Abstract
Previously, we have found that the loss of heterozygosity (LOH) was frequently observed on chromosome 6q in acute/lymphoma-type adult T-cell leukemia (ATL), suggesting a putative tumor-suppressor gene for ATL may be present on chromosome 6q. To further define a region containing this gene, we performed fine-scale deletional mapping of chromosome 6q in 22 acute/lymphomatous ATL samples using 24 highly informative microsatellite markers. LOH was found in 9 samples (40.9%) at 1 or more of the loci examined. Of the 9 samples, 8 shared the same smallest commonly deleted region flanked by D6S1652 and D6S1644 (6q15-21). The genetic distance between these two loci is approximately 4 cM. These results suggest that a putative tumor-suppressor gene on chromosome 6q15-21 probably plays a very important role in the evolution of acute/lymphomatous ATL. Our map provides key information toward cloning the gene.
RECENT STUDIES SUGGEST that functional inactivation of tumor-suppressor genes may play an important role in the pathogenesis of leukemia1 as well as certain cancers.2,3 With tumor-suppressor genes located on autosomes, deletion of the normal copy of the gene that can be frequently detected by analysis for loss of heterozygosity (LOH) of closely linked markers allows expression of any recessive mutation in the other copy. Indeed, tumor-suppressor genes have been identified and characterized from regions showing frequent LOH in tumors.2,4 5
Adult T-cell leukemia (ATL) is one of the peripheral T-cell malignant neoplasms of which human T-cell leukemia virus type I (HTLV-I) is the etiologic agent. Although the evolution from chronic-type to acute/lymphomatous ATL is a common clinical feature, the mechanism of transformation is poorly understood. Recently, substantial evidence has been acquired for a pathogenetic role of tumor-suppressor genes in the progression of ATL; altered expression and structural abnormalitites in the p53,6 p16/INK4A,7-9p18/INK4C,10 and Rb genes,11 in the evolution of acute/lymphoma-type ATL have been observed. However, additional genetic alterations responsible for the transition from chronic to acute/lymphoma type are probably present because abnormalities of candidate tumor-suppressor genes were not identified in a sizable fraction of ATL cases. Furthermore, statistical analysis suggests that ATL arises after approximately five independent genetic events.12
Recent evidence obtained by cytogenetic studies indicates that chromosome 6q is often affected in acute/lymphoma-type ATL,13 14 suggesting the existence of a putative tumor-suppressor gene(s) for ATL on this chromosomal arm. However, the critical region in chromosome 6q has not been mapped. The frequency and region of the deletions are difficult to estimate by conventional cytogenetic analysis because small interstitial deletions are beyond the sensitivity of the technique. We have previously performed allelotype analysis in acute/lymphomatous ATL and have detected frequent LOH on the long arm of chromosome 6. To define a small region on chromosome 6q containing a putative tumor-suppressor gene for ATL, we performed an intensive deletional map using 24 microsatellite markers spanning chromosome 6q in 22 paired samples from acute/lymphoma-type ATL patients.
MATERIALS AND METHODS
Samples.
Twenty-two paired genomic DNA samples were obtained from the patients with acute/lymphoma-type ATL after their informed consent. All the ATL samples were ascertained by determining the monoclonal integration of the HTLV-I proviral genome. The clinical subtypes of ATL were based on the diagnostic criteria proposed by the Lymphoma Study Group of Japan.15 The percentage of contaminating normal cells in the acute/lymphoma-phase samples was at most 30% and usually less than 10%. The corresponding control DNAs were obtained from either their peripheral blood after complete remission (n = 17) or during their chronic phase (n = 5). DNA was extracted by a standard technique with proteinase K digestion and phenol/chloroform extraction.
Allelic loss analysis.
