conceptual flaws with the model. D’Andrea et al relied only on a single strategy (immunoprecipitation followed by immunoblotting) to demonstrate a putative interaction between FANCA and FANCC. We believe that additional methods must be used to validate such interactions, particularly when experiments are being performed in the context of the cellular milieu rather than with purified proteins. Ironically, they always choose to split the interacting partners and present their data on two separate panels, one probed for FANCA and a second for FANCC. There is no attempt to show both interacting partners on the same gel. Although this practice may not be unusual, it frustrates the ability of the reader to assess the integrity of the data independently, and any claims about the stoichiometry of the various subunits remains unfounded. There are also conceptual problems with their model. If complex formation and nuclear translocation “is limited by the expression of other FA gene products,” then overexpressed FANCA should accumulate in the cytoplasm, a prediction that is not supported either by their data1-2 or our own.1-4 The model is also unusual in that most nuclear and nucleolar complexes (eg, the basal transcription machinery, ribonucleoprotein complexes) reach the nucleus as smaller subunits and undergo assembly after translocation.
We suggest that any model on the pathogenesis of FA should take into account the compartmentalization of these proteins based on their function: FANCC works in the cytoplasm, at least partly in collaboration with NADPH cytochrome P450 reductase,1-9whereas FANCA works in the nucleus1-4 in collaboration with currently unknown targets. In a more general sense, although we cannot exclude collaborations among other FA-related proteins, we do not believe that direct or indirect interactions between FANCA and FANCC account for the similarity in the FA phenotype.