The LOH analysis was performed by PCR-amplification of microsatellite sequences as described before.16 The genetic map of chromosome 6 and chromosome 6–specific microsatellite markers, including their primer sequences and sizes used in this study, were compiled from the Geneton human genetic linkage map.17 Some markers have been assigned to the same location in a 0 cM cluster. Primers for polymerase chain reaction (PCR) amplification of microsatellite markers were obtained from Research Genetics (Huntsville, AL). PCR was performed in a final volume of 20 μL containing 25 ng DNA, 1.5 mmol/L MgCl2, 10 pmol/L of each of the primers, 2 nmol/L of each of the four deoxyribonucleotide triphosphates (dNTP; Pharmacia, Stockholm, Sweden), 1 unit of Taq DNA polymerase (GIBCO-BRL, Gaithersburg, MD), and 2 μCi32P-labeled deoxycytidine triphosphates (dCTP) (3000 μCi/mmol; New England Nuclear/Dupont, Boston, MA) with specified buffer provided by the supplier. PCR consisted of 40 seconds at 94°C, 30 seconds at 55° to 57.5°C, and 1 minute at 72°C for 27 to 33 cycles in a Programmable Thermal Controller (MJ Research Inc, Water Town, MA) to examine the products in the linear range of signals. For some of the markers, PCR reaction was performed in a multiplex fashion to ascertain either LOH or duplication of the region; two primer sets were mixed under the conditions described above. PCR products were mixed with a formamide gel-loading solution, heat denatured at 94°C, separated on a denaturing 5% to 8% polyacrylamide gel containing 8.3 mol/L urea, and visualized by autoradiography. Allelic losses were defined by visual comparison of the relative allelic ratios of the normal and tumor samples on the autoradiographs. In some cases of weak radiographic intensity, differences in the alleles in the tumor versus control DNA were analyzed with respect to the number of normal cells compared with malignant cells in representative slides from the tumors. In such cases, the ratio of allele intensities was classified as LOH if it roughly agreed with the percentage of the tumor cells in the sample. When visible reduction of radiographic signal was equivocal, a radioanalytic imaging detector (Ambis; Ambis Inc, San Diego, CA) was used to confirm our interpretation. All positive results were repeated for confirmation.
RESULTS
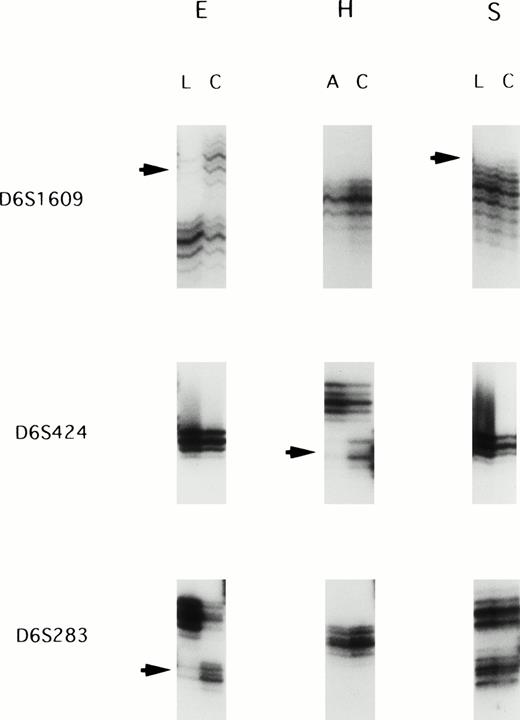
We screened 22 paired ATL samples for LOH with a panel of 24 highly informative microsatellite markers spanning chromosome 6q. All patietns were informative at multiple loci on chromosome 6q. Allelic loss was observed in 9 of 22 cases (40.9%): 4 (samples D, H, L, and T) of the 15 acute leukemias and 5 (samples E, F, G, P, and S) of the 7 lymphoma type. The most frequent LOH (5 of 11 informative cases; 45.5%) was observed at the D6S1601 locus. Figure 1shows examples of allele loss.
Representative autoradiographs showing LOH in patients E, H, and S. Loss of one parental band was observed in the acute/lymphoma ATL samples (arrows). L, DNA samples isolated from the lymphoma cells; C, DNA samples isolated from the corresponding normal peripheral leukocytes after complete remission; A, DNA samples isolated from the leukemic cells in acute type.
Representative autoradiographs showing LOH in patients E, H, and S. Loss of one parental band was observed in the acute/lymphoma ATL samples (arrows). L, DNA samples isolated from the lymphoma cells; C, DNA samples isolated from the corresponding normal peripheral leukocytes after complete remission; A, DNA samples isolated from the leukemic cells in acute type.
Figure 2 shows the deletional map on chromsome 6q as composed from the nine cases that had LOH on the arm. Of the 9 samples, 8 shared the same smallest consensus region, which was approximately 4 cM between markers D6S1652 and D6S1644 located at the 6q15-21 chromosomal band. Allelic loss of the smallest commonly deleted region on 6q was observed in both acute (3 of 15, 30.0%) and lymphoma type (5 of 7, 71.4%) of ATL.
Patterns of LOH on chromosome 6q in ATL. The nine samples that showed LOH at one or more loci are presented. **Represent the smallest region of shared LOH. A partial ideogram of chromosome 6q and the relative positions of the markers used in this study are shown to the left of the diagram. □ Represents informative with retention of both alleles; ▪, informative with LOH;  , not informative.
, not informative.
Patterns of LOH on chromosome 6q in ATL. The nine samples that showed LOH at one or more loci are presented. **Represent the smallest region of shared LOH. A partial ideogram of chromosome 6q and the relative positions of the markers used in this study are shown to the left of the diagram. □ Represents informative with retention of both alleles; ▪, informative with LOH;  , not informative.
, not informative.
DISCUSSION
In ATL, chromosomal regions of nonrandom deletions have been identified by cytogenetics including 6q,13 especially at band 6q21.14 Similarly, we have previously identified by allelotyping using microsatellite markers that chromosomal arm 6q is one of the most frequent sites of LOH in acute/lymphoma-type ATL.18
The aim of the present study was to delineate precisely the critical region that is deleted on the long arm of chromosome 6 to localize further the tumor-suppressor gene involved in ATL. To narrow this region, the LOH on the arm 6q in ATL was mapped using 24 polymorphic markers. We have found that the frequency of LOH on 6q (40.9%) was higher than that reported by cytogenetic analysis (23%).14Thus, cytogenetic studies have probably missed some cases of small interstitial deletions on 6q. Our study showed that eight of the nine tumors with interstitial losses or partial losses of chromosome 6q had a commonly deleted region between D6S1652 and D6S1644 at 6q15-21. The distance between these two loci corresponds to 4 cM of physical distance.
From several LOH studies, chromosome 6q appears to be involved in the pathogenesis of a number of solid tumors including ovarian carcinoma,19-21 breast carcinoma,22,23malignant melanoma,24-27 renal cell carcinoma,28 hepatocellular carcinoma,29salivary gland adenocarcinoma,30 small-cell lung carcinoma,31 prostate carcinoma,32 and parathyroid adenoma.33 However, the precise nature of these molecular deletions has so far not been analyzed in detail. In hematological malignancies, deletions involving the long arm of chromosome 6 are observed primarily in lymphoid malignancies, ie, acute lymphoblastic leukemias (ALL),34, 35 lymphoproliferative disorders (LPD), and non-Hodgkin’s lymphomas (NHL).1Several commonly deleted regions along 6q have been reported in lymphoma and lymphoblastic leukemia including 6q12-21,366q14-21,37 6q21,38 6q21-22,39,406q21-23,41 6q23,38 6q23-24,426q23.1-27,37 and 6q25-27.38 41 However, to date, no altered tumor-suppressor gene responsible for these tumors has been determined. Cloning of the candidate gene(s) will define whether a single or multiple tumor-suppressor gene(s) is clustered on 6q and is commonly involved in these types of tumors.
Deletions of chromosome 6q are correlated with a poor prognosis in NHL.43 Although all of the patients in our series were not treated uniformly, we did not find any significant association between LOH of 6q and the observed proportion of treatment failures probably because the survival time of all the individuals with acute/lymphoma-type ATL was very short.
Taken together, we have identified a commonly deleted region of LOH on chromosome 6q15-21 that may play a pivotal role in development of ATL. Studies are in progress to investigate further this region of interest.
ACKNOWLEDGMENT
The authors thank Kim Burgin and Marge Goldberg for their excellent secretarial help.
Supported by Concern Foundation and the Parker Hughes Fund. H.P.K. is a member of the Jonsson Comprehensive Cancer Center and holds the endowed Mark Goodson Chair of Oncology Research at Cedars-Sinai Medical Center/UCLA School of Medicine.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.